Abstract
Tuberculosis remains one of the leading infectious causes of death worldwide. The emergence of drug-resistant strains of Mycobacterium tuberculosis is a serious public health threat. Resistance to isoniazid (INH) is the most prevalent form of resistance in M. tuberculosis and is mainly caused by mutations in the catalase peroxidase gene (katG). Among high-level INH-resistant isolates (MIC ≥ 2), 89% are associated with a mutation at codon 315 of katG. There is a need to develop rapid diagnostic tests to permit appropriate antibiotic treatment and to improve clinical management. Therefore, a single-tube real-time PCR, using a novel kind of probe (3′-minor groove binder-DNA probe), was developed to detect either the wild-type or the mutant codon directly in Ziehl-Neelsen-positive sputum samples. The detection limit of the assay for purified DNA was 5 fg per well (one mycobacterial genome), and with spiked sputum samples, it was 20 copies per well, corresponding to 103 mycobacteria per ml of sputum. Sputum samples from 20 patients living in Kazakhstan or Moldova and infected with monodrug- or multidrug-resistant M. tuberculosis and 20 sputum samples from patients infected with INH-susceptible M. tuberculosis were tested. The sensitivities and specificities of the probes were 70 and 94% for the wild-type probe and 82 and 100% for the mutant probe. Binding to either probe was nonambiguous. This real-time PCR allows the rapid identification of a mutant katG allele and can easily be implemented in a clinical microbiology laboratory.
Tuberculosis is the second most frequent infectious cause of death worldwide (34), despite the availability of various drugs against the causative agent, Mycobacterium tuberculosis. One of the mainstay drugs for the treatment of tuberculosis is isoniazid (INH). Nowadays, the effectiveness of INH against M. tuberculosis, initially reported in 1952 (4, 18), is hampered due to the presence of INH-resistant M. tuberculosis. Among new cases of tuberculosis, 6.2% of isolates are resistant to INH (ranging from 0% in New Caledonia to 28% in Latvia), and among previously treated cases, 19.6% are resistant (ranging from 0% in Finland to 82% in Uruguay) (6). The emergence of multidrug-resistant strains (defined as resistant to at least INH and rifampin) (7, 11, 13, 24) has further complicated the treatment of tuberculosis. Therefore, the availability of rapid methods for detecting drug resistance in clinical isolates of M. tuberculosis, or preferably directly in patient samples, is required.
The primary mechanism for acquiring resistance in M. tuberculosis is the accumulation of (mostly) point mutations in genes coding for drug targets or drug-converting enzymes (20). In the last decade, mutations in the catalase peroxidase gene (katG) (27, 35) and in a gene encoding the enoyl acyl carrier protein reductase (inhA) (3) have been found to account for 60 to 70 and 10 to 15% of INH-resistant M. tuberculosis isolates, respectively (11). Catalase peroxidase converts INH into its active compound (36), which binds to the inhA-encoded reductase (3), which is part of the fatty acid synthase II (FAS II) complex (16). The predominant mutation of katG is an AGC→ACC point mutation found at codon 315 (S315T). In three studies, INH-resistant isolates from Europe were sequenced, and mutations at codon 315 of katG were further characterized. Among 68 codon 315 mutants, 62 (91%) carried an AGC→ACC (S315T) mutation, 4 (6%) carried an AGC→AAC (S315N) mutation, and 2 (3%) carried an AGC→ACA (S315T) mutation (1, 17, 21). Other rare mutations (AGC→ATC [S315I] and AGC→CGC [S315R]) have been reported in Africa (10).
The prevalence of a mutation at codon 315 of katG was 53% among 295 INH-resistant isolates in The Netherlands (31). However, among isolates for which the MIC was 2 μg/ml or more, the prevalence of this mutation was 89%. In contrast with other katG mutations, the presence of this mutation does not abolish the function of the mycobacterial catalase peroxidase (25, 32), which is a mycobacterial virulence factor (33), and therefore the mutant remains relatively “fit.” Since this mutation is also associated with coresistance to streptomycin and rifampin, it can be considered an excellent target to predict (multidrug) resistance (31).
The aim of this study was to design a real-time PCR assay for detection of INH resistance that can be performed directly on clinical samples. Based on the data mentioned above, we decided to design the assay with two probes: one for detection of the wild-type allele of codon 315 and one for detection of the most common mutant allele, ACC. Detection of point mutations or single-nucleotide polymorphisms in M. tuberculosis is complicated by the high GC content of the genome. Hence, probes are likely to have a high melting temperature, resulting in difficulties with the detection of point mutations. For optimal detection, probes should be as short as possible, because mismatch of a single nucleotide then results in a greater destabilizing effect on probe-amplicon binding and enhances the discriminatory power. In addition, short probes are less influenced by the heterogeneity of the target sequence surrounding the mutation. To fulfill these criteria, a real-time PCR assay was designed using 3′-minor groove binder (MGB) probes. An MGB probe consists of a 14- or 15-bp-long oligonucleotide that carries both a fluorescent dye at its 5′ end and a nonfluorescent quencher and a minor groove binder at its 3′ end, which hybridizes specifically to the amplified target. The MGB part of the probe binds to the minor groove of the double-stranded DNA helix (consisting of the oligonucleotide part of the MGB probe and the complementary target sequence to which it is hybridized) irrespective of the nucleotide sequence and enhances the stability of the probe-amplicon binding. The presence of this MGB increases the melting temperature of the probe-amplicon binding and allows relatively short probes to have a melting temperature of ∼60°C (15).
