Abstract
We have developed a rapid and simplified approach for the strain characterization of Staphylococcus aureus on the basis of multilocus sequence typing (MLST) in which sequence variations in the MLST housekeeping gene loci are detected by restriction fragment pattern analysis rather than sequencing; we refer to this approach as multilocus restriction fragment typing (MLRFT). Briefly, MLRFT for S. aureus involves the PCR amplification of each of the seven MLST housekeeping gene loci by using the same primer pairs used in MLST. The amplicons are then digested directly with one or two restriction enzymes and the restriction fragments are resolved by agarose gel electrophoresis. Projection from published MLST data shows that MLRFT captures about 95% of the genetic diversity detected by MLST. The MLRFT approach was validated with a set of 59 methicillin-susceptible and 44 methicillin-resistant S. aureus isolates from community-acquired and nosocomial sources which had previously been characterized by pulsed-field gel electrophoresis (PFGE). MLRFT resolved the 103 isolates into 15 restriction fragment types, giving a discrimination index of 89.0%. Clonal groupings established by MLRFT correlated well with those established by PFGE. In short, MLRFT provides a convenient alternative to MLST and PFGE because it requires minimal laboratory facilities and is relatively simple and inexpensive to perform.
Staphylococcus aureus has emerged over the past several decades as a leading cause of hospital- and community-acquired infections (9). A significant component in the “success” of S. aureus has been its acquisition of antibiotic resistance factors (1). As new antibiotics have come into use, S. aureus has responded soon after with resistant strains. This phenomenon has made therapy of staphylococcal diseases a global challenge. Penicillin-resistant strains, for example, appeared in hospitalized patients within a short time after the introduction of the antibiotic; over time, penicillin-resistant strains have spread into the community to the extent that penicillin is now only of very limited value as treatment for S. aureus infections. There is concern that methicillin-resistant S. aureus (MRSA) may be following the same path from the hospital to the community (1). Accordingly, there is considerable epidemiological interest in the tracking of strains to gain a more complete picture of the distribution of strains in the population and the dynamics of clonal spread (4).
Molecular typing approaches have been used to great advantage in identifying and monitoring the local and international spread of S. aureus outbreak strains. Pulsed-field gel electrophoresis (PFGE) is generally regarded as the most discriminating technique for strain identification, particularly in the context of identifying strains involved in local outbreaks (14, 16, 18). However, because strain characterization by PFGE is based on pattern matching, the method serves less well for comparison of strains between laboratories (3, 19). Moreover, there is no convenient metric that reliably measures genetic relationships among strains with substantially different PFGE patterns. These features limit the value of PFGE as a tool for investigating the population genetics and global epidemiology of S. aureus. These problems are overcome through the use of multilocus sequence typing (MLST) and spa typing (5, 16). Sequence data are portable; i.e., they can be archived in electronically accessible database repositories and are easily analyzed to provide measures of genetic relationships and population structures. The sequence-based typing approaches generally lack the discrimination power of PFGE for differentiating closely related strains and thus are less useful for the epidemiological investigation of local outbreaks. The two approaches are thus complementary, with sequence-based typing being better at revealing the big picture with regard to strain relationships and PFGE being better at providing fine-scale differentiation.
Unfortunately, neither PFGE nor the sequence-based approaches are conveniently applied in a routine clinical setting. Both require specialized equipment and are relatively costly and time-consuming. The cost factor in particular is a constraint on their use in economically disadvantaged countries, where epidemiological characterization of disease transmission patterns is a major need.
We describe here a rapid, low-cost strain-typing technique based on restriction fragment (RF) pattern analysis of the seven loci used in MLST. Overall, this approach, termed here multilocus RF typing (MLRFT), captures about 95% of the between-strain genetic variability detected by MLST. Moreover, by basing MLRFT on the same seven loci used in MLST, it is possible to systematically link MLRFT results to the MLST sequence database. MLRFT thus has value both as a convenient stand-alone technique for strain typing and as a rapid screening technique to categorize strains for targeted PFGE and/or MLST analysis.
(This work was presented in part at the 101st General Meeting of the American Society for Microbiology, 2001.)
MATERIALS AND METHODS
Bacterial isolates.
