Abstract
Duplicate Staphylococcus aureus isolates were analyzed to determine the impact of multiple isolates from the same patient on annual antibiogram data. During a 6-year period (1996 to 2001), 3,227 patients with 4,844 S. aureus isolates were evaluated. A total of 39% of patients with methicillin-resistant S. aureus (MRSA) (n = 860) and 23% of patients with methicillin-susceptible S. aureus (MSSA) (n = 2,367) infections had duplicate isolates. Cumulative data show that 91% of the patients during this 6-year period with duplicate isolates (2 to 13 duplicates/year) did not switch between MSSA and MRSA but retained the original S. aureus strain whether it was MSSA or MRSA. Rates of MRSA were calculated for each year by using all isolates and then eliminating duplicates. The impact of duplicate MRSA and MSSA isolates was evaluated by using the ratio of isolates per patient such that ratios of >1.0 indicate >1 isolate per patient. The 6-year ratio for MRSA was 1.90 isolates/patient, and the ratio for MSSA was 1.35. A significant difference (P < 0.05) was noted in the MRSA rates in 4 of 6 years when duplicate isolates were removed. Common phenotypic antibiogram patterns were compared for all MRSA isolates during the 6-year period, and 64% were of a single antibiogram phenotype. Eighty-eight percent of patients with duplicate MRSA isolates had phenotypically identical multiple isolates. The rate of MRSA differs when duplicate isolates are removed from the antibiogram data.
Staphylococcus aureus is a common cause of serious infections in the hospital and the community. Methicillin-resistant S. aureus (MRSA) was first detected in the1960s, and since that time it has spread rapidly worldwide, becoming a leading cause of nosocomial infections (5, 7, 15, 16). Currently in the United States, >50% of nosocomial S. aureus infections in intensive care units (ICUs) are due to MRSA (6, 13). An additional concern with MRSA is that many strains have acquired resistance to several classes of antibiotics (3, 11).
The periodic evaluation of rates for MRSA is crucial to both infection control monitoring and decisions regarding empirical therapy. A common method of documenting and monitoring MRSA rates is the antibiogram that reports periodically the rate of antimicrobial susceptibility for each bacterial organism and antibiotic. Generally, an antibiogram is a cumulative profile of antimicrobial susceptibility results for a given time period (10, 12). When properly prepared, antibiograms are important sources of information for healthcare providers. The lack of standardization in the preparation and data assimilation of hospital antibiograms has not been addressed until recently. To assist institutions, the National Committee for Clinical Laboratory Standards (NCCLS) organization has approved guidelines in the last year that are published in a document (M39A) entitled Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data: Approved Guideline (12). This document addresses the issue of repeat isolates and makes recommendations that repeat isolates should be eliminated or reduced to more accurately reflect antimicrobial resistance trends.
The present study has analyzed the number of duplicate isolates of S. aureus from the same patient and determined the impact of these repeat isolates on the annual rate of MRSA over a 6-year period. Duplicate isolates of MRSA were analyzed for changes in the antibiogram phenotype during the 6-year period. The results of this analysis led to an adjustment in laboratory practice for testing S. aureus for antimicrobial susceptibility that resulted in a cost reduction for the laboratory and a more accurate reflection of MRSA rates.
MATERIALS AND METHODS
Susceptibility testing.
All S. aureus isolates were tested with the Vitek system using the GPS-106 card for the years 1999 through 2001 and GPS-101 for the years 1996 through 1998. A repeat isolate of S. aureus from the same patient obtained within 7 days was not tested for antimicrobial susceptibility and is not included in this analysis. Isolates of S. aureus, including MRSA, identified for surveillance purposes only, were not included in the analysis since they were coded and tested by different methods. Isolates of MRSA or MSSA from outpatient locations were not included in this analysis to reflect the rate of methicillin resistance within the hospital units.
Data analysis.