MATERIALS AND METHODS
M. tuberculosis isolates and INH susceptibility.
The INH-susceptible M. tuberculosis strain H37Rv (ATCC 27294) was used as a wild-type control. As mutant controls, four INH-resistant isolates, shown in a previous study to have the AGC→ACC mutation at codon 315 of katG (31), were collected from the National Institute of Public Health and the Environment (RIVM; Bilthoven, The Netherlands). The MIC of INH for three of these isolates (RIVM 16855, 17143, and 9501611) was 5 μg/ml, and the MIC for the fourth (RIVM 9601309) was 10 μg/ml.
For evaluation of the assay, 20 INH-susceptible isolates (MIC < 0.5 μg/ml) from Dutch patients with tuberculosis were collected from the Municipal Medical and Health Service (Amsterdam, The Netherlands). Furthermore, INH-resistant isolates from 12 patients from Moldova and 8 patients from Kazakhstan were obtained from the Gelre Hospital (Apeldoorn, The Netherlands). These isolates had been used in a previous study for the determination of drug susceptibility and genotypic assessment of rifampin resistance (K. E. Templeton, S. Scheltinga, H. R. van Doorn, A. M. Kruijssen, A. van der Zee, H. Goossens, E. J. Kuijper, and E. C. J. Claas, submitted for publication).
INH susceptibility was measured by the MIC method (8). Briefly, isolates were grown in Middlebrook 7H10 medium in the presence of different concentrations (0.1, 0.2, 0.5, 1, 2, 5, 10, 20, and 50 μg/ml) of INH. Isolates were considered resistant if >1% of the inoculum grew in the presence of INH concentrations of 0.5 μg/ml or higher. In one experiment, INH susceptibility was determined by growth on Tarshis medium in the presence of different INH concentrations (0.25, 0.5, 1, 2, 4, 8, 16, 32, and 64 μg/ml) (28).
Other bacterial isolates.
A series of bacterial isolates consisting of organisms causing community- or hospital-acquired pneumonia, nontuberculous mycobacteria, and other human pathogens were used for the determination of the specificity of the assay (Table 1). These isolates were either American Type Culture Collection strains or clinical isolates collected from the Leiden University Medical Center (Leiden, The Netherlands) and had been used previously (30).
TABLE 1.
Results of an MGB assay performed on DNA extracted from 21 nonmycobacterial isolates, 5 nontuberculous mycobacterial isolates, and 1 M. bovis isolate
| Species | ATCC no. | MGB assaya
|
|
|---|---|---|---|
| VIC (wt) | FAM (m) | ||
| Acinetobacter baumannii | 9955 | − | − |
| Actinomyces turicensis | − | − | |
| Bordetella parapertussis | − | − | |
| Bordetella pertussis | 9797 | − | − |
| Chlamydia pneumoniae | − | − | |
| Enterobacter aerogenes | − | − | |
| Enterococcus faecalis | − | − | |
| Enterococcus faecium | − | − | |
| Escherichia coli | 11775 | − | − |
| Haemophilus influenzae | − | − | |
| Klebsiella pneumoniae | 33459 | − | − |
| Moraxella catarrhalis | 25238 | − | − |
| Neisseria meningitidis | − | − | |
| Nocardia abscessus | − | − | |
| Pseudomonas aeruginosa | − | − | |
| Salmonella enteritidis | − | − | |
| Salmonella enteritidis serovar Typhimurium | − | − | |
| Staphylococcus aureus | − | − | |
| Streptococcus pneumoniae | 6305 | − | − |
| Streptococcus pneumoniae | 49619 | − | − |
| Streptococcus pyogenes | 12344 | − | − |
| Mycobacterium avium | − | − | |
| Mycobacterium intracellulare | − | − | |
| Mycobacterium kansasii | − | − | |
| Mycobacterium malmoense | − | − | |
| Mycobacterium xenopi | − | − | |
| Mycobacterium bovis | + | − | |
Hybridization with a VIC-labeled MGB probe specific for M. tuberculosis katG with a wild-type (wt) codon 315 sequence or hybridization with a FAM-labeled MGB probe specific for M. tuberculosis katG with an AGC→ACC mutation (m) at codon 315 is indicated by + (hybridization) or − (no hybridization).
Sputum samples and sputum preparation.
