Abstract
More sensitive assays for human immunodeficiency virus type 1 (HIV-1) RNA are needed to detect, quantify, and characterize persistent viremia in patients who are receiving antiretroviral therapy and whose plasma HIV-1 RNA levels are suppressed to less than 50 to 75 copies/ml. We therefore developed an internally controlled real-time reverse transcriptase-initiated PCR assay that quantifies HIV-1 RNA concentrations down to 1 copy per ml of plasma. This assay with single-copy sensitivity (the single-copy assay) generates a reproducible linear regression plot of input copy number versus threshold cycle by using HIV-1 RNA transcripts at copy numbers ranging from 1 to 106 per reaction mixture. The single-copy assay was compared to the ultrasensitive AMPLICOR HIV-1 MONITOR assay and a more sensitive modification of the ultrasensitive assay by repeatedly testing a low-copy-number panel containing 200 to 0.781 copies of HIV-1 RNA per ml of plasma. This comparison showed that the single-copy assay had a greater sensitivity than the other assays and was the only assay that detected HIV-1 RNA at levels as low as 0.781 copies/ml. Testing of plasma samples from 15 patients who were receiving antiretroviral therapy and who had <75 HIV-1 RNA copies/ml revealed persistent viremia in all 15 patients, with HIV-1 RNA levels ranging from 1 to 32 copies/ml (median, 13 copies/ml). The greater sensitivity of the single-copy assay should allow better characterization of persistent viremia in patients who are receiving antiretroviral therapy and whose HIV-1 RNA levels are suppressed to below the detection limits of present assays.
Untreated human immunodeficiency virus type 1 (HIV-1) infection is characterized by an early acute phase with high levels of viremia that leads to a clinically asymptomatic phase of variable duration, followed by immunodeficiency (15, 23). Throughout infection an extraordinarily large number of viral replication cycles occurs, and this high replicative capacity of HIV-1 leads to both varied genetic pools and high viral loads (2, 8).
Although the time course of HIV-1 disease is highly variable, the outcome is not. Almost all untreated patients infected with HIV-1 die of AIDS within approximately 2 to 20 years. The course of HIV-1 disease has been slowed in recent years by the use of potent antiretroviral treatment; however, the ability of HIV-1 to develop drug resistance presents the major impediment to long-term effective anti-HIV therapy (11, 14, 29).
The concentration of HIV-1 RNA in the plasma of HIV-infected individuals is an important predictor of disease outcome and a marker of antiretroviral drug efficacy (9, 18, 19, 22, 27). Of the Food and Drug Administration (FDA)-approved tests for HIV-1 RNA in plasma, the most sensitive have detection limits of 50 copies/ml (21, 30), and treatment is generally considered initially successful if the level of viremia can be reduced to below this level. However, reduction of the HIV-1 RNA load to <50 copies/ml does not guarantee long-term success, and a rebound of drug resistance can occur (20, 26), implying that HIV-1 replication and evolution may be continuing while patients are receiving therapy. Application of experimental assays with sensitivities in the range of 10 copies/ml has demonstrated that many patients whose levels of viremia are suppressed to <50 copies/ml have persistently detectable HIV-1 RNA (7). The source and dynamics of this residual viremia are unknown. Viremia could arise either from activation of virus expression from latently infected cell reservoirs or from ongoing cycles of viral replication, for example, in a sanctuary site where there is suboptimal drug penetration (1, 6, 31).
To better characterize the residual viremia in patients receiving antiretroviral therapies recommended at present, we have developed a quantitative real-time, reverse transcriptase (RT)-initiated PCR (RT-PCR) assay that detects and quantifies HIV-1 RNA at levels below those that can be quantified by approved assays. This assay uses improved nucleic acid isolation and purification techniques, larger plasma sample volumes, and a real-time PCR method to detect and quantify HIV-1 RNA in plasma down to 1 copy/ml. To monitor the recovery of HIV-1 from patient plasma, samples are spiked with an internal virion standard, consisting of the replication-competent avian sarcoma)-leukosis retroviral vector RCAS BP(A) (RCAS), derived from an unrelated retrovirus based on Rous sarcoma virus (RSV).
(Ann P. Wiegand performed this study as partial fulfillment of the requirements for a degree in biomedical science in the Graduate School of Hood College, Frederick, Md.)
MATERIALS AND METHODS
Low-copy-number HIV-1 RNA panel.