The susceptibility data were extracted from the laboratory information system (MISYS) and transferred into a Microsoft Access database. A report was created in MISYS Cache SQL by using function RDB to transfer the data to result tables. Selected fields were specified by using the MCRES_VIEW to gather data on S. aureus cultures from MISYS. The fields specified were date of admission, date of culture, source of culture, location of patient in the hospital, patient identification number, unique laboratory accession number, and the results of all antimicrobial agents that were tested for each isolate. These data were then transferred to the Microsoft Access database, which allowed for more detailed grouping and sorting of the antibiotic results. Analysis for rates of MRSA was calculated for a 6-year period between 1996 and 2001. Comparisons were made between the rates of MRSA noted in all hospital units versus the rates of MRSA reported from ICUs or non-ICUs. Some patients were hospitalized both in the ICUs and in non-ICU locations during a single hospital stay; thus, the patient and isolates were designated based on the units where the first MRSA or MSSA was isolated. The chi-square test was used to determine the statistical significance of repeat isolates on annual susceptibility rates.
Phenotypic analysis.
Common phenotypic antibiogram patterns of MRSA were examined and compared cumulatively for the 6-year period. When we evaluated phenotypes, isolates exhibiting intermediate resistance to antimicrobial agents were considered resistant. The most common phenotype of MRSA was designated the major phenotype, while the remaining minor phenotypes were given numbered designations (types 2 to 7).
RESULTS
We examined 4,844 S. aureus isolates from 3,227 patients in a university hospital over a 6-year period from 1996 to 2001. Cumulative data for this 6-year period showed that 879 patients had duplicate isolates during the 6-year antibiogram period. The majority (91%) of patients with duplicate S. aureus isolates had multiple isolates that were all MRSA or MSSA. During this 6-year period, 6% of patients (n = 56) originally presented with an MSSA and then had an MRSA isolated. This calculates to <10 patients per year. This change from MSSA to MRSA occurred during the same hospital admission in 27 patients (48%), with an average time between isolates of 2.5 weeks (range, 1 to 4 weeks). Twenty-nine patients (52%) changed from MSSA to MRSA isolates during different hospital admissions, with an average time between isolates of 2.5 months (range, 1 to 40 weeks). Interestingly, during the 6 years a few patients (n = 16) originally presented with an MRSA and subsequently had an MSSA strain isolated. The majority of these patients (76%) changed from an MRSA to an MSSA during different hospital admissions, with the average time between isolates of 5.6 months (range, 2 to 40 weeks). The majority of patients with duplicate isolates had either all MRSA or MSSA isolates, and the impact of these duplicate isolates was analyzed for each year (Table 1).
TABLE 1.
Analysis of MRSA and MSSA ratio of isolates per patient from 1996 to 2001
| Yr and strain type | All hospital unitsa |
ICUsb |
Non-ICUsc |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of: |
Ratiod | No. of: |
Ratiod | No. of: |
Ratiod | ||||
| Patients | Isolates | Patients | Isolates | Patients | Isolates | ||||
| 1996 | |||||||||
| MRSA | 144 | 298 | 2.07 | 42 | 94 | 2.24 | 102 | 204 | 2.00 |
| MSSA | 450 | 631 | 1.40 | 79 | 114 | 1.44 | 371 | 517 | 1.39 |
| 1997 | |||||||||
| MRSA | 133 | 269 | 2.02 | 38 | 86 | 2.26 | 95 | 183 | 1.93 |
| MSSA | 455 | 613 | 1.35 | 81 | 120 | 1.48 | 374 | 493 | 1.32 |
| 1998 | |||||||||
| MRSA | 143 | 236 | 1.65 | 48 | 76 | 1.58 | 95 | 160 | 1.68 |
| MSSA | 424 | 589 | 1.39 | 70 | 97 | 1.39 | 354 | 492 | 1.39 |
| 1999 | |||||||||
| MRSA | 136 | 248 | 1.82 | 37 | 66 | 1.78 | 99 | 182 | 1.84 |
| MSSA | 391 | 515 | 1.32 | 53 | 78 | 1.47 | 338 | 437 | 1.29 |
| 2000 | |||||||||
| MRSA | 112 | 195 | 1.74 | 29 | 53 | 1.83 | 83 | 142 | 1.71 |
| MSSA | 257 | 345 | 1.34 | 55 | 72 | 1.31 | 202 | 273 | 1.35 |
| 2001 | |||||||||
| MRSA | 192 | 391 | 2.04 | 47 | 105 | 2.23 | 145 | 286 | 1.97 |
| MSSA | 390 | 514 | 1.32 | 52 | 83 | 1.60 | 338 | 431 | 1.26 |
| All years | |||||||||
| MRSA | 860 | 1,637 | 1.90 | 241 | 480 | 1.99 | 619 | 1,157 | 1.87 |
| MSSA | 2,367 | 3,207 | 1.35 | 390 | 564 | 1.45 | 1,977 | 2,643 | 1.34 |
Analysis of patients with S. aureus (both MRSA and MSSA) from all units in the hospital, including ICUs but not including outpatient clinics or 1-day surgery.