Sputum samples from 16 patients with pneumonia or bronchitis, not suspect for tuberculosis, were collected at the clinical microbiology laboratory of the Academic Medical Center in Amsterdam, The Netherlands. These samples were pooled, examined microscopically for the presence of acid-fast bacilli, and used for spiking experiments.
Ziehl-Neelsen (ZN)-positive sputum samples, containing the M. tuberculosis isolates described above, were obtained from patients with tuberculosis in The Netherlands (n = 20), Kazakhstan (n = 8), and Moldova (n = 12).
All sputum samples were decontaminated by the N-acetyl-l-cysteine (NALC)-NaOH method (14). Briefly, the sputum samples were incubated for 15 min on a shaker with an equal volume of 0.75 M NaOH, 50 mM sodium citrate, and 0.5% (wt/vol) (Merck, Darmstadt, Germany), after which they were neutralized with phosphate buffer (Merck). The sputum samples were centrifuged at 4,000 × g for 20 min, and the pellets were resuspended in 1 ml of 0.067 M phosphate buffer (Merck).
Culture.
The M. tuberculosis H37Rv strain was cultured in liquid Dubos medium. Isolates from sputum samples were grown on Löwenstein-Jensen solid medium.
Spiking experiments.
M. tuberculosis H37Rv was grown in liquid Dubos medium to an optical density, assessed nephelometrically, corresponding to 108 mycobacteria/ml. Tenfold dilutions (containing 107 to 102 mycobacteria/ml) were made in TETB buffer (10 mM Tris-HCl [pH 8.3], 1 mM EDTA, 0.1% Tween 80 [Merck], and 0.5% bovine serum albumin [Sigma-Aldrich, St. Louis, Mo.]). Five-milliliter aliquots of pooled sputum samples were infected 1:100 with these suspensions prior to decontamination.
DNA isolation.
From 100 μl of decontaminated sputum samples, nucleic acids were isolated using guanidinium thiocyanate as described by Boom et al. (5). Nucleic acids were eluted with 100 μl of RNase-free water (Invitrogen, Carlsbad, Calif.). From liquid Dubos medium M. tuberculosis cultures, nucleic acids were isolated as described by Ausubel et al. (2). Briefly, the bacteria were killed by heating them at 80°C for 20 min and then incubated with 1 mg of lysozyme (Sigma-Aldrich)/ml at 37°C for 1 h. The bacterial suspension was further incubated with 1% sodium dodecyl sulfate (Serva Electrophoresis GmbH, Heidelberg, Germany) and 0.1 mg of proteinase K (Roche, Palo Alto, Calif.)/ml at 65°C for 10 min. Lysis was completed by incubating the suspension with 1% N-cetyl-N,N,N-trimethyl ammonium bromide (Merck) at 65°C for 10 min. DNA was extracted from the lysed bacteria by chloroform-isoamyl alcohol (Merck) and subsequently precipitated with isopropanol (Merck). From solid cultures (on Löwenstein-Jensen medium), nucleic acids were isolated by suspending 10 mg of wet bacterial cells in 250 μl of H2O and heating them at 100°C for 30 min (37). DNA was extracted using the Capture Column DNA extraction kit (Gentra Systems, Minneapolis, Minn.) following the manufacturer's instructions.
Primers and probes.
Primers and MGB probes (Table 2) were designed using Primer Express version 2.0 software (Applied Biosystems Inc., Foster City, Calif.) and the sequence of M. tuberculosis katG (GenBank accession no. NC000962). The specificities of the probes and primers for bacteria belonging to the M. tuberculosis complex were assessed by BLASTn analysis (http://www.ncbi.nlm.nih.gov/blast). The possible presence of secondary structures was determined using mfold (http://www.bioinfo.rpi.edu/applications/mfold/old/dna http://www.ncbi.nlm.nih.gov/blast). The possible presence of secondary structures was determined using mfold (http://www.bioinfo.rpi.edu/applications/mfold/old/dna) (26).
TABLE 2.
Primers and probes used in this study
| Oligo- nucleotide | Position and directiona | Sequenceb |
|---|---|---|
| Primer S | 888 F | 5′-GGGCTTGGGCTGGAAGAG-3′ |
| Primer AS | 986 R | 5′-ACAGGATCTCGAGGAAACTGTTGT-3′ |
| Wild-type probe | 936 F | 5′-VIC-GATCACCAGCGGCA- MGB-TAMRA-3′ |
| Mutant probe | 936 F | 5′-FAM-GATCACCACCGGCAT- MGB-TAMRA-3′ |
Position of the 5′ nucleotide and direction of the primer relative to the sequence of the gene indicated as F (forward) or R (reverse).
Sequence of the oligonucleotide and presence of fluorophores (VIC and FAM), MGB molecules, and quenchers (TAMRA).
PCR.