HIV-1 virions of known concentration (Virology Quality Assurance Laboratory [VQA], Rush-Presbyterian-St. Luke's Medical Center, Chicago, Ill.) were serially diluted in HIV-1-seronegative human plasma to predicted final RNA concentrations of 200, 100, 50, 12.5, 6.25, 3.125, 1.56, or 0.781 copies/ml. The dilutions were performed once, and sufficient numbers of aliquots from each dilution were stored at −80°C to complete the testing described. In addition to analysis by the assay with single-copy sensitivity (the single-copy assay), dilutions were tested by using the AMPLICOR HIV-1 MONITOR ultrasensitive assay (version 1.0; Roche Diagnostics, Branchburg, N.J.), which was performed according to the instructions of the manufacturer by a laboratory (University of Pittsburgh) certified by the VQA program of the AIDS Clinical Trials Group, or by the modified AMPLICOR ultrasensitive assay with 2.0 ml of plasma, which was performed by the same laboratory as described previously (7, 32).
Clinical specimens.
All clinical samples were collected from volunteers and patients attending the Critical Care Medical Department of the National Institute of Allergy and Infectious Diseases at the Clinical Center of the National Institutes of Health (NIH), Bethesda, Md., and was approved by the Institutional Review Board of the National Institute of Allergy and Infectious Diseases, NIH. All participants provided written informed consent. Blood samples were collected from 20 HIV-1-seronegative volunteers; 15 HIV-1-infected individuals who were receiving antiretroviral treatment and whose plasma HIV-1 RNA levels were <75 copies/ml, as determined by the Versant HIV-1 RNA (version 3.0) assay (a branched DNA [bDNA] assay; Bayer AG, Leverkusen, Germany) (3, 5); and 20 untreated HIV-1-infected individuals. Plasma was separated from whole blood by centrifugation within 4 h of collection, and aliquots were stored at −80°C until use.
Preparation of standards for HIV-1 RNA quantification.
Plasmid pQP1, which contains a 1,420-bp fragment of HIV-1 HXB2 gag (SacI to BglII) downstream of the T7 promoter, was linearized with EcoRI, purified with a PCR purification kit (Qiagen, Valencia, Calif.), and transcribed with T7 RNA polymerase by using the RiboMax Express Large Scale RNA production system (Promega, Madison, Wis.) (24, 25). The template DNA was degraded with 5 U of RNase-free DNase, and the RNA transcripts were purified twice with an RNeasy kit (Qiagen). The RNA was quantified spectrophotometrically at 260 nm, diluted to 106 copies/μl, divided into aliquots, and stored at −80°C. Diluted HIV-1 transcripts (106 to 0.3 copies/10 μl) were used to generate a standard curve for each single-copy assay.
Preparation of standards for RCAS quantification.
A region of RSV gag was amplified from RCASBP(A)gfp plasmid (a gift of S. Hughes, National Cancer Institute) by using primers 1661F (CATTGACTGCTTTAGGCAGA) and 2215R (AACAGCGCGGTGATATAC) (10). The resulting 554-bp PCR product was cloned into the vector pPCR-Script Amp (Stratagene, La Jolla, Calif.). The plasmids were purified on columns with a Qiagen kit and linearized with EcoRI. The resulting DNA template was quantified and then transcribed with T3 RNA polymerase by using the MAXIscript T3 kit (Ambion, Austin, Tex.). The RNA product was diluted and stored at −80°C. Diluted RCAS transcripts (106 to 10 copies/10 μl) were used to generate a standard curve for each RCAS real-time PCR assay.
RCAS internal virion standard.
High-titer RCAS stocks were obtained by transfecting DF-1 cells with RCASBP(A)gfp plasmid and harvesting the supernatant at the peak of production, as determined by measuring viral RT activity (28). RCAS stocks were diluted to 1.5 × 106 copies/ml of viral RNA (as measured by the real-time RT-PCR assay) with RPMI 1640 medium containing 10% fetal calf serum. A total of 200 μl of the diluted RCAS stock was added to each plasma sample as an internal control for virion recovery following centrifugation, RNA extraction, and determination of RT-PCR efficiency.
Viral RNA extraction.