Analysis of patients with S. aureus (both MRSA and MSSA) infection only from ICUs that included surgical ICUs, medical or trauma ICUs, burn units, pediatric ICUs, and neonatal ICUs.
Analysis of patients with S. aureus (both MRSA and MSSA) from all units in the hospital, excluding the ICUs listed in columns 5 to 7.
The ratio expressed is the number of isolates detected divided by the number of patients during the analysis period.
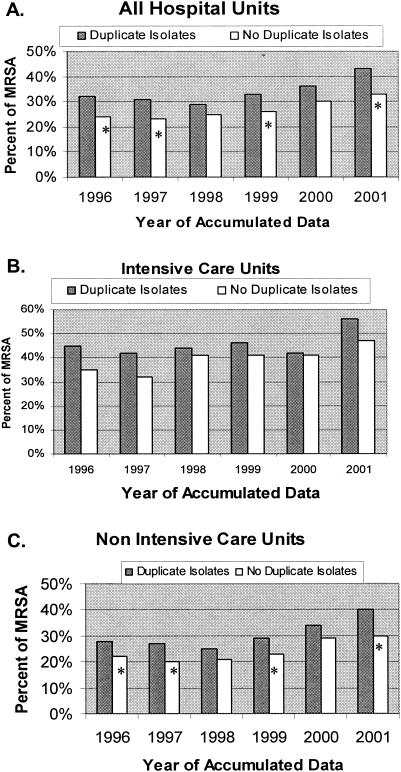
In Table 1, the numbers of MRSA and MSSA isolates and patients are divided by hospital location, such as ICUs, non-ICUs, and all hospital units, for each individual year studied. The data were evaluated as the ratio of isolates per patient in a 1-year period as described by Stelling (J. M. Stelling, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 394, 2002). For example, a ratio of 1.0 indicated one isolate for each patient during a 1-year period. This allows for the analysis and standardization of the number of isolates per patient. The average ratio of MRSA isolates per patient for all years and all hospital units was 1.90 (range, 1.65 to 2.07) versus the average ratio for MSSA of 1.35 (range, 1.32 to 1.40) (Table 1). The average ratio of MRSA in ICUs for all 6 years was 1.99 (range, 1.58 to 2.26), while the average ratio of MSSA in ICUs was 1.45 (range, 1.31 to 1.60). Similar ratios were noted for non-ICUs over the 6 years. The higher number of duplicate isolates from patients with MRSA significantly impacted the overall hospital susceptibility rates reported for 4 of 6 years (P < 0.05) (Fig. 1A). Interestingly, when we analyzed data from ICUs only, we found that duplicate MRSA isolates increased the MRSA annual susceptibility rates but that this increase was not statistically significant for any single year (P = 0.06 to 0.99) (Fig. 1B). However, when we evaluated the total from 6 years of ICU data, the MRSA rates were significantly different (P = 0.002) if the values for the duplicate isolates of MRSA and MSSA were removed. In non-ICU hospital units, the annual susceptibility rate was increased each year due to duplicate MRSA isolates, and in 4 of 6 years this increase was significant (P < 0.05) (Fig. 1C).
