Abstract
Collected between 1993 and 2002 at a Taiwanese university hospital, 77 group A streptococcus isolates associated with scarlet fever were grouped by emm typing and pulsed-field gel electrophoresis. The predominance of an emm1 clone before 1996 and the presence of genetically diverse emm1 and emm4 strains thereafter were found.
The resurgence of invasive infections caused by group A streptococcus (GAS) has been noted worldwide since the mid-1980s (5, 7, 8, 11, 13, 26). Epidemiologic studies of GAS have relied historically on M and T serotyping (10, 16, 18); however, many strains cannot be typed because of the lack of appropriate type-specific antisera, and comprehensive typing serum sets are not available for most of the laboratories in the world. Recently, sequence analysis of the 5′ end of the emm gene that encodes the M protein has been widely used to characterize GAS strains (2-4, 9). Since the 5′ emm sequences can be efficiently and reliably used to predict M serotypes (2, 4), the typing system has become a useful molecular epidemiologic tool with which to survey and monitor GAS isolate diversity.
Among the GAS M serotypes, M1, M2, M3, M4, M6, M18, and M22 have been associated with scarlet fever epidemics (1, 12, 15, 19, 21-24). Scarlet fever remains a notifiable disease in Taiwan. An outbreak of scarlet fever occurring at a hospital day care center during a 2-month period in 1996 was reported to be caused by one GAS clone (12); however, genetic relationships among GAS strains associated with other scarlet fever epidemics in Taiwan and emm types of these strains are still unknown. The purpose of the present study was to investigate the molecular epidemiology of GAS isolates associated with scarlet fever in southern Taiwan. The prevalence and mechanisms of erythromycin resistance in these isolates were also determined.
GAS isolates.
Among clinical isolates of GAS randomly collected between 1993 and 1995 and consecutively collected between 1997 and 2002 at the National Cheng Kung University Hospital, Tainan City, Taiwan, a total of 77 isolates were found to be associated with scarlet fever and were investigated in this study. All of the isolates studied were recovered from throat swabs of different patients. Isolates collected between 1993 and 1998 have been tested previously for their resistance phenotypes and genotypes, and among these isolates, erythromycin-resistant isolates have been examined for their pulsed-field gel electrophoresis (PFGE) patterns (30).
emm typing.
PCR amplification of the emm gene was performed with previously described primers (2). The amplicons were sequenced on an ABI PRISM 310 sequencer analyzer (Applied Biosystems, Foster City, Calif.) with the primer pair for PCR. The emm sequences obtained were compared with those deposited in the GenBank database. An isolate was considered to be of a given emm type if it had ≥95% identity over the first 160 bases obtained (4, 9).
PFGE.
PFGE was carried out with a contour-clamped homogeneous electric field system (CHEF Mapper XA; Bio-Rad Laboratories, Hercules, Calif.) as described previously (6). Genomic DNAs were prepared as described by Piggot et al. (25) and were digested overnight with 10 U of SmaI (New England Biolabs, Beverly, Mass.). Restricted DNA samples were electrophoresed through a 1% agarose gel in 0.5× Tris-borate-EDTA buffer at 6 V/cm with switch times ranging from 2.16 to 44.69 s and a lineal switch time ramp for 27 h. Bacteriophage lambda DNA concatemers (Gibco BRL, Gaithersburg, Md.) were used as size standards. The PFGE patterns obtained were interpreted in accordance with the criteria of Tenover et al. (28).
Susceptibility testing and determination of erythromycin resistance phenotypes.
MICs of penicillin, erythromycin, and clindamycin (Sigma Chemical Co., St. Louis, Mo.) were determined by the agar dilution method in accordance with the recommendations of the NCCLS (20). Resistance phenotypes of erythromycin-resistant isolates were determined by the double-disk test with erythromycin and clindamycin (Becton Dickinson, Cockeysville, Md.) disks as described previously (27). Blunting of the growth inhibition zone around clindamycin in the area between the two disks indicated an inducible type of macrolide, lincosamide, and streptogramin B (MLS) resistance (27, 29), and resistance to both disks indicated a constitutive type of MLS resistance (17, 27). The M phenotype was characterized by resistance to erythromycin and susceptibility to clindamycin with no blunting of the growth inhibition zone (27).