The 50-μl reaction mixture for PCR contained 25 μl of Hot-Start Master Mix (Qiagen, Hilden, Germany), 5 μl of extracted DNA, 1 μM primers S and AS, and 200 nM wild-type and mutant probes (Applied Biosystems Inc.). Thermocycling and detection were performed using the iCycler iQ Real-Time PCR Detection system (Bio-Rad, Hercules, Calif.). The thermocycling protocol was 95°C for 10 min followed by 50 cycles of 92°C for 15 s, 60°C for 1 min and 72°C for 30 s. Fluorescence data collection took place during the final 30 s of the 60°C step. Data were analyzed using the Bio-Rad iCycler iQ Real-Time Detection system software version 3.0a as provided by the manufacturer.
Agarose gel electrophoresis.
Agarose gel electrophoresis was carried out as described by Ausubel et al. (2) on 2% agarose gels.
Fluorescence-based nucleotide sequence analysis.
The katG region comprising the mutation at codon 315 was amplified using primers S and AS (Table 2). The nucleotide sequences of these amplicons were determined by a PCR-based sequence reaction using the PRISM BigDye Terminators version 3.0 cycle sequencing kit and analyzed on the PRISM 3100 Genetic Analyzer (Applied Biosystems Inc.).
RESULTS
Real-time PCR.
Real-time PCR was performed on the iCycler IQ Real-Time Detection system. Standard concentrations of MgCl2, primers, and probes as supplied by the manufacturer were used. Better sensitivity was obtained when an additional 72°C elongation phase of 30 s during each cycle was added to the thermocycling protocol, which consisted of 15 s at 92°C and 1 min at 60°C per cycle. The PCR amplification was monitored by measuring fluorescence during the annealing phase of each cycle. Fluorescence was generated by the release of the 5′ reporter dyes, which were cleaved from the probes by the 5′→3′ exonuclease activity of the Taq polymerase. After cleavage, the dyes (VIC and FAM [6-carboxyfluorescein]) were no longer quenched by the TAMRA (6-carboxytetramethylrhodamine) at the 3′ end of the probes, and fluorescence was emitted and measured.
In vitro sensitivity and specificity.
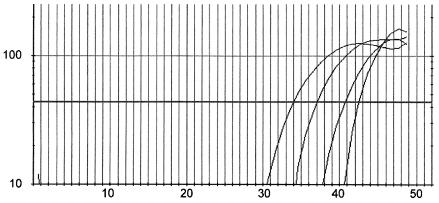
The sensitivity of the assay in water with mycobacterial DNA from the wild-type M. tuberculosis H37Rv or from mutant isolates was 5 fg (corresponding to one mycobacterial genome) of DNA per reaction. No cross-reactivity of the wild-type probe with mutant DNA, or vice versa, was observed (data not shown). The sensitivity of the PCR in spiked pooled sputum samples was 20 mycobacterial cells per reaction (corresponding to 1,000 mycobacteria per ml of sputum) for M. tuberculosis H37Rv (Fig. 1).
FIG. 1.
Fluorescence of a VIC MGB probe upon hybridization with a 122-bp-long real-time PCR fragment of wild-type katG amplified from sputum samples artificially infected with M. tuberculosis H37Rv. On the x axis, the cycle number is depicted, with the number of relative fluorescence units on the y axis. From left to right, the curves represent PCRs on sputum containing 106, 105, 104, and 103 mycobacteria/ml.
The specificity was assessed using DNAs from 21 frequent human pathogens associated mainly with community- or hospital-acquired pneumonia. Furthermore, five nontuberculous mycobacterial isolates and an isolate of Mycobacterium bovis were used (Table 1). No hybridization with either probe was observed when large amounts (>20 ng/ml) of nonmycobacterial DNA or nontuberculous mycobacterial DNA were added to the PCR mixture. The PCR was positive for M. bovis, which belongs to the M. tuberculosis complex (data not shown).
Sequence analysis.
The sequences of codon 315 of the isolates in the sputum samples which were to be used for clinical evaluation of the real-time PCR were determined by fluorescence-based nucleotide sequence analysis of DNA extracted from solid cultures of the sputum samples. The sequencing results are shown in Table 3. The strain from one sputum sample (Table 3, sputum K6) was not available, and sequencing was performed directly on DNA extracted from the sputum.
TABLE 3.