FIG. 1.
MRSA detection rates. Bars: ░⃞, rate of MRSA with duplicate isolates included; □, recalculated rates after all duplicate isolates are removed. P values were calculated by chi-square analysis. A P value of <0.05 is considered significant and is indicated by an asterisk.
Several patients with MRSA had as many as 13 duplicate isolates of MRSA within a 1-year period (data not shown). Sixty-two patients during the 6-year period had ≥5 MRSA isolates (range, 5 to 13) in any 1-year period. Of these 62 patients, 14 (23%) were located in an ICU. Thirty-five patients had ≥5 isolates of MSSA (range, 5 to 12) within a 1-year period. Seven (20%) of these patients with duplicate MSSA isolates were in an ICU.
Table 2 shows a comparison of the antibiogram phenotypes for all of the MRSA isolates obtained during the 6-year period, including duplicate isolates. The majority of the MRSA isolates (64%) had the same antibiogram phenotype, and this phenotype was designated the major phenotype. The other common MRSA phenotypes are shown in Table 2 as types 2 through 7 and are designated minor phenotypes. Phenotype 2 (8% of all MRSA types) was similar to the major type except for its resistance to gentamicin and trimethoprim-sulfamethoxazole (Table 2, boldface). Phenotype 3 comprised 7% of all the MRSA isolates, and this phenotype varied from the major phenotype in that it was resistant to gentamicin. Phenotype 4 comprised 5% of the total MRSA and varied from the major type in that it was susceptible to clindamycin. Phenotypes 5 and 6 each accounted for 3% of the total MRSA isolates. Phenotype 5 varied from the major type by exhibiting susceptibility to clindamycin and levofloxacin-ciprofloxacin. Phenotype 6 was similar to the major type except for resistance to tetracycline. Phenotype 7 was susceptible to all of the antimicrobial agents tested except oxacillin and made up 2% of all the MRSA. There were 26 other phenotypes that varied from the phenotypes listed in Table 2, and each consisted of ≤1% of the total MRSA isolates during a 6-year period. Of these 26 unusual phenotypes, 12 were isolated only once or from only one patient during the 6 years.
TABLE 2.
Common antibiogram phenotypes of MRSA isolates from 1996 through 2001
| Type | % of all MRSA isolates | Phenotype for antimicrobial agenta: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ERY | CLI | GEN | LEV-CIPb | OXA | RIF | SXT | TET | VAN | ||
| Major type | 64 | R | R | S | R | R | S | S | S | S |
| Type 2 | 8 | R | R | R | R | R | S | R | S | S |
| Type 3 | 7 | R | R | R | R | R | S | S | S | S |
| Type 4 | 5 | R | S | S | R | R | S | S | S | S |
| Type 5 | 3 | R | S | S | S | R | S | S | S | S |
| Type 6 | 3 | R | R | S | R | R | S | S | R | S |
| Type 7 | 2 | S | S | S | S | R | S | S | S | S |
All isolates were tested by using the Vitek system. ERY, erythromycin; CLI, clindamycin; GEN, gentamicin; LEV-CIP, levofloxacin-ciprofloxacin; OXA, oxacillin; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline; VAN, vancomycin. Phenotypes were determined by comparing the results of each antimicrobial susceptibility for the 6-year period. Isolates with intermediate results were considered resistant. Differences from the major phenotype are indicated in boldface.
Ciprofloxacin was tested for the years 1996 to 1998, and levofloxacin was tested for the years 1999 to 2001.
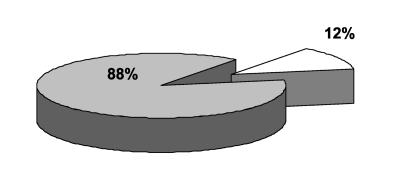
Figure 2 shows that 295 of 335 (88%) patients with duplicate MRSA isolates had the same antibiogram type in each repeat isolate during a 6-year period. Of 40 patients, 37 (93%) with different MRSA phenotypes among the duplicate isolates had only a single isolate that differed from the other duplicates. The patients with different duplicate isolates tended to have more isolates per patient with a ratio of 4.42, whereas the ratio of isolate to patient in the group with identical duplicate MRSA isolates was 3.19.