For samples expected to have <1,000 copies of HIV-1 RNA per ml, 7 ml of patient plasma to which 200 μl of RCAS stock (300,000 copies of RNA) had been added was ultracentrifuged at 170,000 × g for 30 min at 4°C in a Sorvall T-1270 rotor. For samples expected to have HIV-1 RNA concentrations >1,000 copies/ml, a smaller volume of plasma (typically 1 to 2 ml) was diluted to 7 ml with Tris-buffered saline and processed in the same manner described above for 7 ml of plasma. After ultracentrifugation, the supernatant was removed and the virion pellet was treated with 100 μl of 5 mM Tris-HCl (pH 8.0) containing 200 μg of proteinase K for 30 min at 55°C. The pellet was then treated with 400 μl of 5.8 M guanidinium isothiocyanate containing 200 μg of glycogen, and the lysate was transferred to a microcentrifuge tube. After the addition of 500 μl of 100% isopropanol, the lysate was centrifuged at 21,000 × g for 15 min. The pellet was washed with 70% ethanol, and the nucleic acids were resuspended in 55 μl of 5 mM Tris-HCl (pH 8.0) containing 1 μM dithiothreitol and 1,000 U of an RNase inhibitor (RNase out; Invitrogen, Carlsbad, Calif.). Control experiments showed that the recovery of HIV-1 and RCAS RNA was independent of the plasma volume over the range of volumes used (1 to 7 ml). For each sample, three separate 10-μl aliquots of the resuspended nucleic acids were used for the real-time PCR for HIV-1 and two separate 5-μl aliquots were used for the real-time PCR for RCAS.
Real-time PCR assay.
The real-time assay involved a two-step, two-enzyme RT-PCR protocol. Reverse transcription reactions (30 μl) were performed in 96-well plates and contained the following components at the indicated final concentrations or amounts in sterile molecular-grade water: random hexamers (0.15 μg/reaction; Promega), MgCl2 (5 mM), deoxynucleoside triphosphates (0.5 mM), RNase out (20 U), dithiothreitol (0.67 mM), Taqman buffer A (Applied Biosystems, Foster City, Calif.) diluted to 1×, and RT (20 U of Superscript II RT; Invitrogen). After 15 min at 25°C, the reverse transcription reaction mixture was incubated at 42°C for 40 min. Following completion of the reverse transcription step, the reaction mixture was heated to 85°C for 10 min and then held at 25°C for 30 min, after which the plate was cooled to 4°C.
Twenty microliters of the PCR mixture was added to the cDNA reaction products (final volume, 50 μl) containing the following components at the indicated final amounts or concentrations in sterile molecular-grade water: PCR buffer II (Applied Biosystems) diluted to 1×, MgCl2 (4 mM), AmpliTaq Gold (1.25 U, Applied Biosystems), and the primer-probe set for HIV-1 quantification designed to bind to a conserved region of gag: primers 6F (5′-CATGTTTTCAGCATTATCAGAAGGA-3′) and 84R (5′-TGCTTGATGTCCCCCCACT-3′) (600 nM) and probe 5′FAM-CCACCCCACAAGATTTAAACACCATGCTAA-Q 3′ (100 nM), where FAM indicates a reporter 6-carboxyfluorescein group and Q indicates a 6-carboxytetramethylrhodamine group quencher conjugated through a linker arm nucleotide, as described previously (17). The primer-probe set used for internal standard quantification was selected to bind to a conserved region of RSV gag: primers 1849F (5′-GTCAATAGAGAGAGAGGGATGGACAAA-3′) and 1896R (5′-TCCACAAGTGTAGCAGAGCCC-3′) (600 nM) and probe 5′FAM-TGGGTCGGGTGGTCGTGCC-Q 3′ (100 nM).
Following thermal activation of the AmpliTaq Gold (95°C, 10 min), 45 cycles of PCR amplification (with each cycle consisting of 95°C for 15 s and 60°C for 1 min) were performed. For each run, two standard curves, one from diluted HIV-1 transcripts and one from diluted RCAS transcripts, were generated. For each experiment new dilutions were prepared from thawed single-use aliquots of transcript stocks stored at −80°C. Threshold cycle (Ct) values were plotted as a function of the input transcript copy number, and linear regression was performed with ABI 7700 Sequence Detection software (Applied Biosystems). For each specimen, three replicate reactions were performed for HIV-1 quantification, with 18% of the total RNA extracted from the original sample used in each reaction. Two replicate reactions were performed for internal standard RCAS quantification, with 9% of the total RNA extracted from the original sample used in each reaction. One reaction mixture for each specimen was processed and amplified without the addition of RT as a control to detect HIV-1 DNA in the source specimen. Additional controls lacking RNA templates were run for both the HIV-1 and the RCAS reactions to test for contamination with the PCR product during sample processing or assay setup. The numbers of copies of HIV-1 and RCAS RNA in the test samples were calculated by interpolation of the experimentally determined Ct value for the test sample by using the transcript-derived linear regression as a standard curve and were rounded to the nearest integer value. For HIV-1 quantification, the calculated number of copy equivalents per reaction mixture was expressed as the number of copies per milliliter of the starting plasma sample. Assay acceptability was contingent on the R2 value for the HIV-1 and RCAS linear regressions, which was >0.95, and the average measured copy equivalent per reaction mixture for the RCAS internal standard was greater than 15,000 copies (55% recovery). Acceptance of the results for samples with RCAS levels greater than 15,000 copies ensured that the measured HIV-1 RNA levels were within a factor of 2 of the actual HIV-1 RNA levels. RCAS levels were >15,000 copies for 95% of the plasma samples assayed. Negative and positive (12.5 copies/reaction mixture) plasma sample controls were included with each assay run.