A sample population consisting of 59 methicillin-susceptible S. aureus (MSSA) isolates and 44 MRSA isolates previously characterized by PFGE was analyzed in this study; the isolates were selected to represent a diversity of PFGE types. Most of the isolates, 59 MSSA isolates and 17 MRSA isolates, were nasal isolates collected in a community-based study of the urban poor population in San Francisco, Calif., between August 1999 and April 2000 (2). Of the remainder of the isolates, 12 MRSA isolates originated from an outpatient, population-based prevalence study of nasal carriage among injection drug users (1); and 16 MRSA isolates were nosocomial isolates from blood (n = 4), wounds or abscesses (n = 8), respiratory sources (n = 3), and urine (n = 1) collected between 1995 and 2000 at San Francisco General Hospital.
Methicillin resistance and antibiotic susceptibility testing.
The mecA gene was detected as described previously (11). Susceptibility to methicillin was determined as described previously (10). Susceptibility to ampicillin, ciprofloxacin, tetracycline, gentamicin, erythromycin, trimethoprim-sulfamethoxazole, clindamycin, linezolid, and vancomycin was determined on Mueller-Hinton agar (BBL, Cockeysville, Md.) by the conventional Kirby-Bauer disk diffusion method. Antibiograms were interpreted in accordance with National Committee for Clinical Laboratory Standards guidelines (document M100-S3) (12).
PFGE typing.
The S. aureus isolates were typed by PFGE of SmaI- digested chromosomal DNA (2). A standard reference sample was run on each gel. Isolates were scored as described by Tenover et al. (18); we classified a PFGE group as isolates with patterns that differed by less than six bands.
MLRFT.
S. aureus DNA was prepared for PCR by boiling. Briefly, cells were scraped off an overnight blood agar plate with a sterile loop, washed twice in 1.5 ml of 1× Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.5]), resuspended in 0.5 ml of H2O, and boiled for 15 min. The cell debris was pelleted by centrifugation at 8,000 × g for 5 min, and the supernatant containing the released DNA was transferred to a fresh microcentrifuge tube. PCR amplifications were done in 15-μl reaction volumes containing 1.0 μl of the boiled whole-cell lysate, 3.75 pmol of each of the forward and reverse primers, 200 μM each deoxynucleoside triphosphate, 0.1 mg of acetylated bovine serum albumin per ml, 0.75 U of Taq polymerase, and 1.5 μl of 10× buffer B with 1.5 mM MgCl2 supplied with the polymerase (Promega Corporation, Madison, Wis.). The primers and PCR cycling conditions used for MLRFT are the same as those described by Enright et al. (5) and are updated on the S. aureus MLST website (http://www.mlst.net).
Amplicons were directly subjected to digestion with restriction endonucleases by adding 7 μl of DNA amplicon to 7 μl of a reaction mixture containing 1.4 μl of 10× digestion buffer, 0.1 mg of acetylated bovine serum albumin per ml, and 3 U of restriction enzyme; the digestion conditions otherwise followed the recommendations of the manufacturer. Complete digestion was achieved without prior purification of the PCR amplicon. The restriction enzyme or combination of restriction enzymes used for each locus are listed in Table 1. Tsp509I was purchased from New England Biolabs, Inc. (Beverly, Mass.), and all others were purchased from Promega Corporation).
TABLE 1.
Restriction endonucleases used in MLRFT
| Locus | Size of product (bp) | No. of MLST alleles/locusa | Restriction enzyme(s) | No. of MLRFT alleles/locus |
|---|---|---|---|---|
| arcC | 570 | 37 | HinfI | 3 |
| aroE | 536b | 66 | AluI and CfoI | 5 |
| glpF | 543c | 44 | Tsp509I | 9 |
| gmk | 488 | 35 | CfoI | 5 |
| pta | 575 | 44 | RsaI | 7 |
| tpi | 475 | 58 | BbuI and MboI | 4 |
| yqiL | 598 | 52 | VspI and DdeI | 4 |
The MLST allele count from the MLST database (http://www.mlst.net) as of February 3, 2003.
The aroE amplification product contains an AluI restriction site between the 3′ end of the trimmed MLST sequences and the aroE-Dn primer-binding site. This AluI site is polymorphic but is not used for allele differentiation to retain concordance with the MLST database.
The glpF amplification product contains two nonpolymorphic Tsp509I restriction sites between the primer-binding sites and the trimmed MLST sequence, one at each end.
RFs were separated by electrophoresis on 4.0% Metaphor agarose containing a 0.1× concentration of GelStar nucleic acid stain (Cambrex Bio Science Rockland, Inc.). RFs were sized against a 50-bp DNA ladder (Life Technologies, GIBCO BRL, Gaithersburg, Md.). The gels were visualized either as digital images on a FluorImagerSI (Molecular Dynamics, Calif.) or on film under UV illumination.