MIC and presence of katG codon 315 mutation among 20 INH-resistant isolates of M. tuberculosisa
| Origin | Sputum | INH MIC (μg/ml) | MGB assayb
|
Codon 315 p. sequence | |
|---|---|---|---|---|---|
| VIC (wt) | FAM (m) | ||||
| Moldova | M1 | >10 | − | + | ACC |
| M2 | 10 | − | + | ACC | |
| M3 | 10 | + | − | ACC | |
| M4 | >10 | + | − | AGC | |
| M5 | >10 | − | + | ACC | |
| M6 | >10 | − | + | ACC | |
| M7 | 5 | − | − | AAC | |
| M8 | 1 | − | + | ACC | |
| M9 | 5 | − | + | ACC | |
| M10 | 10 | − | + | ACC | |
| M11 | 5 | − | + | ACC | |
| M12 | >10 | − | + | ACC | |
| Kazakhstan | K1 | 5 | + | − | AGC |
| K2 | 10 | − | − | ACC | |
| K3 | 5 | − | + | ACC | |
| K4 | 5 | − | + | ACC | |
| K5 | 0.4 | − | + | ACC | |
| K6 | 5 | + | − | (AGC) | |
| K7 | >10 | − | + | ACC | |
| K8 | 8 | − | + | ACC | |
Determined by real-time PCR using an MGB assay and by DNA sequencing (codon 315 sequence). The MGB assay was performed on sputum after decontamination and nucleic acid extraction; DNA sequencing was performed on DNAs isolated from cultured colonies or, in one case, from sputum (indicated by parentheses).
Hybridization with a VIC-labeled MGB probe specific for M. tuberculosis katG with a wild-type (wt) codon 315 sequence or hybridization with a FAM-labeled MGB probe specific for M. tuberculosis katG with an AGC→ACC mutation (m) at codon 315 is indicated by + (hybridization) or − (no hybridization).
Evaluation using sputum samples from tuberculosis patients.
The presence of wild-type and mutant alleles of codon 315 of katG of M. tuberculosis in sputum samples from 8 patients from Kazakhstan and 12 patients from Moldova infected with M. tuberculosis was studied. The MICs of INH for all isolates were at least 0.4 μg/ml. In 14 sputum samples, the mutant allele was detected, and in 4 sputum samples, the wild-type allele was detected (Table 3). Hybridization with either the mutant or the wild-type probe was unambiguous. In two sputum samples, no hybridization with either probe was detected. PCR products from these two sputum samples were subjected to agarose gel electrophoresis. In DNA amplified from one of the samples, a fragment of the predicted length was seen. Sequencing of this fragment showed that the isolate present in the sputum carried the AGC→AAC mutation (Table 3, sputum M7). No fragments were amplified from the other sputum sample.
The results of the real-time PCR were compared to the sequencing results: 18 sequences were concordant with the results of the katG 315 MGB assay, and 2 sequence results were discordant with the MGB assay (Table 3, M3 and K2).
As wild-type controls, sputum samples from 20 patients infected with INH-susceptible M. tuberculosis were used. In 13 sputum samples, the wild-type allele was detected, and in 7 sputum samples, no hybridization was detected; hybridization with the mutant probe was not detected in any samples (data not shown). Also, when DNAs from the seven negative sputum samples were used for a conventional PCR with the same primers and agarose gel electrophoresis, no amplification product could be observed.
The sensitivity of the assay compared with ZN staining can be calculated as follows: 14 mutants + 4 wild types out of 20 ZN-positive sputum samples and 13 wild types out of 20 ZN-positive sputum samples = 78%. The sensitivities and specificities of the probes compared to DNA sequencing are 82 and 100% for the mutant probe and 70 and 94% for the wild-type probe when tested on ZN-positive sputum samples (Table 4).
TABLE 4.
Sensitivities and specificities of mutant and wild-type probes compared to DNA-sequencing results
| Probe | MGB assay resultsa | Codon 315 sequence data
|
Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|
| No. mutant (total, 17) | No. wild type (total, 23) | ||||
| Mutant | + | 14 | 0 | 82 | 100 |
| − | 3 | 23 | |||
| Wild type | + | 1 | 16 | 70 | 94 |
| − | 16 | 7 | |||
+, hybridization; −, no hybridization.
DISCUSSION
A rapid assessment of drug sensitivity is crucial for clinical management of patients infected with M. tuberculosis. Here, the development of an assay for detection of a point mutation associated with INH resistance in M. tuberculosis directly in sputum, in real-time PCR format, is described. The assay consists of the following three steps: sputum samples are decontaminated using the NALC-NaOH method (14), DNA is extracted using the Boom method (5), and PCR amplification is performed on an iCycler iQ Real-Time PCR Detection system simultaneously with the detection of hybridization with an MGB probe. The primers and probes are specific for bacteria belonging to the M. tuberculosis complex. Prior to the application of these MGB probes, we tried to develop a similar assay using molecular-beacon probes. However, the design of these molecular beacons was very difficult because of the high GC content of the mycobacterial genome. When the beacons were tested, the discriminatory power was low (data not shown). MGB probes have the advantage of an MGB group that binds nonspecifically to double-stranded DNA and allows the probes to be shorter and thus more discriminatory. This is the first assay that uses these probes in a bacteriological context.