Other HIV-1 RNA assays.
HIV-1 RNA measurements by a bDNA assay (Versant HIV-1 RNA [version 3.0] assay; Bayer Corp.) were performed according to the specifications of the manufacturer at a manufacturer-certified site by using the semiautomated Bayer System 340 unit. Versant HIV-1 RNA (version 3.0) was recently approved by FDA for clinical use. The limit of quantification of that assay is 75 copies of HIV-1 RNA/ml of plasma.
RESULTS
Reproducibility of single-copy assay.
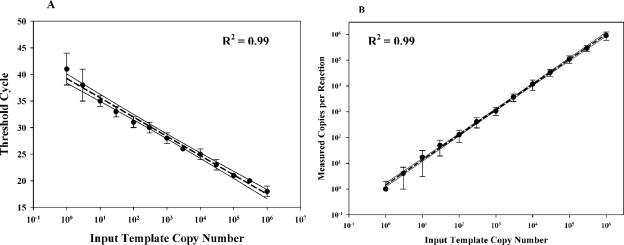
To assess the reproducibility of the single-copy assay, we performed 10 independent assay runs using HIV-1 RNA transcript amounts ranging from 106 to 0.3 copies/reaction mixture. A plot of Ct values versus the input transcript copy number shows that the assay could readily detect a single copy of HIV-1 RNA, yet it had a linear dynamic range to at least 106 copies/reaction mixture (Fig. 1A). Figure 1B demonstrates the excellent agreement between the expected and the measured values for 10 independent serial dilutions of the control RNA transcripts (R2 = 0.99). In reactions with 3 RNA copies or less, the measured values were consistent with that expected from the Poisson distribution (data not shown).
FIG. 1.
(A) Plot of input control template (RNA transcript) copy number versus Ct value; (B) plot of input control template copy number versus measured copy number per reaction mixture. The hashed lines represent regression plots of the measured data, and the solid lines represent the 95% confidence intervals for the measured values.
Comparison of assay performance.
To compare the performance of the single-copy assay with those of other commonly used HIV-1 RNA assays, a test panel with HIV-1 virions of known concentration produced in vitro and diluted in human plasma (200 to 0.781 RNA copies/ml) was assayed by the following methods: (i) the AMPLICOR HIV-1 MONITOR ultrasensitive procedure (version 1.0), (ii) a modified AMPLICOR HIV-1 MONITOR ultrasensitive procedure that uses 2 ml of plasma (7, 32), and (iii) the single-copy assay. Table 1 shows the results for the low-copy-number panel tested four times by each assay. The AMPLICOR HIV-1 MONITOR ultrasensitive assay consistently detected and quantified HIV-1 RNA at levels from 200 to 50 copies/ml. At 25 copies/ml, only two of four assays detected HIV-1 RNA, and at less than 25 copies/ml, the assay was consistently negative (the optical density [OD] for the undiluted sample was below the background OD). The modified AMPLICOR HIV-1 MONITOR ultrasensitive assay showed greater sensitivity, consistently detecting HIV-1 RNA at 6.25 copies/ml, although the mean value for the four assays was very close to the mean value for the samples with 12.5 copies/ml (10 and 12 copies/ml, respectively), suggesting that the limit of quantification was 12.5 copies/ml. With HIV-1 RNA at less than 6.25 copies/ml, the modified ultrasensitive assay was consistently negative (the OD for the undiluted sample was below the background OD). The single-copy assay consistently detected HIV-1 RNA in all samples (samples with 200 to 0.781 copies/ml), and the mean values for the four assays indicate that HIV-1 RNA could be quantified to 1 copy per ml, although the standard deviation increased for samples with 1.56 and 0.781 copies/ml. For samples with these low levels, however, greater variation is expected because of stochastic influences.
TABLE 1.