RF allele assignment was made by visual comparison of the RF banding patterns against the predicted banding patterns derived from primary MLST sequence data (Table 1). A translation table for interconverting MLRFT and MLST data and an MLRFT allele definition table are located at http://socrates.berkeley.edu/∼microbes/sensabaugh/research/MLRFT.htm. For each locus, unique predicted RF banding patterns were assigned letter codes (see the website with the interconversion and definition tables mentioned above). RF types (RFTs) were defined by the combination of alleles at the seven loci (e.g., RFT-BBBBBAB in the order RFT-arcC-aroE-glpF-gmk-pta-tpi-yqiL).
MLST and spa typing.
MLST was performed with at least one isolate from each RFT; in addition, spa typing was performed with two isolates from each of the five most common RFTs with two or more MRSA isolates. MLST and spa typing were performed as described previously (5, 16).
Statistical analysis.
Discrimination indices (DIs) and confidence intervals were calculated on the basis of Simpson's index of diversity, as described by Grundmann et al. (8).
RESULTS
Development of MLRFT.
The allelic sequences at the seven loci used for MLST (http://www.mlst.net) were surveyed to identify highly discriminating polymorphic restriction sites. Because the sequences in the MLST database are trimmed to a length shorter than the distance between the primer-binding sites, primary gene sequence data (http://www.ncbi.nlm.nih.gov) were used to fill in between the trimmed sequences and the primer-binding sites. It was possible to identify for each locus a single restriction enzyme or compatible pair of restriction enzymes projected to provide a high level of allelic differentiation (Table 1). These were tested with our sample population of 103 isolates and were found to yield reproducible and unambiguous RF patterns. All allelic RF patterns observed in our sample were consistent with the RF patterns projected from the sequence data. The side-by-side reproducibility of the RF allelic patterns generated in different test runs is illustrated in Fig. 1.
FIG. 1.
MLRFT patterns of three loci. The reproducibility of the patterns in duplicate experiments is demonstrated.
For convenience, each distinguishable RF pattern at a locus has been assigned a letter designation. Given that the sizes of the RFs for every allele at each locus in the MLST sequence database can be predicted, each sequence-defined allele can readily be assigned an RF allele designation. For example, at the gmk locus, sequence-defined alleles 1, 9, 12, and 14 have identical sites cut by CfoI, yielding a three-band pattern with bands at 334, 105, and 49 bp (Fig. 1); these four sequence-defined alleles are all given the RF allele designation A. Following gel electrophoresis, allelic assignments can be made simply and unambiguously by visually comparing the band positions with a 50-bp DNA ladder. The stoichiometric yield of RFs enables unambiguous allelic assignment.
Preliminary assessment of MLRFT discrimination power.
MLRFT detects fewer alleles per locus than MLST (5.3 versus 48 alleles, on average; Table 1). To compare the discrimination power of MLRFT and MLST for strain differentiation, we translated the sequence-based alleles detected in the original MLST study of Enright et al. (5) into the RF patterns that would have been observed if MLRFT had been performed with this test population. In that study, 155 isolates were resolved by MLST into 53 sequence types (STs), corresponding to a DI of 94.3%. By projecting the typing results obtained by MLRFT, this same test population would be resolved into 32 RFTs, giving a DI of 89.1%; thus, in terms of the DI, MLRFT captured 94.5% of the genetic variability detected by MLST.
Characterization of a San Francisco test sample population.
To assess the performance of MLRFT in practice, we tested a sample population of 103 isolates (59 MSSA and 44 MRSA isolates) selected to represent the diversity of S. aureus PFGE patterns in the Molecular Epidemiology Research Laboratory at San Francisco General Hospital; these isolates had been additionally characterized for their susceptibilities to a range of antibiotics. Resistance to methicillin was confirmed by PCR-based detection of the mecA gene. MLRFT differentiated the 103 isolates into 15 RFTs with a DI of 89.0%, a value comparable to the projected 89.1% DI for the test population evaluated by MLST, as noted above (Table 2).
TABLE 2.