The MGB assay was able to amplify a katG fragment when 5 fg of purified mycobacterial DNA per reaction was added (corresponding to one mycobacterial genome). The detection limit for mycobacterial DNA isolated from artificially infected sputum samples was 20 genomes per reaction, corresponding to 103 mycobacteria per ml of sputum. This was assessed only for the wild-type H37Rv strain because of the high risk of working with suspensions containing 108 high-level INH-resistant M. tuberculosis organisms per ml. However, it is likely that the detection limit for M. tuberculosis with mutant DNA is similar, as amplifications of both wild-type and mutant M. tuberculosis DNAs from water were also similar. The assay was negative when tested on large amounts of nonmycobacterial DNA or DNA from nontuberculous mycobacteria.
The assay was evaluated by testing ZN-positive sputum samples from 20 patients from Kazakhstan and Moldova. The katG fragment could be amplified from 18 sputum samples: 14 were shown to have the codon 315 mutant genotype, and the other 4 carried the wild-type allele. In one of the two remaining sputum samples, in which no hybridization could be detected, we found the rare AGC→AAC mutation by means of agarose gel electrophoresis and sequencing, a mutation that has occasionally been found in other studies (1, 17, 21).
The real-time PCR results for 13 (65%) of the sputum samples from 20 patients from The Netherlands infected with INH-susceptible M. tuberculosis were positive for the wild-type allele, whereas 7 samples remained negative. When DNA from these negative sputum samples was used for a conventional PCR with the same primers and agarose gel electrophoresis, no amplification product was observed. This indicates that the sensitivity of real-time PCR for the detection of mycobacteria in sputum is lower than those of diagnostic tests such as ZN staining. The lower sensitivity of real-time PCR on sputum samples from The Netherlands compared with samples from Kazakhstan and Moldova is probably due to the stage of the mycobacterial disease and the number of mycobacteria present in the sputum. In The Netherlands, patients are diagnosed at an early stage of the disease, whereas patients from Kazakhstan and Moldova usually present with a distinct syndrome of pulmonary tuberculosis. Unfortunately, quantitative ZN data from sputum samples were not available to elucidate the significance of the number of acid-fast stained rods present in sputum samples for the sensitivity of the real-time PCR. In addition, the possibility that the wild-type probe differs in sensitivity from the mutant probe cannot be excluded. Despite the fact that the wild-type probe was tested only on sputum artificially infected with M. tuberculosis H37Rv, we consider this possibility not very likely, since the probes had identical sensitivities when tested on mycobacterial DNA in water.
The overall sensitivity of the assay was 75% (17 out of 20 sputum samples from Kazakhstan and Moldova and 13 out of 20 susceptible controls). Combined with agarose gel electrophoresis, the sensitivity was 78%. These results are lower than the results for diagnostic PCR-based assays for tuberculosis (12). However, these assays usually have targets that are present in high numbers, like IS6110 or 16S rRNA.
The results were compared with sequencing of DNAs isolated from cultures of M. tuberculosis or from sputum. The sensitivities and specificities of the probes were calculated from the sequence data as 70 and 94% for the wild-type probe and 82 and 100% for the mutant probe (Table 4). These sensitivity data correspond to the findings of Espasa et al., who found a sensitivity of 78% for a mutant Taqman probe with 52 sputum samples with INH-resistant strains and 67% for a wild-type probe with 15 samples (M. Espasa, J. M. Gonzales, F. Alcaide, J. Lonca, X.-M. Manterola, E. Verdu, and P. Coll, Abstr. 12th Eur. Congr. Clin. Microbiol. Infect. Dis., Clin. Microbiol. Infect. 8:O131, 2002). The sequence data were concordant with the results of the assay for all but two of the sputum samples. In one sputum sample (Table 3, M3), we found the wild-type codon with our assay and the mutant codon by sequencing DNA from culture. When we performed our assay on this DNA, we also found the mutant codon. This might be explained by the presence of a heterogeneous and heteroresistant infection with M. tuberculosis, in which a patient is simultaneously infected with two different isolates of M. tuberculosis with different resistance characteristics (23). In another sputum sample (Table 3, K2), the assay was negative and no fragment was seen on an agarose gel, but with sequence analysis of DNA from culture, the mutant codon was obtained. This could be due either to the presence of inhibitory substances in the sputum or to an amount of mycobacteria that was below the threshold of our assay.
In four sputum samples (Table 3, M8, K5, K7, and K8), mycobacteria carrying the mutation were detected, while the INH MIC was <2 μg/ml. This is contradictory to results from an earlier study (31), in which it was shown that the presence of this mutation was linked with an MIC of ≥2 μg/ml. However, when tested again, colonies cultured from two of these sputum samples (Table 3, K7 and K8) grew on Tarshis medium containing >10 and 8 μg of INH/ml, respectively. Isolates from the two other patients were not available for further testing.
Various assays for detection by real-time PCR of point mutations associated with resistance to INH or rifampin in M. tuberculosis have been described, most of them making use of molecular beacons (9, 22, 29). However, none of these assays has been successfully used directly on clinical (sputum) samples, with the exception of the Taqman probe assay developed by Espasa et al., which produced results similar to those of our assay (Espasa et al., Clin. Microbiol. Infect. 8:O131, 2002). Mokrousov et al. have described a PCR-based method (multiplex allele-specific PCR) that was used on positive auramine-stained slides (19).