Comparison of four different assays for HIV-1 RNAa
| No. of copies/ml predicted by VQA (control) | No. of copies/ml
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMPLICOR ultrasensitive assay
|
Modified AMPLICOR ultrasensitive assay
|
Single-copy assay
|
||||||||||||||||
| Run 1 | Run 2 | Run 3 | Run 4 | Mean | SD | Run 1 | Run 2 | Run 3 | Run 4 | Mean | SD | Run 1 | Run 2 | Run 3 | Run 4 | Mean | SD | |
| 200 | 173 | 176 | 170 | 127 | 162 | 23 | 190 | 149 | 140 | 199 | 170 | 29 | 187 | 200 | 189 | 232 | 202 | 30 |
| 100 | 90 | 137 | 150 | 90 | 117 | 31 | 87 | 45 | 107 | 88 | 82 | 26 | 85 | 84 | 75 | 139 | 96 | 29 |
| 50 | 24 | 70 | 60 | 59 | 53 | 20 | 69 | 27 | 61 | 44 | 50 | 19 | 32 | 33 | 39 | 62 | 42 | 14 |
| 25 | 31 | 28 | Negb | Neg | 30 | 2.1 | 27 | 18 | 28 | 17 | 23 | 5.8 | 15 | 25 | 11 | 30 | 20 | 8.8 |
| 12.5 | Neg | Neg | Neg | Neg | Neg | Neg | 9 | 11 | 15 | 11 | 12 | 2.5 | 11 | 8 | 10 | 12 | 10 | 1.7 |
| 6.25 | Neg | Neg | Neg | Neg | Neg | Neg | 17 | 12 | 5 | 7 | 10 | 5.4 | 2 | 9 | 8 | 8 | 6.8 | 3.2 |
| 3.125 | Neg | Neg | Neg | Neg | Neg | Neg | 10 | Neg | Neg | Neg | 10 | 0 | 2 | 5 | 4 | 3 | 3.5 | 1.3 |
| 1.56 | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | 2 | 1 | 1 | 6 | 2.5 | 2.4 |
| 0.78 | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | 0.3 | 1 | 0.8 | NDc | 0.7 | 0.4 |
An amount of preassayed virus with the corresponding number of copies of RNA indicated was mixed with normal plasma.
Neg, the OD was not above background.
ND, not determined (due to RCAS value <15,000 copies/reaction mixture).
Internal standard values.
To monitor the recovery of virion RNA through the ultracentrifugation and extraction procedures and the efficiency of the RT-PCR for each sample, an internal virion standard consisting of RCAS, an unrelated retrovirus vector based on RSV (10), was added to each sample at the start of the assay (i.e., before ultracentrifugation). Table 2 shows the values for the RCAS internal standard for 22 different plasma samples from five patients. Recovery of the internal standard was determined in duplicate for each sample assayed, and these values were averaged. The average RCAS value obtained for all 22 samples tested was 25,716 ± 4,057 copies/reaction mixture, or about 95% ± 15% of the RCAS RNA added. The coefficient of variation (CV) ranged from 13 to 37% for samples derived from individual patients, with an average CV for the five patients of 16%. These results demonstrate the completeness of recovery and the stability of the internal standard during the extraction procedure, allowing rejection of the results for assays in which recovery and/or reverse transcription of the HIV-1 RNA is likely to be incomplete. We reject or reanalyze any sample yielding a value for the RCAS internal standard of <15,000 copies/reaction mixture (∼50% of input).
TABLE 2.
Recovery of the internal virion standarda
| Patient | Measured value for RCAS internal standard (no. of copies/reaction mixture)
|
CV (%) | |
|---|---|---|---|
| Avg valuec | SD | ||
| A (8)b | 32,838 (121) | 9,152 | 28 |
| B (8) | 24,694 (91) | 8,098 | 33 |
| C (2) | 22,811 (84) | 3,004 | 13 |
| D (2) | 23,613 (87) | 8,686 | 37 |
| E (2) | 24,624 (91) | 6,481 | 26 |
| Avg | 25,716 (95.2) | 4,057 | 16 |
A cell culture supernatant containing 300,000 copies of RCAS RNA was added to each patient plasma sample. Nine percent of the total sample was assayed following centrifugation.
Values in parentheses are the number of samples analyzed.
Two values for the internal standard were measured for each sample analyzed. The percent recovery is shown in parentheses.
Assay of patient samples.
To test the utility of the single-copy assay for analysis of patient samples and to compare it to existing assays, we examined plasma samples from HIV-1-seronegative and -seropositive individuals. To assess whether the single-copy assay yields false-positive results, plasma samples from 20 seronegative volunteers were analyzed. Each of the 20 samples gave negative results by the single-copy assay; that is, the calculated HIV-1 RNA values were less than 1 copy in the 3.8 ml of plasma analyzed by each assay (data not shown).