FIG. 2.
Proportion of patients (n = 335) with duplicate MRSA isolates during a 6-year period with identical duplicate antibiogram phenotypes. The shaded section depicts patient population with identical isolates, and the clear section represents the populations with nonidentical duplicate isolates.
Based on these results, the testing of multiple MRSA isolates from the same patient was abbreviated by modifying the protocol used to detect duplicate MRSA. For a 90-day period repeat S. aureus isolates from known MRSA patients were flagged by the laboratory computer and were not tested using the Vitek ($2.50 per card) but were screened only for susceptibility to oxacillin and vancomycin by using antibiotic agar screen plates ($0.20 each). Upon the request by the attending physician, any MRSA isolate during this period would be fully tested on the Vitek. After 90 days the S. aureus isolates were retested with the Vitek. This modification in protocol was designed in conjunction with the medical staff and was validated for 90 days before implementation. This screening only applied to patients infected with MRSA. All patients with MSSA had duplicate isolates tested by using the Vitek to detect the MRSA. This change reduced the cost associated with testing duplicate isolates by $1,200 annually due to decreased labor and material costs. In addition, this practice reduced the impact of duplicate MRSA isolates on the calculated rate of MRSA. Screened results are coded differently and not included into the annual antibiogram data by the laboratory computer. Since we have implemented this change (1.5 years), there have been no requests by clinicians for testing the duplicate MRSA with the Vitek. The first year of this practice resulted in a ratio of MRSA isolate(s) per patient of 1.41, a result similar to the ratio of MSSA isolates per patient (1.32). During this year the calculated rate of MRSA with duplicates was 39%, which was not significantly different (P = 0.99) than the MRSA rate without duplicates (37%). Thus, it seems that adoption of this screening protocol for duplicate MRSA within a 90-day period diminished the impact of the duplicate MRSA on the calculated susceptibility rates.
DISCUSSION
Numerous factors influence MRSA rates within an institution. Comorbidities, advanced age, and the number of invasive procedures are associated with acquiring MRSA (8, 18). The transfer of an infected patient from a different institution can introduce MRSA into a hospital. At other times, MRSA is spread from patient to patient or selected out by use of antimicrobials (2, 17, 20, 22). Several studies have shown that MRSA infections cost more than MSSA infections (1, 14, 17). Infections with MRSA are linked with an excess length of hospital stay (14). Nosocomial bloodstream infections and mortality associated with S. aureus pneumonia was noted to be greater when patients were infected with MRSA (4, 17). Institutions have become concerned with ensuring that the rate of MRSA is accurate because of the morbidity, mortality, and cost associated with MRSA infections. Assessing and correcting the data for duplicate isolates will reduce the calculated rate of MRSA and allow institutions to more accurately evaluate the impact of MRSA.
We performed a retrospective analysis of S. aureus isolates tested for antimicrobial susceptibility during a 6-year period to determine the impact of duplicate isolates on the annual rate of MRSA. These data showed that multiple repeat isolates of MRSA from the same patient are more frequent than multiple repeat isolates of MSSA in each year of a 6-year period. Of a total of 860 patients infected with MRSA, 335 (39%) had one or more duplicate isolates (Table 1), resulting in an average isolate to patient ratio of 1.90 for the 6-year period. In contrast, during the same time period duplicate isolates of MSSA were cultured from the same patient (n = 2,367) 23% of the time, with an average isolate/patient ratio of 1.35. These data show that the overall annual rate of MRSA can differ when duplicate isolates are used in calculating antibiogram data. A significant difference was noted in MRSA rates (P < 0.05) in 4 of 6 years when we evaluated data with or without duplicate isolates from all hospital units (Fig. 1). When duplicate MRSA isolates from patients in the ICUs were removed, there was a small decrease in annual MRSA rates. This difference was not statistically significant for individual years due to the small number of isolates and patients in the ICU compared to the number in all hospital units. When we evaluated cumulative data from all 6 years of the study, the ICU rates were significantly (P = 0.002) altered if duplicate S. aureus isolates were removed.