Detection of erythromycin resistance genes.
Detection of erythromycin resistance genes in erythromycin-resistant isolates by PCR was performed with the oligonucleotide primer pairs specific to ermB, ermTR, and mefA (14, 30). The expected sizes of PCR products were 640 bp for ermB, 348 bp for mefA, and 530 bp for ermTR.
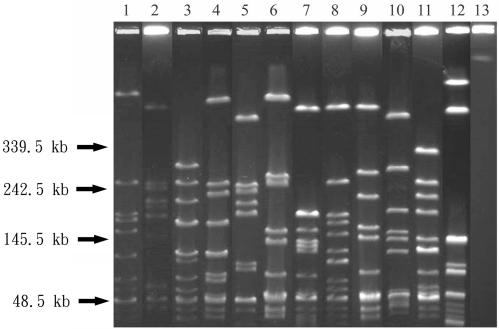
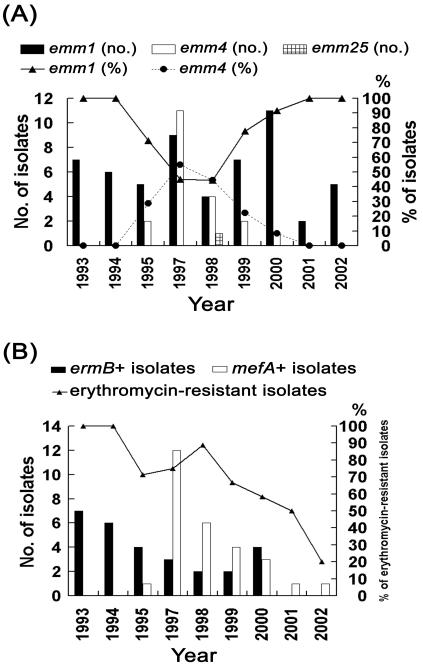
Three emm types were obtained for the 77 isolates (Table 1). The most prevalent emm sequence type was emm1 with 56 (72.7%) isolates, followed by emm4 with 20 (26.0%) isolates. PFGE yielded 13 different patterns among these isolates, and each pattern from a representative isolate is shown in Fig. 1. The most prevalent PFGE patterns represented were pattern J with 25 (32.5%) isolates, pattern D with 13 (16.9%) isolates, and pattern G with 8 (10.4%) isolates. The emm1 type, prevalent throughout the study period, was predominant among the isolates collected between 1993 and 1995 and after 1998 (Fig. 2A). Only three PFGE patterns were obtained for the isolates collected between 1993 and 1995 (Table 1). All of the emm1 isolates collected during this period except one showed PFGE pattern J, suggesting the occurrences of scarlet fever outbreaks caused by a single emm1 clone between 1993 and 1995 in southern Taiwan. After 1996, genomic diversity was found among the emm1 isolates, which displayed nine PFGE patterns (A, E, F, G, H, I, J, L, and M). None of these patterns were found to be prominent. Serotype M1 isolates have caused large outbreaks of scarlet fever in Germany (15). The increase in the numbers of serotype M1 isolates associated with invasive GAS infections has been reported around the world since the mid-1980s (5, 7, 8, 11, 13, 26). The serotype has also been reported to be more prevalent among invasive isolates than among noninvasive isolates in Taiwan (13). Whether the association of emm1 and scarlet fever outbreaks found in the present study is due to a high prevalence of emm1 among GAS strains in Taiwan or to specific pathogenic factors is not known and needs further investigation.
TABLE 1.
Susceptibility to erythromycin, resistance genotypes, and strain typing of the 77 GAS isolates associated with scarlet fever
| Year (total no. of isolates) and susceptibility to erythromycin(no. of isolates) | emm resistance gene/PFGE pattern (no. of isolates) |
|---|---|
| 1993-1995 (20) | |
| Susceptible (2) | emm4/K (2) |
| Resistant (18) | |
| ermB (17) | emm1/J (17) |
| mefA (1) | emm1/A (1) |
| 1997-1999 (38) | |
| Susceptible (9) | emm1/E (1), emm1/G (1), emm1/H (3), emm1/I (1), emm4/C (3) |
| Resistant (29) | |
| ermB (7) | emm1/A (1), emm1/E (1), emm1/I (2), emm1/J (3) |
| mefA (22) | emm1/A (4), emm1/E (1), emm1/H (1), emm1/J (1), emm4/C (2), emm4/D (12), emm25/B (1) |
| 2000-2002 (19) | |
| Susceptible (10) | emm1/E (2), emm1/G (7), emm1/L (1) |
| Resistant (9) | |
| ermB (4) | emm1/J (4) |
| mefA (5) | emm1/F (1), emm1/M (3), emm4/D (1) |
FIG. 1.