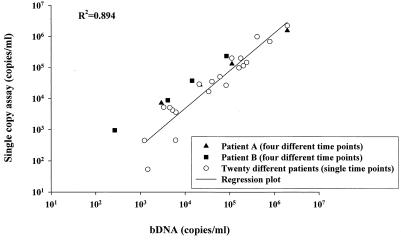
The PCR primers and probe used in the assay were selected on the basis of the regions that are highly conserved among HIV-1 subtype B sequences (12). To ensure that the assay was capable of detecting and accurately quantifying HIV-1 RNA in a wide range of U.S. patients, we assayed by the single-copy assay samples from 22 HIV-infected, antiretroviral agent-naïve patients who were monitored at the NIH Clinical Center and compared the results to those obtained by the broadly reactive bDNA assay (version 3.0) (Fig. 2). Included in this study were four plasma samples drawn from each of two patients before and after they started antiretroviral therapy. There was excellent agreement between the HIV-1 RNA values obtained by the two assays (R2 = 0.894), implying that the single-copy assay can be used to quantify HIV-1 RNA in most HIV-1-infected individuals in the United States. Nevertheless, as for other HIV-1 RNA assays, it would not be appropriate to test posttherapy samples without testing a pretherapy sample to ensure that a particular patient's virus can be quantified by the assay used.
FIG. 2.
Comparison of bDNA assay values versus single-copy assay values for 22 individual patients with bDNA assay values of >1,000 copies/ml. Twenty individual patient plasma samples (open circles) were analyzed by both assays at a single time point. For two of the patients, levels were measured at four times (one time before and three times after treatment initiation) (filled symbols). For all patient samples, 1 to 2 ml of patient plasma was analyzed by the single-copy assay.
Analysis of plasma samples with <75 HIV-1 RNA copies/ml.
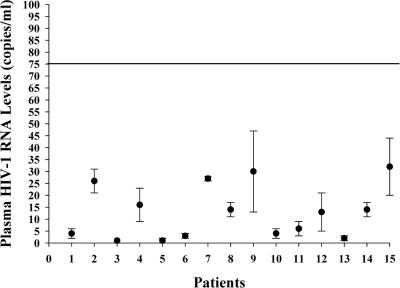
We next explored whether samples from patients receiving antiretroviral therapy had persistent quantifiable viremia. Plasma samples from 15 patients who were receiving antiretroviral therapy and who had <75 HIV-1 RNA copies/ml by the bDNA assay (version 3.0) were analyzed by the single-copy assay (Fig. 3). The results show that HIV-1 RNA could be detected in the plasma samples from all 15 patients, with the values ranging from 1 to 32 copies/ml and with the median being 13 copies/ml.
FIG. 3.
Single-copy assay measurements of HIV-1 RNA loads in 15 patient plasma samples found to contain <75 copies/ml by the bDNA assay. Each point represents the mean and standard deviation of at least triplicate values for the sample from each patient. Pretherapy samples were available for patients 1 to 4.
DISCUSSION
We have developed an internally controlled real-time RT-PCR assay which can detect HIV-1 RNA at concentrations as low as 1 copy per reaction mixture. This assay should prove useful in detecting, quantifying, and characterizing the persistence of viremia in patients who are receiving different antiretroviral treatment regimens and whose HIV-1 RNA levels are suppressed to <50 to 75 copies/ml. Sample volumes of 1 to 7 ml can be used. Although sensitivity is maximal with 7 ml of plasma, when only the smaller volumes are available, the assay still has a sensitivity significantly greater than those of other methods. Plasma samples are spiked with a known quantity of an internal standard virion to monitor viral RNA extraction and recovery. To our knowledge, this is the first HIV-1 RNA assay to use an internal standard virion control that allows one to detect problems with virion recovery that would otherwise go unnoticed. At present we use the internal standard to detect HIV-1 in the small number (ca. 5%) of plasma samples for which problems with virion recovery are encountered. We are investigating its use to correct the observed HIV-1 RNA copy numbers for virion recovery. In repeated testing of a panel of samples with low HIV-1 RNA copy numbers, our assay proved to be more sensitive than the FDA-approved AMPLICOR HIV-1 MONITOR ultrasensitive assay (version 1.0), the recently approved Versant bDNA assay (version 3.0), and a modified AMPLICOR HIV-1 MONITOR assay that uses 2 ml of plasma.