The real-time PCR described here can be very useful in rapid laboratory diagnosis of INH-resistant M. tuberculosis in ZN-positive sputum samples. When combined with agarose gel electrophoresis, all known mutations of codon 315 of katG can be detected within 2 or 3 days. However, in >95% of all cases, the time required to produce diagnostic results can be shortened to hours, compared to weeks for culture diagnosis.
Acknowledgments
This work was supported by a grant from the Foundation Microbiology Leiden.
REFERENCES
- 1.Abate, G., S. E. Hoffner, V. O. Thomsen, and H. Miorner. 2001. Characterization of isoniazid-resistant strains of Mycobacterium tuberculosis on the basis of phenotypic properties and mutations in katG. Eur. J. Clin. Microbiol. Infect. Dis. 20:329-333. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Banerjee, A., E. Dubnau, A. Quemard, V. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. de Lisle, and W. R. Jacobs, Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230. [DOI] [PubMed] [Google Scholar]
- 4.Berstein, J., W. A. Lott, B. A. Steinberg, and H. L. Yale. 1952. Chemotherapy of experimental tuberculosis. Am. Rev. Tuberc. 65:357-364. [DOI] [PubMed] [Google Scholar]
- 5.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espinal, M. A., A. Laszlo, L. Simonsen, F. Boulahbal, S. J. Kim, A. Reniero, S. Hoffner, H. L. Rieder, N. Binkin, C. Dye, R. Williams, M. C. Raviglione, et al. 2001. Global trends in resistance to antituberculosis drugs. N. Engl. J. Med. 344:1294-1303. [DOI] [PubMed] [Google Scholar]
- 7.Frieden, T. R., T. Sterling, A. Pablos-Mendez, J. O. Kilburn, G. M. Cauthen, and S. W. Dooley. 1993. The emergence of drug-resistant tuberculosis in New York City. N. Engl. J. Med. 328:521-526. [DOI] [PubMed] [Google Scholar]
- 8.Gangadharam, P. (ed.). 1984. Drug resistance in mycobacteria. CRC Press, Boca Raton, Fla.
- 9.Garcia de Viedma, D., M. del Sol Diaz Infantes, F. Lasala, F. Chaves, L. Alcala, and E. Bouza. 2002. New real-time PCR able to detect in a single tube multiple rifampin resistance mutations and high-level isoniazid resistance mutations in Mycobacterium tuberculosis. J. Clin. Microbiol. 40:988-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haas, W. H., K. Schilke, J. Brand, B. Amthor, K. Weyer, P. B. Fourie, G. Bretzel, V. Sticht-Groh, and H. J. Bremer. 1997. Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob. Agents Chemother. 41:1601-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heym, B., N. Honore, C. Truffot-Pernot, A. Banerjee, C. Schurra, W. R. Jacobs, Jr., J. D. van Embden, J. H. Grosset, and S. T. Cole. 1994. Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: a molecular study. Lancet 344:293-298. [DOI] [PubMed] [Google Scholar]
- 12.Ieven, M., and H. Goossens. 1997. Relevance of nucleic acid amplification techniques for diagnosis of respiratory tract infections in the clinical laboratory. Clin. Microbiol. Rev. 10:242-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kritski, A. L., M. J. Marques, M. F. Rabahi, M. A. Vieira, E. Werneck-Barroso, C. E. Carvalho, G. D. N. Andrade, R. Bravo-de-Souza, L. M. Andrade, P. P. Gontijo, and L. W. Riley. 1996. Transmission of tuberculosis to close contacts of patients with multidrug-resistant tuberculosis. Am. J. Respir. Crit. Care Med. 153:331-335. [DOI] [PubMed] [Google Scholar]
- 14.Kubica, G. P., W. E. Dye, M. L. Cohn, and G. Middlebrook. 1963. Sputum digestion and decontamination with N-acetyl-l-cysteine-sodium hydroxide for culture of mycobacteria. Am. Rev. Respir. Dis. 87:775-779. [DOI] [PubMed] [Google Scholar]
- 15.Kutyavin, I. V., I. A. Afonina, A. Mills, V. V. Gorn, E. A. Lukhtanov, E. S. Belousov, M. J. Singer, D. K. Walburger, S. G. Lokhov, A. A. Gall, R. Dempcy, M. W. Reed, R. B. Meyer, and J. Hedgpeth. 2000. 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 28:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrakchi, H., G. Laneelle, and A. Quemard. 2000. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology 146:289-296. [DOI] [PubMed] [Google Scholar]
- 17.Marttila, H. J., H. Soini, E. Eerola, E. Vyshnevskaya, B. I. Vyshnevskiy, T. F. Otten, A. V. Vasilyef, and M. K. Viljanen. 1998. A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob. Agents Chemother. 42:2443-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Middlebrook, G. 1952. Sterilization of tubercle bacilli by isonicotinic acid hydrazide and the incidence of variants resistant to the drug. Am. Rev. Tuberc. 65:765-767. [PubMed] [Google Scholar]
