Abstract
Rapid identification of patients infected with clarithromycin-resistant Helicobacter pylori without the need for culture can help to avoid useless prescriptions of clarithromycin. We developed and tested a routine real-time quantitative PCR assay dedicated to that purpose. One hundred ninety-six consecutive gastric biopsy specimens were examined by culture, histology performed by a trained physician, and rapid PCR with the LightCycler apparatus. Infection was defined as (i) positivity of culture, (ii) positivity of histology, or (iii) positivity of PCR if confirmed by positivity of a concomitant indirect test (serology or urea breath test). Susceptibility to clarithromycin was tested by E-test and PCR. The prevalence of infection was 33.7% (66 of 196 samples). The sensitivities of culture, histology, and PCR were 90.9% (60 of 66 samples), 87.9% (58 of 66 samples), and 97.0% (64 of 66 samples), respectively. The specificity of PCR was 94.6% (123 of 130 samples). The linearity of the PCR results was achieved over a 6-log range of input DNA, and we were able to accurately quantify as few as 300 bacteria and to qualitatively detect as few as 30 bacteria per DNA sample. For clarithromycin susceptibility testing, there was 98.2% (55 of 56 samples) concordance between E-test and PCR. Forty-eight strains were clarithromycin susceptible, and 9 strains were clarithromycin resistant. The single discrepancy concerned a culture which was a mixture of mutant and wild type, with a susceptible-to-resistant ratio of 11.5: the resistant population was detected by E-test but not by PCR. Our PCR assay is accurate for fast detection of H. pylori as well as of clarithromycin resistance and is also able to objectively determine bacterial density.
Many diagnostic assays have been developed for Helicobacter pylori: culture, histology, rapid urease test, urea breath test, serology, stool antigen test, and molecular-based tests (24). Culture has the great advantage of permitting subsequent determination of the antimicrobial susceptibility of the strain isolated, in particular to macrolides. Indeed, the macrolide drug clarithromycin is the key component of many combination therapies used to eradicate H. pylori, and macrolide resistance is the most important cause of treatment failure (13, 22). However, disadvantages of culture include special conditions for specimen transportation, the use of complicated media with special conditions for maintenance, the need for special incubation conditions, and the length of time necessary to obtain a result (20).
Clarithromycin resistance in H. pylori is due to the lack of binding of the macrolides to the 23S rRNA components of the bacterial ribosome due to modification of the target site by occurrence of a single spontaneous point mutation in the peptidyltransferase region of the 23S rRNA gene (17, 25). Mutations A2142G and A2143G are the most often observed, with the A2142C mutation being much rarer (25). Other mutations (A2115G, G2141A, and T2717C) have been described but appear to be exceptional (10, 14). The small number of mutations implicated in macrolide resistance of H. pylori makes molecular detection of resistance, notably by PCR, realistic. Several techniques, notably some relying on PCR and particularly on real-time PCR, have been developed for bacterial detection or macrolide susceptibility testing (4, 19). Some of the advantages of real-time PCR are rapidity, low rate of contaminations because of product analysis in a closed-tube system, and possibility of accurate quantification of the DNA target (26).
Two studies have used real-time PCR to detect and quantify H. pylori directly from gastric biopsy specimens. He et al. amplified a fragment of the ureC gene (12) by this technique with the LightCycler apparatus (26). However, specificity of PCR was perhaps not optimal in this work because it was found that a great number of culture-negative and histology-negative biopsy specimens were strongly positive by PCR. Kobayashi et al. amplified a fragment of the 16S rRNA gene and compared real-time PCR results obtained by using the TaqMan apparatus to those of culture, histology, urea breath test, and rapid urease test (15). They found that TaqMan PCR had the highest sensitivity and specificity, 100% for both. In these two studies, the authors found that real-time PCR was able to reliably quantify the gastric mucosal density of H. pylori.
Concerning the detection of clarithromycin resistance by real-time PCR, three works have shown that amplification of a critical fragment of the 23S rRNA gene by LightCycler PCR (LC-PCR) permits an accurate determination of genotypic clarithromycin resistance (5, 16, 18). But in two of them, the reliability of concomitant detection of the bacterium by real-time PCR was not assessed (16, 18). In the third study (5), the authors compared LC-PCR for H. pylori detection only to the rapid urease test with the Helicocheck kit and found 100% concordance between the two tests, which is somewhat surprising given that the sensitivity of the rapid urease test is good but not excellent (8).