A possible concern with PCR-based assays is the genetic variation of HIV-1 in the genomic regions complementary to the primer or probe. Such variation might lead to variations in the ability to detect HIV-1 RNA or various sensitivities of detection from one patient sample to the next. To investigate this possibility, we compared the results obtained by the single-copy assay with those obtained by the broadly reactive bDNA assay (version 3.0) by testing plasma samples from 22 antiretroviral agent-naïve patients at the NIH Clinical Center with levels of viremia ranging from 103 to more than 106 copies of RNA/ml of plasma. This comparison showed an excellent correlation between the results obtained by the single-copy assay and those obtained by the more broadly based bDNA assay (Fig. 2). This finding implies that natural sequence variation may not be a major problem, at least among subtype B viruses. Furthermore, a survey of 36 subtype B sequences in the Los Alamos National Laboratory HIV database (13) revealed that 21 sequences are perfectly complementary to the two primers and the probe that we designed, whereas the rest of the sequences vary by one or two bases. This analysis implies that the assay is tolerant of small numbers of mismatches between the primers and the virus. Sequencing of the amplicon region is being undertaken to test whether the differences observed between the two assays for a few samples are due to such sequence variations. As expected, comparisons among viruses of other subtypes reveals larger numbers of potential mismatches, implying that the assay may not work as well with non-subtype B viruses and that primer and probe sequences would need to be redesigned for this purpose.
We detected residual viremia in all 15 plasma samples from a series of 15 patients with <75 HIV-1 RNA copies/ml by the bDNA assay; the levels ranged from 1 to 32 copies/ml (median, 13 copies/ml). This finding confirms predictions from previous studies that viremia persists in plasma at levels below the sensitivities of available assays (4, 16). Although a recent study concluded that intermittent viremia (>50 copies/ml) in patients receiving combination antiretroviral therapy was not associated with virologic failure (7), it is not known whether persistent low-level viremia will allow the emergence of drug-resistant viral populations and lead to virologic failure. It is also not known whether the level of persistent viremia will correlate with the likelihood of virologic failure or whether it reflects ongoing virus replication. The single-copy assay described here should help address these important questions. In conclusion, we have developed a virion-controlled real-time RT-PCR assay which can quantify HIV-1 RNA at concentrations as low as 1 copy/reaction mixture. This assay may provide new insights into studies on viral dynamics, evolution, and the source of persistent replication in patients who are receiving antiretroviral therapy and who have viral loads less than 50 to 75 copies/ml.
Acknowledgments
We thank Mike Piatak and Jeff Lifson for valuable help and discussions and for supplying plasmid pQP1 of HIV-1, and we thank Steve Hughes and Andrea Ferris for supplying plasmid RCAS and DF1 cell culture supernatants.
This study was supported in part by the Adult AIDS Clinical Trials Group funded by the National Institute of Allergy and Infectious Diseases (5 U01 A1046383-04 and 203VC001).
REFERENCES
- 1.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 3.Collins, M. L., B. Irvine, D. Tyner, E. Fine, C. Zayati, C. Chang, T. Horn, D. Ahle, J. Detmer, L. P. Shen, J. Kolberg, S. Bushnell, M. S. Urdea, and D. D. Ho. 1997. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 25:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dornadula, G., H. Zhang, B. VanUitert, J. Stern, L. Livornese, Jr., M. J. Ingerman, J. Witek, R. J. Kedanis, J. Natkin, J. DeSimone, and R. J. Pomerantz. 1999. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA 282:1627-1632. [DOI] [PubMed] [Google Scholar]
- 5.Elbeik, T., E. Charlebois, P. Nassos, J. Kahn, F. M. Hecht, D. Yajko, V. Ng, and K. Hadley. 2000. Quantitative and cost comparison of ultrasensitive human immunodeficiency virus type 1 RNA viral load assays: Bayer bDNA quantiplex versions 3.0 and 2.0 and Roche PCR Amplicor monitor version 1.5. J. Clin. Microbiol. 38:1113-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 7.Havlir, D. V., R. Bassett, D. Levitan, P. Gilbert, P. Tebas, A. C. Collier, M. S. Hirsch, C. Ignacio, J. Condra, H. F. Gunthard, D. D. Richman, and J. K. Wong. 2001. Prevalence and predictive value of intermittent viremia with combination HIV therapy. JAMA 286:171-179. [DOI] [PubMed] [Google Scholar]
- 8.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 9.Hughes, M. D., V. A. Johnson, M. S. Hirsch, J. W. Bremer, T. Elbeik, A. Erice, D. R. Kuritzkes, W. A. Scott, S. A. Spector, N. Basgoz, M. A. Fischl, R. T. D'Aquila, et al. 1997. Monitoring plasma HIV-1 RNA levels in addition to CD4+ lymphocyte count improves assessment of antiretroviral therapeutic response. Ann. Intern. Med. 126:929-938. [DOI] [PubMed] [Google Scholar]
- 10.Hughes, S. H., J. J. Greenhouse, C. J. Petropoulos, and P. Sutrave. 1987. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Japour, A., P. Chatis, H. Eigenrauch, and C. Crumpacker. 1991. Detection of human immunodeficiency virus type 1 clinical isolates with reduced sensitivity to zidovudine and dideoxyinosine by RNA-RNA hybridization. Proc. Natl. Acad. Sci. USA 88:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, F. McCutchan, P. A. Marx, J. W. Mellors, J. I. Mullins, J. Sodroski, and S. Wolinksy. 2000. Human retroviruses and AIDS 2000: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.M.