PFGE profiles of SmaI-digested genomic DNAs from 13 representative isolates of GAS. Lanes 1 to 13, PFGE patterns A to M.
FIG. 2.
(A) Changes in the numbers and percentages of GAS isolates of different emm types during the period studied. (B) Changes in the percentage of erythromycin-resistant GAS isolates and in the numbers of ermB-positive and mefA-positive isolates during the study period.
The emm4 type was only found in the isolates collected between 1995 and 2000 and was most prevalent in 1997 and 1998 (Fig. 2A). Only three PFGE patterns were obtained for the 20 emm4 isolates, of which 13 (65.0%) exhibited PFGE pattern D (Table 1). These data suggest the occurrence of scarlet fever outbreaks caused by an emm4 clone between 1997 and 1999 in southern Taiwan. Scarlet fever epidemics associated with M4 strains have also been reported in the United Kingdom and elsewhere (8, 24).
All 77 GAS isolates were susceptible to penicillin (MICs, <0.03 μg/ml), and 56 (72.7%) isolates were resistant to erythromycin (MIC, ≥1 μg/ml). Our previous study has demonstrated a high rate (63.2%) of erythromycin resistance in GAS isolates collected between 1993 and 1998 (30). The percentage of erythromycin resistance during the period was 83.6% for the isolates associated with scarlet fever and 56.8% for the isolates from non-scarlet fever patients (unpublished data). The percentages of erythromycin resistance in GAS associated with scarlet fever were found to decline after 1998 in the present study (Fig. 2B). The increase in the erythromycin-susceptible isolates might be in part due to the occurrence of an outbreak caused by an emm1 clone with PFGE pattern G (Table 1). Whether the decrease in the number and percentage of erythromycin-resistant isolates represents a trend in erythromycin resistance in GAS associated with scarlet fever or is just a temporal variation in this area deserves further investigation. If such a trend exists, erythromycin may become the drug of choice for the treatment of scarlet fever in penicillin-hypersensitive patients in Taiwan once again.
The erythromycin resistance phenotypes of the 56 erythromycin-resistant isolates were determined by the double-disk test with erythromycin and clindamycin disks. Twenty-eight (50.0%) isolates were of the erythromycin-resistant M phenotype, 28 (50.0%) isolates were of the constitutive MLS resistance phenotype, and none of the isolates exhibited the inducible MLS resistance phenotype. All 28 constitutively MLS-resistant isolates showed high-level resistance to both erythromycin (MICs, ≥128 μg/ml) and clindamycin (MICs, ≥128 μg/ml). All 28 isolates demonstrating the M phenotype of erythromycin resistance showed low-level resistance to erythromycin (MICs, 2 to 32 μg/ml) and were susceptible to clindamycin (MIC, 0.03 to 0.13 μg/ml). Genetic determinants of erythromycin resistance were investigated in all erythromycin-resistant isolates by means of PCR experiments with specific primer sets for ermB, ermTR, and mefA. As predicted, all 28 constitutively MLS-resistant isolates carried the ermB gene and all 28 isolates demonstrating the M phenotype of erythromycin resistance carried the mefA gene. The resistance genotypes showed a different chronological distribution. The ermB-positive isolates were predominant between 1993 and 1995, and the mefA-positive isolates were predominant between 1997 and 1999 (Fig. 2B). The numbers of mefA-positive isolates appeared to decline year by year after 1997. Only three PFGE patterns were obtained for the 28 ermB-positive isolates, and 25 (89.3%) of them showed PFGE pattern J (Table 1). The data indicate that a genetically related ermB-positive clone cause outbreaks during 1993 and 1995, and the clone still existed more recently. Genetic diversity was found among the mefA-positive isolates, which revealed nine PFGE patterns (A, B, C, D, E, F, H, J, and M). However, 13 (46.4%) of the 28 mefA-positive isolates showed pattern D. Thus, the predominance of mefA between 1996 and 1999 was in part due to the prevalence of a genetically related clone.