Discrimination power of MLRFT
| Test population | No. of isolates | DI (% [95% confidence interval])
|
No. of types
|
||
|---|---|---|---|---|---|
| MLST | MLRFT | MLST | MLRFT | ||
| Enright et al. (5) | 155 | 94.3 (92.8, 95.8) | 89.1 (85.8, 92.3) | 53 | 32 |
| This study | 103 | 89.0 (86.5, 91.5) | 15 | ||
The distribution of isolates in the 15 RFTs is shown in Table 3. Sixty-two (59.6%) of 103 isolates belonged to four RFTs. These RFTs included 38 (86%) of the 44 MRSA isolates. The remaining six MRSA isolates were scattered among four additional RFTs. Because of the translational property between MLRFT and MLST, three of the common MRSA RFTs could be provisionally identified as belonging to STs corresponding to known MRSA clonal lineages: RFT-CAAACAC to the archaic/Iberian/clone V group, RFT-AAACCAA to the New York/pediatric/Japan group, and RFT-BBBBBAB to the epidemic MRSA type 16 (EMRSA-16) group (6, 13). The fourth prominent MRSA type, RFT-CAFBCDB, did not correspond to any RF allelic profiles projected from any of the major MRSA clonal lineages determined by MLST.
TABLE 3.
Distribution of RFT in a San Francisco test population and concordance with PFGE groupings, MLST, and spa typing
| MLRFTa | No. of isolates | No. of MRSA isolates | PFGE clone group | PFGE subtype | MLSTb | spa type | Associated epidemic clonal group (clonal complex)c |
|---|---|---|---|---|---|---|---|
| AAAAAAA | 5 | 1 | K | 4 | ST-1 | ||
| AAAACBC | 1 | 0 | L | 1 | ST-25 | ||
| AAABCBC | 2 | 0 | U | 1 | ST-22 | CC22 | |
| AAACAAC | 1 | 0 | J | 1 | ST-20 | ||
| AAACCAA | 13 | 7 | D | 9 | ST-5 | TJMBMDMBMK | NY/pediatric/Japan (CC5) |
| AAACCAC | 6 | 3 | B | 3 | ST-2 | UH2GFGMDMGGM | |
| ABDBCAB | 7 | 1 | X | 5 | ST-45 | ||
| ACAACAA | 3 | 0 | V | 3 | ST-15 | ||
| BBBBBAB | 6 | 6 | A | 4 | ST-36 | WGKAKAOMQQQ | EMRSA-16 (CC30) |
| BBBBBAB | 11 | 5 | Z | 5 | ST-30 | WGKAKAOMQQQ | CC30 |
| CAAAAAA | 5 | 0 | Q | 4 | ST-109 | ||
| CAAAABC | 4 | 0 | R | 1 | ST-97 | ||
| CAAACAC | 21 | 10 | C | 9 | ST-8 | YHGCMBQBLO | Clone V (CC8) |
| CAABCCB | 2 | 1 | N | 1 | ST-121 | ||
| CAACAAA | 5 | 0 | M | 3 | ST-188 | ||
| CAFBCDB | 11 | 10 | P | 4 | ST-59 | ZDGDGDEB |
An RFT was defined by the combination of alleles at the seven loci (e.g., RFT-BBBBBAB in the order RFT-arcC-aroE-glpF-gmk-pta-tpi-yqiL).
The ST was defined according to the MLST database (http://www.mlst.net).
MLST and spa typing results are shown also in Table 3. By using these two sequence-based typing systems, the provisional classification of RFT-CAAACAC was refined with its identification as a clone V strain (ST-8), an MRSA lineage associated with disease in human immunodeficiency virus-infected patients (15, 17). RFT-AAACCAA was confirmed to be the NY/Pediatric/Japan clone (ST-5). RFT-CAFBCDB is ST-59; only one ST-59 MRSA isolate was reported in the MLST database and was recovered in Georgia in 1993 (6). The two RFT-BBBBBAB isolates with highly divergent PFGE patterns belonged to two closely related MLST types, ST-36 (PFGE group A) and ST-30 (PFGE group Z).
The 103 isolates in our sample population had a broad range of epidemiologic and antibiotic resistance patterns. All isolates were susceptible to linezolid and vancomycin. Both methicillin-resistant and methicillin-susceptible isolates were detected among the isolates in eight RFT groups. For example, penicillin-susceptible S. aureus strains collected from the nares of healthy San Franciscans, MRSA strains from the nares of injection drug users, and multidrug-resistant MRSA strains from patients in the intensive care and gynecology units at San Francisco General Hospital were in the RFT-AAACCAA group. However, two RFTs exhibited a clear association with specific antimicrobial susceptibility patterns. Ten of 11 isolates in the RFT-CAFBCDB group were resistant to methicillin, a much higher proportion than that found in any of the other RFT groups containing MRSA isolates. Furthermore, these MRSA strains exhibited resistance to only a few antibiotics (to erythromycin in 7 of 10 isolates, to tetracycline in 1 of 10 isolates); this is in contrast to the typical pattern of multidrug resistance seen in hospital MRSA strains (1). The RFT-CAAACAC group showed an association with trimethoprim-sulfonamide resistance (11 of 12 isolates).