In the present work, our aim was to evaluate whether a simpler system based entirely on LC-PCR amplification of the 23S rRNA gene directly from gastric biopsy specimens could accurately perform these three tasks: detect the bacterium, quantify the bacterial density, and determine the susceptibility of the strain to clarithromycin. To do that, we developed a quantitative LC-PCR assay and compared its performances to those of culture (with clarithromycin susceptibility testing by the E-test method) and histology. The bacterial density determined by LC-PCR was compared with the semiquantitative results obtained by culture and histology.
MATERIALS AND METHODS
Study design.
Biopsy samples were obtained over a 16-month period (July 2000 to October 2001) from 196 dyspeptic patients referred for endoscopy at Henri-Mondor hospital. The mean age of the patients was 51.6 years (range, 19 to 90 years), and 63% of the patients were men. They had either nonulcer dyspepsia (91 patients, 46%), gastroduodenal ulcer (83 patients, 42%), MALT lymphoma of the stomach (19 patients, 10%), or gastric cancer (3 patients, 2%). Three endoscopic biopsy specimens taken from the antrum (3 cm from the pylorus) and three taken from the fundus were sent in formalin for histology. In addition, one biopsy specimen from the antrum and one from the fundus were immediately frozen and kept at −80°C for culture and LC-PCR. Some patients benefited from additional diagnostic tests (serology or urea breath test). Clarithromycin susceptibility testing was performed by the E-test method and by LC-PCR as described below. Infection by H. pylori was defined as (i) positivity of culture, (ii) positivity of histology performed by a trained histologist, or (iii) positivity of LC-PCR and of a concomitant indirect test (serology or urea breath test). The bacterial density was evaluated semiquantitatively by culture and histology and quantitatively by LC-PCR, as described below.
Culture and antibiotic susceptibility testing.
Two biopsy specimens from each patient (one from the antrum and one from the fundus) were immersed together into 0.5 ml of glycerol-supplemented brain-heart infusion broth and homogenized in individual microtubes. One half of the homogenate was used for culture, and the other half was kept at −80°C until completion of LC-PCR. Cultures were performed with a commercialized serum-supplemented selective medium, Pylori agar (bioMérieux, Marcy l'Etoile, France), under microaerophilic conditions at 37°C for a maximum of 6 days. H. pylori isolates were defined as gram-negative spiral-shaped bacilli that were oxidase positive and rapidly (less than 1 h) urease positive. To estimate the bacterial density, a scoring system of 1 to 4 was established as follows: 1, 1 to 10 colonies on the plate, 2, 10 to 100 colonies on the plate; 3, 100 to 1,000 colonies on the plate; 4, more than 1,000 colonies on the plate.
MICs of clarithromycin were determined by the E-test method (AB Biodisk, Solna, Sweden), according to the instructions of the manufacturer, by using Mueller-Hinton agar (Oxoid, Lyon, France) supplemented with 10% horse blood (bioMérieux) and a cell suspension calibrated at 3 McFarland units. Plates were read after 3 days of incubation at 37°C. Strains were considered resistant to clarithromycin if the MIC was >0.5 μg/ml (11).
Histological analysis with grading of H. pylori density.
Formalin-fixed and paraffin-embedded gastric biopsy specimens were routinely processed. Gastritis activity was graded on a four-point scale of none (grade 0), mild (grade 1), moderate (grade 2), and severe (grade 3) according to the guidelines of the Sydney system (21). The presence of H. pylori was assessed on modified Giemsa-stained sections. The bacterial density was estimated on a scoring system of 1 to 3 as follows: 1, less than 10 bacteria per gland, 2, 10 to 20 bacteria in at least one gland, and 3, more than 20 bacteria in at least one gland or over the superficial epithelium. The overall bacterial density quantified by histology was estimated by a global score, which was the sum of the density grades in antrum and corpus, and thus ranged from 0 to 6.