- 19.Mokrousov, I., T. Otten, M. Filipenko, A. Vyazovaya, E. Chrapov, E. Limeschenko, L. Steklova, B. Vyshnevskiy, and O. Narvskaya. 2002. Detection of isoniazid-resistant Mycobacterium tuberculosis strains by a multiplex allele-specific PCR assay targeting katG codon 315 variation. J. Clin. Microbiol. 40:2509-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris, S., G. H. Bai, P. Suffys, L. Portillo-Gomez, M. Fairchok, and D. Rouse. 1995. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J. Infect. Dis. 171:954-960. [DOI] [PubMed] [Google Scholar]
- 21.Musser, J. M., V. Kapur, D. L. Williams, B. N. Kreiswirth, D. van Soolingen, and J. D. van Embden. 1996. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J. Infect. Dis. 173:196-202. [DOI] [PubMed] [Google Scholar]
- 22.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 23.Rinder, H., K. T. Mieskes, and T. Loscher. 2001. Heteroresistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 5:339-345. [PubMed] [Google Scholar]
- 24.Ristow, M., M. Mohlig, M. Rifai, H. Schatz, K. Feldmann, and A. Pfeiffer. 1995. New isoniazid/ethionamide resistance gene mutation and screening for multidrug-resistant Mycobacterium tuberculosis strains. Lancet 346:502-503. [DOI] [PubMed] [Google Scholar]
- 25.Rouse, D. A., J. A. DeVito, Z. Li, H. Byer, and S. L. Morris. 1996. Site-directed mutagenesis of the katG gene of Mycobacterium tuberculosis: effects on catalase-peroxidase activities and isoniazid resistance. Mol. Microbiol. 22:583-592. [DOI] [PubMed] [Google Scholar]
- 26.SantaLucia, J., Jr. 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 95:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoeckle, M. Y., L. Guan, N. Riegler, I. Weitzman, B. Kreiswirth, J. Kornblum, F. Laraque, and L. W. Riley. 1993. Catalase-peroxidase gene sequences in isoniazid-sensitive and -resistant strains of Mycobacterium tuberculosis from New York City. J. Infect. Dis. 168:1063-1065. [DOI] [PubMed] [Google Scholar]
- 28.Tarshis, M. S., and A. W. Frisch. 1951. Blood media for the cultivation of Mycobacterium tuberculosis. Am. J. Clin. Pathol. 21:101-113. [DOI] [PubMed] [Google Scholar]
- 29.Torres, M. J., A. Criado, J. C. Palomares, and J. Aznar. 2000. Use of real-time PCR and fluorimetry for rapid detection of rifampin and isoniazid resistance-associated mutations in Mycobacterium tuberculosis. J. Clin. Microbiol. 38:3194-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Zanden, A. G., E. M. te Koppele-Vije, N. Vijaya Bhanu, D. van Soolingen, and L. M. Schouls. 2003. Use of DNA extracts from Ziehl-Neelsen-stained slides for molecular detection of rifampin resistance and spoligotyping of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:1101-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Soolingen, D., P. E. de Haas, H. R. van Doorn, E. Kuijper, H. Rinder, and M. W. Borgdorff. 2000. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in the Netherlands. J. Infect. Dis. 182:1788-1790. [DOI] [PubMed] [Google Scholar]
- 32.Wengenack, N. L., J. R. Uhl, A. L. St. Amand, A. J. Tomlinson, L. M. Benson, S. Naylor, B. C. Kline, F. R. Cockerill III, and F. Rusnak. 1997. Recombinant Mycobacterium tuberculosis KatG(S315T) is a competent catalase-peroxidase with reduced activity toward isoniazid. J. Infect. Dis. 176:722-727. [DOI] [PubMed] [Google Scholar]
- 33.Wilson, T. M., G. W. de Lisle, and D. M. Collins. 1995. Effect of inhA and katG on isoniazid resistance and virulence of Mycobacterium bovis. Mol. Microbiol. 15:1009-1015. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization. 2001. World health report 2001. World Health Organization, Geneva, Switzerland.
- 35.Zhang, Y., B. Heym, B. Allen, D. Young, and S. Cole. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Y., and D. B. Young. 1993. Molecular mechanisms of isoniazid: a drug at the front line of tuberculosis control. Trends Microbiol. 1:109-113. [DOI] [PubMed] [Google Scholar]
- 37.Zwadyk, P., Jr., J. A. Down, N. Myers, and M. S. Dey. 1994. Rendering of mycobacteria safe for molecular diagnostic studies and development of a lysis method for strand displacement amplification and PCR. J. Clin. Microbiol. 32:2140-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]