In conclusion, emm1 was found to be prevalent throughout the past 10 years and the outbreaks of scarlet fever caused by emm4 isolates were found to occur between 1995 and 2000 in the present study. The outbreaks in the early 1990s were mostly associated with an ermB-positive erythromycin-resistant emm1 clone, and those occurring thereafter were associated with genetically diverse clones. Moreover, erythromycin-resistant isolates seem to be decreasing. This paper is the first report of the genotypes of GAS associated with scarlet fever in Taiwan. Our data provide a basis for future studies on changes in the epidemiology of GAS, outbreak investigation, and development of preventive measures and of recommendations for treatment strategies in Taiwan.
Acknowledgments
This work was partially supported by grants DOH 91-DC-1010 from the Department of Health, NSC90-2320-B-006-088 from the National Science Council, and NHRI-EX91-9027 SP from the National Health Research Institutes, Taiwan.
REFERENCES
- 1.Anthony, B. F., T. Yamauchi, J. S. Penso, I. Kamei, and S. S. Chapman. 1974. Classroom outbreak of scarlet fever and acute glomerulonephritis related to type 2 (M-2, T-2) group A streptococcus. J. Infect. Dis. 129:336-340. [DOI] [PubMed] [Google Scholar]
- 2.Beall, B., R. R. Facklam, and T. Thompson. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall, B., R. R. Facklam, T. Hoenes, and B. Schwartz. 1997. Survey of emm gene sequences and T-antigen types from systemic Streptococcus pyogenes infection isolates collected in San Francisco, California; Atlanta, Georgia; and Connecticut in 1994 and 1995. J. Clin. Microbiol. 35:1231-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beall, B., R. R. Facklam, J. A. Elliott, A. R. Franklin, T. Hoenes, D. Jackson, L. Laclaire, T. Thompson, and R. Viswanathan. 1998. Streptococcal emm types associated with T-agglutination types and the use of conserved emm gene restriction fragment patterns for subtyping group A streptococci. J. Med. Microbiol. 47:893-898. [DOI] [PubMed] [Google Scholar]
- 5.Carapetis, J., R. Robins-Browne, D. Martin, T. Shelby-James, and G. Hogg. 1995. Increasing severity of invasive group A streptococcal disease in Australia: clinical and molecular epidemiological features and identification of a new virulent M-nontypeable clone. Clin. Infect. Dis. 21:1220-1227. [DOI] [PubMed] [Google Scholar]
- 6.Chu, G., D. Vollrath, and R. W. Davis. 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234:1582-1585. [DOI] [PubMed] [Google Scholar]
- 7.Cleary, P. P., E. L. Kaplan, J. P. Handley, A. Wlazlo, M. H. Kim, A. R. Jauser, and P. M. Schlievert. 1992. Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet 339:518-521. [DOI] [PubMed] [Google Scholar]
- 8.Colman, G., A. Tanna, A. Efstratiou, and E. T. Gaworzewska. 1993. The serotypes of Streptococcus pyogenes present in Britain during 1980-1990 and their association with disease. J. Med. Microbiol. 39:165-178. [DOI] [PubMed] [Google Scholar]
- 9.Facklam, R., B. Beall, A. Efstratiou, V. Fischetti, D. Johnson, E. Kaplan, P. Kriz, M. Lovgren, D. Martin, B. Schwartz, A. Totolian, D. Bessen, S. Hollingshead, F. Rubin, J. Scott, and G. Tyrrell. 1999. emm typing and validation of provisional M types for group A streptococci. Emerg. Infect. Dis. 5:247-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischetti, V. A. 1989. Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev. 2:285-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holm, S. E., A. Norrby, A.-M. Bergholm, and M. Norgren. 1992. Aspects of pathogenesis of serious group A streptococcal infections in Sweden, 1988-1989. J. Infect. Dis. 166:31-37. [DOI] [PubMed] [Google Scholar]