Detection of H. pylori in gastric biopsy specimens using the Light-Cycler.
LC-PCR was targeted at the 23S rRNA gene, and the hybridization probe method was used for detection of the amplified product. The primers used were HP23S1 (5′-GGA GCT GTC TCA ACC AGA GAT TC-3′) (nucleotide positions 2071 to 2093) and HP23S2 (5′-CGC ATG ATA TTC CC[AG] TTA GCA G-3′) (nucleotide positions 2181 to 2201), and the result was a 132-bp product. The two hybridization probes used were called HP23S3 and HP23S4. HP23S3 (5′-GGA GCT GTC TCA ACC AGA GA[Red640]T TC-3′) had the same sequence as primer HP23S1 and was internally labeled with Red640. HP23S4 (5′-GGA ATT TTC ACC TCC ACT ACA ATT TCA CTG[Fluo]-3′) (nucleotide positions 2201 to 2230) was 3′ labeled with fluorescein and located just downstream of HP23S3 on the other strand.
H. pylori DNA was extracted from bacterial colonies by using the QIAamp tissue kit (Qiagen, Courtaboeuf, France) and was quantified by UV spectrophotometry at 260 nm. We estimated that one bacterium corresponded to 1.815 fg of DNA, assuming the mean H. pylori genome size is 1,655,849 ppb (1, 23). After DNA extraction, one 10-fold serial dilution of H. pylori DNA was made, with bacterial concentrations ranging from 3 to 3 × 108 bacteria per 5 μl. A series of 10-fold dilutions of H. pylori DNA was included in each amplification run. DNA from 0.25 ml of gastric biopsy homogenates was extracted by using the High Pure PCR template preparation kit (Roche Molecular Biochemicals, Meylan, France).
PCR was performed in a final volume of 25 μl with the DNA master hybridization probes kit (Roche Molecular Biochemicals), 5 and 10 pmol of oligonucleotide primers HP23S1 and HP23S2, respectively, 5 pmol of hybridization probes HP23S3 and HP23S4, and 5 μl of extracted DNA sample. Carryover was prevented by using heat-labile uracil-DNA glycosylase (Roche Molecular Biochemicals). Amplification was performed for 50 cycles of denaturation (95°C, 10 s), annealing (55°C, 10 s), and extension (72°C, 15 s).
A single fluorescence reading for each sample was taken at the annealing step. Quantitative results were expressed by determination of the threshold of detection, or the crossing point (Cp), which marked the cycle when the fluorescence of a given sample significantly exceeded the baseline signal. They were expressed as fractional cycle numbers. Each sample was tested in duplicate, and the final Cp was the mean of the two results. Then the Cp's were plotted against the known concentration of the bacterium to obtain the standard curve. The bacterial count for a given bacterial sample was calculated by interpolation from this standard curve. To compare the densities obtained by PCR to those of histology and culture, the values obtained by quantitative LC-PCR were converted into decimal logarithmic values.
Melting analysis for detection of mutations in the 23S rRNA gene.
PCR was performed as described above except that the following set of internal hybridization probes was used: HP23S7, 5′-GTG GAG GTG AAA ATT CCT CCT ACC CG5(Fluorescein) (nucleotide positions 2106 to 2131), and HP23S8, 5′-(Red640)GGC AAG ACG GAA AGA CCC C(Phosphate)-3′ (nucleotide positions 2133 to 2151). After completion of the amplification process, the reaction mixture was denatured at 95°C for 1 min, held at 45°C for 15 s, and then slowly heated to 75°C at a ramp rate of 0.1°C/s. During this process, fluorescence was continuously monitored, and melting curves were constructed automatically with software attached to the LightCycler. Melting curves were converted to melting peaks by plotting the negative derivative of the fluorescence with respect to temperature (−dF/dT).
Statistics.
Quantitative parameters were expressed as means ± standard errors of the means. Contingency analysis was performed by using the Fisher exact test. Correlations between the different scores were made by the Spearman nonparametric test. P values of less than 0.05 were considered significant.
RESULTS
Development of LC-PCR.