Comparison of MLRFT and PFGE.
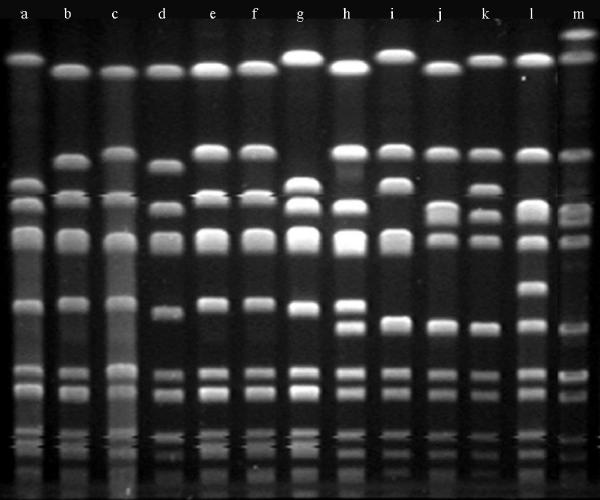
That the isolates in our test population had been previously characterized by PFGE allowed a direct comparison of MLRFT and PFGE for strain differentiation. The 103 isolates had been initially differentiated into 17 PFGE groups containing 62 subtypes. Thirteen of the RFT groups corresponded one to one with a previously assigned PFGE group (Table 3). The remaining two RFT groups, CAAACAC and BBBBBAB, each contained isolates of two previously assigned PFGE groups. On the basis of the MLRFT results, the PFGE groupings within each of these RFTs were reevaluated. Given that the initial PFGE groupings were based on results from different gels analyzed in different molecular epidemiologic studies, isolates in each of the two RFT groups were reanalyzed side by side on the same gels. These results demonstrated that the isolates in the RFT-CAAACAC group were possibly related (less than six band differences) and in fact should have been classified within a single PFGE group (Fig. 2). However, for the RFT-BBBBBAB group, the results confirmed the initial PFGE differentiation of the isolates into two groups, PFGE groups A and Z (data not shown). As noted above, we performed MLST with the group A and the group Z isolates by MLST to clarify their genetic relationship (Table 3). The PFGE group Z isolate was ST-30, a clonal lineage increasingly associated with methicillin resistance (E. D. Charlebois, D. R. Bangsberg, N. Moss, and F. Perdreau-Remington, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., p. 120, 2001). The PFGE group A isolate was ST-36, which is associated with the epidemic EMRSA-16 group from the United Kingdom. The difference between ST-30 and ST-36 is a single nucleotide substitution at the pta locus, and Enright et al. (6) have suggested that MSSA ST-30 may be ancestral to MRSA ST-36. Thus, MLRFT correctly identified the genetic relationship between ST-30 and ST-36 isolates, even though it did not differentiate them.
FIG. 2.
Heterogeneity of SmaI PFGE patterns in group RFT-CAAACAC. Lanes a to h, MRSA isolates; lanes i to m, MSSA isolates. The MRSA isolates and MSSA isolates were collected in different epidemiologic studies and were originally assigned to different PFGE groups. MLRFT with these isolates suggested a common clonal lineage; reanalysis by PFGE by running these samples side by side confirmed the MLRFT results.
DISCUSSION
We demonstrate here that MLRFT is a useful tool for the characterization of S. aureus strains. It possesses two of the important virtues of MLST, the strain characterization approach upon which it is based: it is highly discriminating and it is portable. Moreover, because both MLRFT and MLST rely on the same sequence database, it is possible to work back and forth between the typing systems. That MLRFT captures about 95% of the discrimination power of MLST may appear counterintuitive, given that MLRFT detects far fewer alleles per locus than MLST. The answer is likely that although recombination in S. aureus is uncommon, it has occurred with sufficient frequency within and between loci to generate a very large number of allelic combinations (7). Given only the alleles detectable by MLRFT, it is possible in principle to distinguish more than 75,000 RFTs. The portability of MLST derives from its basis in primary sequence data. The portability of MLRFT derives from the fact that cleavage at defined restriction sites in defined sequence segments yields reproducible RF banding patterns that are amenable to a uniform scoring method and nomenclature. Compared to MLST, MLRFT possesses the additional virtues of being rapid and simple. MLRFT is also low cost, about $15 per isolate for reagents and disposable materials, compared to a like amount for PFGE and about $120 for MLST. These virtues should make the technique widely applicable, particularly in clinical research settings for strain screening purposes and in the developing world, where the sequencing technology required for MLST is not readily available.