Of interest was the finding that 64% of all the MRSA isolates, including both duplicates and nonduplicates, during this period had the same antibiogram phenotype (Table 2). Over the 6-year period there was no change in the most common MRSA phenotype. There were a few additional MRSA phenotypes identified but each of the other types comprised <10% of the total and, as a result, impact the annual antibiogram susceptibility rates minimally. In addition, the majority of patients (88%) with duplicate MRSA isolates had multiple isolates with identical phenotypic antibiograms (Fig. 2). These duplicate isolates from the same patient consisted of both a major phenotype or most-common phenotype and several minor phenotypes or less-prevalent phenotypes. One reason for duplicate testing of bacterial isolates from the same patient is to monitor for the development of antimicrobial resistance. Since most of the duplicate MRSA isolates were identical, including these duplicate isolates in the annual antibiogram rate may impact the calculated rate of susceptibility to other antimicrobial agents. However, this pattern noted with MRSA may not apply to other bacteria that develop resistance at a much more rapid rate. The gram-negative pathogens Pseudomonas aeruginosa, Acinetobacter baumanii, Klebsiella species, or Enterobacter species are a few noted examples (9, 19, 21, 23). Thus, different bacterial species should be evaluated to determine the appropriate testing and data analysis strategy.
Additional issues in the analysis of duplicate isolates were recognized as we evaluated these data. There appears to be little information regarding a consensus for the time period between testing antimicrobial susceptibility on the same bacterial species from the same patient. A certain number of patients stay in the hospital for extended periods of time, and duplicate isolates are tested after 3, 4, 7, or 10 days (21, 23). In this study the laboratory policy was to repeat susceptibility tests on the duplicate S. aureus isolate from the same patient only after 7 days. Other laboratories note different time frames, such as 3 to 10 days, for repeat testing (21, 23). The new NCCLS guidelines (12) recommend using the first isolate from a patient in the antibiogram during each analysis period regardless of the total number of isolates tested or changes in phenotype. The data from the present study show that of 3,227 patients, 917 had duplicate isolates during a 6-year period. Most patients (91%) with duplicate S. aureus isolates had repeat isolates that were all the same; thus, duplicates were either all MRSA or all MSSA. It was uncommon (6%) for a patient to be infected with an MSSA and then have an MRSA strain isolated and even more uncommon (2%) for a patient to be infected with an MRSA and then have an MSSA strain isolated. Based on the present study, the NCCLS M39 guideline seems to be appropriate for MRSA, since we demonstrated here that there was little change in phenotype in duplicate isolates from the same patient (Fig. 2). However, it may not apply to other bacterial species that can acquire resistance within a few days (19, 21, 23). Further studies are needed to evaluate the impact of duplicate isolates from other bacterial species on antibiogram data. The detection of new phenotypes can be used to monitor the development of antimicrobial resistance, which is often used to determine empirical therapy. Thus, additional information on these issues would help in developing antibiogram data that can facilitate comparisons between institutional, regional, and national data and provide assurance that the data accurately reflect current resistance patterns.
Analysis of cumulative antibiogram data is valuable in monitoring the development of resistance and in establishing empirical therapy. Removing duplicate isolates is only one factor in evaluating resistance rates. There is some value in the determination of both isolate-specific and patient-specific data. The ratio of isolate per patient as proposed by Stelling (42nd ICAAC) was used in this study to analyze duplicate isolates. Dividing the number of isolates of each bacterial species by the number of patients in each analysis period defines this ratio. If the ratio is 1, then there was one isolate per patient for that time period. When >1, duplicate isolates were included in the data. Using this ratio was helpful here in evaluating S. aureus, but the usefulness of adding this ratio to antibiograms is currently unknown.