- 13.Kuiken, C. L., B. Foley, B. Hahn, P. A. Marx, F. McCutchan, J. W. Mellors, S. Wolinksy, and B. Korber. 2001. HIV sequence compendium 2001. Los Alamos National Laboratory, Los Alamos, N.M.
- 14.Larder, B. A., and S. D. Kemp. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246:1155-1158. [DOI] [PubMed] [Google Scholar]
- 15.Levy, J. A. 1993. Pathogenesis of human immunodeficiency virus infection. Microbiol. Rev. 57:183-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewin, S. R., M. Vesanen, L. Kostrikis, A. Hurley, M. Duran, L. Zhang, D. D. Ho, and M. Markowitz. 1999. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J. Virol. 73:6099-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak, K. J., S. J. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 18.Mellors, J. W., A. Munoz, J. V. Giorgi, J. B. Margolick, C. J. Tassoni, P. Gupta, L. A. Kingsley, J. A. Todd, A. J. Saah, R. Detels, J. P. Phair, and C. R. Rinaldo, Jr. 1997. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann. Intern. Med. 126:946-954. [DOI] [PubMed] [Google Scholar]
- 19.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 20.Montaner, J. S., M. Harris, T. Mo, and P. R. Harrigan. 1998. Rebound of plasma HIV viral load following prolonged suppression with combination therapy. AIDS 12:1398-1399. [DOI] [PubMed] [Google Scholar]
- 21.Mulder, J., N. McKinney, C. Christopherson, J. Sninsky, L. Greenfield, and S. Kwok. 1994. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J. Clin. Microbiol. 32:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Brien, W. A., P. M. Hartigan, E. S. Daar, M. S. Simberkoff, J. D. Hamilton, et al. 1997. Changes in plasma HIV RNA levels and CD4+ lymphocyte counts predict both response to antiretroviral therapy and therapeutic failure. Ann. Intern. Med. 126:939-945. [DOI] [PubMed] [Google Scholar]
- 23.Pantaleo, G., C. Graziosi, and A. S. Fauci. 1993. New concepts in the immunopathogenesis of human immunodeficiency virus infection. N. Engl. J. Med. 328:327-335. [DOI] [PubMed] [Google Scholar]
- 24.Piatak, M., Jr., K. C. Luk, B. Williams, and J. D. Lifson. 1993. Quantitative competitive polymerase chain reaction for accurate quantitation of HIV DNA and RNA species. BioTechniques 14:70-81. [PubMed] [Google Scholar]
- 25.Piatak, M., Jr., M. S. Saag, L. C. Yang, S. J. Clark, J. C. Kappes, K. C. Luk, B. H. Hahn, G. M. Shaw, and J. D. Lifson. 1993. Determination of plasma viral load in HIV-1 infection by quantitative competitive polymerase chain reaction. AIDS 7(Suppl. 2):S65-S71. [DOI] [PubMed] [Google Scholar]
- 26.Raboud, J. M., J. S. Montaner, B. Conway, S. Rae, P. Reiss, S. Vella, D. Cooper, J. Lange, M. Harris, M. A. Wainberg, P. Robinson, M. Myers, and D. Hall. 1998. Suppression of plasma viral load below 20 copies/ml is required to achieve a long-term response to therapy. AIDS 12:1619-1624. [DOI] [PubMed] [Google Scholar]
- 27.Riddler, S. A., and J. W. Mellors. 1997. HIV-1 viral dynamics and viral load measurement: implications for therapy. AIDS Clin. Rev. 1997-1998:47-65. [PubMed] [Google Scholar]
- 28.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 29.Shafer, R. W., M. A. Winters, S. Palmer, and T. C. Merigan. 1998. Multiple concurrent reverse transcriptase and protease mutations and multidrug resistance of HIV-1 isolates from heavily treated HIV-1 infected patients. Ann. Intern. Med. 128:906-911. [DOI] [PubMed] [Google Scholar]
- 30.Sun, R., J. Ku, H. Jayakar, J. C. Kuo, D. Brambilla, S. Herman, M. Rosenstraus, and J. Spadoro. 1998. Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 36:2964-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 32.Yerly, S., O. T. Rutschmann, M. Opravil, F. Marchal, B. Hirschel, L. Perrin, et al. 1999. Cell-associated HIV-1 RNA in blood as indicator of virus load in lymph nodes. J. Infect. Dis. 180:850-853. [DOI] [PubMed] [Google Scholar]