MLRFT clearly lacks the capacity of PFGE to make fine distinctions between strains, a property that makes PFGE the tool of choice for the tracking of local S. aureus outbreaks. In contrast to MLRFT, which detects genetic variations that accrue relatively slowly in housekeeping genes, PFGE indexes variations that accumulate more rapidly, such that clonal relationships can become obscured after a short transmission period. Despite this difference, however, the agreement of the MLRFT and PFGE groupings was excellent; the sole discordance was seen with the two distinct PFGE groups (groups A and Z), which were not differentiated by MLRFT. However, the genetic relationship between the divergent PFGE groups identified by MLRFT was readily established by MLST.
The shortcoming of PFGE typing is that strain classification is based on banding pattern recognition, which is dependent on electrophoretic and gel parameters; this complicates the comparison of results between laboratories and, as demonstrated in this study, within a laboratory over time. MLRFT complements PFGE by providing a stable genetic framework for grouping isolates that allows the association of PFGE profiles that might have evolved beyond the suggested limit of five to six band differences (18).
In summary, we have described a, MLRFT approach for S. aureus strain typing. This approach can be used to rapidly index informative genetic variations present at MLST housekeeping gene loci. The low cost and simplicity of this strain typing technique allow the processing of large numbers of clinically and epidemiologically relevant S. aureus isolates. When the portability of MLRFT data across different laboratories is factored in, it is clear that MLRFT is a powerful method for monitoring changes in strain distributions and probing more basic epidemiologic questions that may give new insights into the epidemiology of S. aureus.
Acknowledgments
This work was supported in part by a Faculty Research Bridging Grant (G.F.S.) and by an unrestricted grant from the Pfizer Corporation, New York, N.Y. (F.P.-R.).
REFERENCES
- 1.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charlebois, E. D., D. R. Bangsberg, N. J. Moss, M. R. Moore, A. R. Moss, H. F. Chambers, and F. Perdreau-Remington. 2002. Population-based community prevalence of methicillin-resistant Staphylococcus aureus in the urban poor of San Francisco. Clin. Infect. Dis. 34:425-433. [DOI] [PubMed] [Google Scholar]
- 3.Cookson, B. D., P. Aparicio, A. Deplano, M. Struelens, R. Goering, and R. Marples. 1996. Inter-centre comparison of pulsed-field gel electrophoresis for the typing of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 44:179-184. [DOI] [PubMed] [Google Scholar]
- 4.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin- resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 10.McDougal, L. K., and C. Thornsberry. 1984. New recommendations for disk diffusion antimicrobial susceptibility tests for methicillin-resistant (heteroresistant) staphylococci. J. Clin. Microbiol. 19:482-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami, K., W. Minamide, K. Wada, E. Nakamura, H. Teraoka, and S. Watanabe. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 1996. Performance standards for antimicrobial disk susceptibility tests. Document M100-S3. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 14.Peacock, S. J., G. D. de Silva, A. Justice, A. Cowland, C. E. Moore, C. G. Winearls, and N. P. Day. 2002. Comparison of multilocus sequence typing and pulsed-field gel electrophoresis as tools for typing Staphylococcus aureus isolates in a microepidemiological setting. J. Clin. Microbiol. 40:3764-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, et al. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 16.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shopsin, B., B. Mathema, X. Zhao, J. Martinez, J. Kornblum, and B. N. Kreiswirth. 2000. Resistance rather than virulence selects for the clonal spread of methicillin-resistant Staphylococcus aureus: implications for MRSA transmission. Microb. Drug Resist. 6:239-244. [DOI] [PubMed] [Google Scholar]
- 18.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Belkum, A., W. van Leeuwen, M. E. Kaufmann, B. Cookson, F. Forey, J. Etienne, R. Goering, F. Tenover, C. Steward, F. O'Brien, W. Grubb, P. Tassios, N. Legakis, A. Morvan, N. El Solh, R. de Ryck, M. Struelens, S. Salmenlinna, J. Vuopio-Varkila, M. Kooistra, A. Talens, W. Witte, and H. Verbrugh. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]