An additional issue is that annual antibiogram data is often generated electronically, and current laboratory computer systems are unable to generate this isolate/patient ratio. Laboratory information systems do not distinguish the duplicate isolates that occur during the analysis period. Many laboratory information systems are unable to deal with the complex issue of duplicate isolates from the same patient and at the same time detect new phenotypic resistance patterns in consecutive isolates. An additional concern is that it is difficult to accurately separate the overall hospital rates for MRSA verses ICU rates since patients move in and out of these units during a single hospital stay. At present, the process of identifying and sorting duplicate isolates and antibiogram types is tedious and requires enormous amounts of time and resources. Therefore, many hospitals have difficulty following the new NCCLS M39 guideline because of the requisite time and resource commitment. The data analysis in the present study took approximately 6 to 10 h for each year analyzed and was only evaluating a single bacterial species. The development of new computerized programs is essential to be able to evaluate duplicate isolates accurately.
One solution to this issue of duplicate MRSA isolates is to adopt the alternate testing method to screen the duplicate isolates by using an oxacillin and a vancomycin agar screen rather than by performing a comprehensive susceptibility panel. This strategy was successful in reducing the impact of duplicate MRSA isolates on the calculated rate of MRSA. It also resulted in a decreased cost for the laboratory and may be an option that is useful until more sophisticated computer programs become available.
Removing duplicate isolates of S. aureus can change an institution's antibiogram. This is one of the recommendations in the new NCCLS document, M39-A (12), designed to improve the quality of antibiogram data. Adherence to the guidelines should result in greater utility and standardization of antibiograms in tracking resistance, improving antibiotic selection, and comparing data locally, regionally, and nationally. Thus, it is important to establish within each institution how duplicate isolates from the same patient are handled and to acknowledge the effect these duplicate isolate have on annual antimicrobial susceptibility rates.
Acknowledgments
We appreciate the contribution of Michael Martin in the Information Technology section of the Clinical Laboratories at the University of Kansas Hospital in converting the data from the laboratory information system into the Microsoft Access database.
REFERENCES
- 1.Abramson, M. A., and D. J. Sexton. 1999. Nosocomial methicillin-resistant and methicillin-susceptible Staphylococcus aureus primary bacteremia: at what costs? Infect. Control Hosp. Epidemiol. 20:408-411. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, B. M., R. Lindemann, K. Bergh, B. I. Nesheim, G. Syversen, N. Solheim, and F. Laugerud. 2002. Spread of methicillin-resistant Staphylococcus aureus in a neonatal intensive unit associated with understaffing, overcrowding and mixing of patients. J. Hosp. Infect. 50:18-24. [DOI] [PubMed] [Google Scholar]
- 3.Berger-Bachi, B. 2002. Resistance mechanisms of gram-positive bacteria. Int. J. Med. Microbiol. 292:27-35. [DOI] [PubMed] [Google Scholar]
- 4.Blot, S. I., K. H. Vandewoude, E. A. Hoste, and F. A. Colardyn. 2002. Outcome and attributable mortality in critically ill patients with bacteremia involving methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Arch. Intern. Med. 162:2229-2235. [DOI] [PubMed] [Google Scholar]
- 5.File, T. M., Jr. 1999. Overview of resistance in the 1990s. Chest 115:3S-8S. [DOI] [PubMed] [Google Scholar]
- 6.Fridkin, S. K. G. R. P. 1999. Antimicrobial resistance in intensive care units. Clin. Chest Med. 20:303-316. [DOI] [PubMed] [Google Scholar]