- 12.Hsueh, P. R., L. J. Teng, P. I. Lee, P. C. Yang, L. M. Huang, S. C. Chang, C. Y. Lee, and K. T. Luh. 1997. Outbreak of scarlet fever at a hospital day care centre: analysis of strain relatedness with phenotypic and genotypic characteristics. J. Hosp. Infect. 36:191-200. [DOI] [PubMed] [Google Scholar]
- 13.Hsueh, P. R., J. J. Wu, P. J. Tsai, J. W. Liu, Y. C. Chuang, and K. T. Luh. 1998. Invasive group A streptococcal disease in Taiwan is not associated with the presence of streptococcal pyrogenic exotoxin genes. Clin. Infect. Dis. 26:584-589. [DOI] [PubMed] [Google Scholar]
- 14.Kataja, J., P. Huovinen, M. Skurnik, The Finnish Study Group for Antimicrobial Resistance, and H. Seppälä. 1999. Erythromycin resistance genes in group A streptococci in Finland. Antimicrob. Agents Chemother. 43:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köhler, W., D. Gerlach, and H. Knöll. 1987. Streptococcal outbreaks and erythrogenic toxin type A. Zentbl. Bakteriol. Hyg. A. 266:104-111. [DOI] [PubMed] [Google Scholar]
- 16.Lancefield, R. C. 1962. Current knowledge of the type specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed] [Google Scholar]
- 17.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, J. M., E. R. Wald, and M. Green. 1998. Field inversion gel electrophoresis as a typing system for group A streptococcus. J. Infect. Dis. 177:504-506. [DOI] [PubMed] [Google Scholar]
- 19.Musser, J. M., K. Nelson, R. K., Selander, D. Gerlach, J. C. Huang, V. Kohler, and S. Kanjulal. 1993. Temporal variation in bacterial disease frequency: molecular population genetic analysis of scarlet fever epidemics in Ottawa and in eastern Germany. J. Infect. Dis. 167:759-762. [DOI] [PubMed] [Google Scholar]
- 20.NCCLS. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. NCCLS, Wayne, Pa.
- 21.Ohga, S., K. Okada, K. Mitsui, T. Aoki, and K. Ueda. 1992. Outbreaks of group A beta-hemolytic streptococcal pharyngitis in children: correlation of serotype T4 with scarlet fever. Scand. J. Infect. Dis. 24:599-605. [DOI] [PubMed] [Google Scholar]
- 22.Parker, M. T. 1967. International survey of the distribution of serotypes of Streptococcus pyogenes (group A streptococcus). Bull. W. H. O. 37:513-527. [PMC free article] [PubMed] [Google Scholar]
- 23.Perea-Mejíam, L. M., A. E. Inzunza-Montiel, and A. Cravioto. 2002. Molecular characterization of group A streptococcus strains isolated during a scarlet fever outbreak. J. Clin. Microbiol. 40:278-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perks, E. M., and R. T. Mayon-White. 1983. The incidence of scarlet fever. J. Hyg. 91:203-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piggot, P. J., M. Amjad, J. J. Wu, H. Sandoval, and J. Castro. 1990. Genetic and physical maps of Bacillus subtilis 168, p. 493-543. In C. R. Harwood and S. M. Cutting (ed.), Molecular biology methods for Bacillus. John Wiley & Sons Ltd., West Sussex, England.
- 26.Schwartz, B., R. R. Facklam, and R. F. Breiman. 1990. Changing epidemiology of group A streptococcal infection in the USA. Lancet 336:1167-1171. [DOI] [PubMed] [Google Scholar]
- 27.Seppälä, H., A. Nissinen, Q. Yu, and P. Huovinen. 1993. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J. Antimicrob. Chemother. 32:885-891. [DOI] [PubMed] [Google Scholar]
- 28.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisblum, B. 1985. Inducible resistance to macrolides, lincosamides and streptogramin type B antibiotics: the resistance phenotype, its biological diversity, and structural elements that regulate expression—a review. J. Antimicrob. Chemother. 16(Suppl. A):63-90. [DOI] [PubMed] [Google Scholar]
- 30.Yan, J. J., H. M. Wu, H. M. Fu, C. T. Lee, and J. J. Wu. 2000. Polyclonal widespread of mefA-containing isolates among erythromycin-resistant group A streptococci in southern Taiwan. J. Clin. Microbiol. 38:2475-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]