- 7.Haddadin, A. S., S. A. Fappiano, and P. A. Lipsett. 2002. Methicillin resistant Staphylococcus aureus (MRSA) in the intensive care unit. Postgrad. Med. J. 78:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill, P. C., M. Birch, S. Chambers, D. Drinkovic, R. B. Ellis-Pegler, R. Everts, D. Murdoch, S. Pottumarthy, S. A. Roberts, C. Swager, S. L. Taylor, M. G. Thomas, C. G. Wong, and A. J. Morris. 2001. Prospective study of 424 cases of Staphylococcus aureus bacteraemia: determination of factors affecting incidence and mortality. Intern. Med. J. 31:97-103. [PubMed] [Google Scholar]
- 9.Jarlier, V., T. Fosse, and A. Philippon. 1996. Antibiotic susceptibility in aerobic gram-negative bacilli isolated in intensive care units in 39 French teaching hospitals (ICU study). Intensive Care Med. 22:1057-1065. [DOI] [PubMed] [Google Scholar]
- 10.Lamp, K. 1996. Antibiograms. Pharm. Pract. Manag. Q. 16:52-56. [PubMed] [Google Scholar]
- 11.Livermore, D. M. 2000. Antibiotic resistance in staphylococci. Int. J. Antimicrob. Agents 16(Suppl. 1):S3-S10. [DOI] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 2002. Analysis and presentation of cumulative susceptibility test data. Proposed guideline. NCCLS document M-39. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.National Nosocomial Infections Surveillance System. 2002. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 to June 2002, issued August 2002. Am. J. Infect. Control 30:458-475. [DOI] [PubMed] [Google Scholar]
- 14.Niederman, M. S. 2001. Impact of antibiotic resistance on clinical outcomes and the cost of care. Crit. Care Med. 29:N114-N120. [DOI] [PubMed] [Google Scholar]
- 15.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 16.Panlilio, A. L., D. H. Culver, R. P. Gaynes, S. Banerjee, T. S. Henderson, J. S. Tolson, and W. J. Martone. 1992. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975-1991. Infect. Control Hosp. Epidemiol. 13:582-586. [DOI] [PubMed] [Google Scholar]
- 17.Rello, J., A. Torres, M. Ricart, J. Valles, J. Gonzalez, A. Artigas, and R. Rodriguez-Roisin. 1994. Ventilator-associated pneumonia by Staphylococcus aureus: comparison of methicillin-resistant and methicillin-sensitive episodes. Am. J. Respir. Crit. Care Med. 150:1545-1549. [DOI] [PubMed] [Google Scholar]
- 18.Safdar, N., and D. G. Maki. 2002. The commonality of risk factors for nosocomial colonization and infection with antimicrobial-resistant Staphylococcus aureus, enterococcus, gram-negative bacilli, Clostridium difficile, and Candida. Ann. Intern. Med. 136:834-844. [DOI] [PubMed] [Google Scholar]
- 19.Shannon, K. P., and G. L. French. 2002. Antibiotic resistance: effect of different criteria for classifying isolates as duplicates on apparent resistance frequencies. J. Antimicrob. Chemother. 49:201-204. [DOI] [PubMed] [Google Scholar]
- 20.Shopsin, B., B. Mathema, X. Zhao, J. Martinez, J. Kornblum, and B. N. Kreiswirth. 2000. Resistance rather than virulence selects for the clonal spread of methicillin-resistant Staphylococcus aureus: implications for MRSA transmission. Microb. Drug Resist. 6:239-244. [DOI] [PubMed] [Google Scholar]
- 21.Thomson, R. B., Jr., T. M. File, Jr., and R. A. Burgoon. 1989. Repeat antimicrobial susceptibility testing of identical isolates. J. Clin. Microbiol. 27:1108-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vriens, M. R., A. C. Fluit, A. Troelstra, J. Verhoef, and W. C. van der. 2002. Is methicillin-resistant Staphylococcus aureus more contagious than methicillin-susceptible S. aureus in a surgical intensive care unit? Infect. Control Hosp. Epidemiol. 23:491-494. [DOI] [PubMed] [Google Scholar]
- 23.White, R. L., L. V. Friedrich, D. S. Burgess, E. W. Brown, and L. E. Scott. 2001. Effect of removal of duplicate isolates on cumulative susceptibility reports. Diagn. Microbiol. Infect. Dis. 39:251-256. [DOI] [PubMed] [Google Scholar]