Concerning amplification of H. pylori DNA obtained from a pure culture, linearity was achieved over a 6-log range of input DNA amounts, from 54 ng to 540 fg of H. pylori DNA or 3 × 108 to 300 bacteria, with 300 bacteria corresponding to 600 23S rRNA gene copies given that there are two gene copies in the genome of H. pylori (1, 23). PCR was also positive for 5.4 fg of H. pylori DNA (30 bacteria or 60 copies of the 23S rRNA gene fragment) but with a sharp drop in amplification efficiency. The amplification efficiencies obtained with DNA prepared from gastric biopsy specimens were identical to those obtained with DNA samples prepared from bacterial colonies. Thus, the sensitivity of our assay, for DNA samples prepared either from cultures or from gastric biopsy specimens, could quantitatively reach 300 bacteria or 600 copies of the amplicon and qualitatively reach 30 bacteria or 60 copies. The reproducibility of our assay was evaluated with 10-fold serial dilutions of purified H. pylori DNA, with no significant difference between runs.
Detection of H. pylori.
Of the biopsy specimens tested, 33.7% (66 of 196) were classified H. pylori positive (Table 1), with infection defined as positivity of culture, of histology performed by a trained physician, or of PCR if an indirect test was concomitantly positive (serology or urea breath test). There were nine patients with positivity by PCR only, and four of these benefited from one or two indirect diagnostic tests. Two were recognized as H. pylori positive: one patient with gastric cancer tested positive by breath test and serology, the other presented with duodenal ulcer not linked to consumption of aspirin or nonsteroidal anti-inflammatory agents, tested positive by serology, and was highly positive by quantitative PCR (236,130 bacteria in the DNA sample). The last two patients had a negative breath test and were considered noninfected. The remaining five patients, who were not subjected to an indirect test, were also considered noninfected. Even with our definition of infection, the sensitivity and negative predictive value of our PCR assay were excellent (97.0 and 98.4%, respectively) and higher than those of culture (Table 1). The specificity and predictive positive value of PCR were also very good, 94.6 and 90.1%, respectively (Table 1).
TABLE 1.
Comparison of performances of PCR-based detection of H. pylori with culture and histology performed by a trained physiciana
| Parameter | Result (%) (no. positive/no. negative) for:
|
||
|---|---|---|---|
| Culture | Histology | PCR | |
| Sensitivity | 90.9 (60/66) | 87.9 (58/66) | 97.0 (64/66) |
| Specificity | 94.6 (123/130) | ||
| Positive predictive value | 90.1 (64/71) | ||
| Negative predictive value | 95.6 (130/136) | 94.1 (128/136) | 98.4 (123/125) |
Infection was defined as (i) positivity of culture, (ii) positivity of histology, or (iii) positivities of PCR and a concomitant indirect test (serology or urea breath test).
Quantification of the mucosal bacterial density.
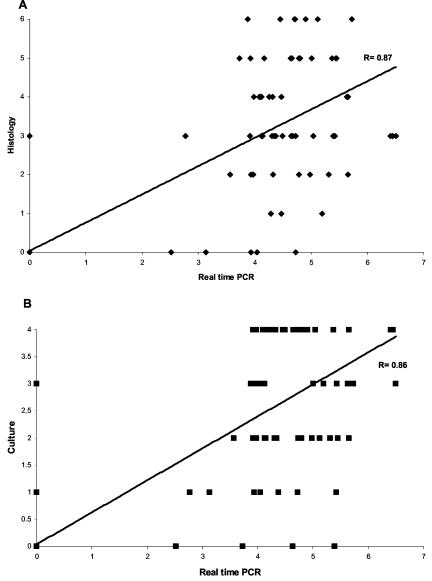
Mucosal bacterial density quantification was performed by culture, histology, and LC-PCR. The bacterial density by this latter technique could be evaluated for 63 of the 64 PCR-positive infected patients and ranged from 587 to 3,204,000 bacteria in the DNA sample (mean bacterial density, 222,782 bacteria). Concerning the seven cases of isolated positivity of PCR, the bacterial densities for three of them could not be precisely determined because fewer than 300 bacteria were detected. The bacterial densities for the four other patients were quantifiable but low: 330, 360, 620, and 1,398 (mean bacterial density, 677). The PCR-determined bacterial density was found to be statistically related to semiquantitative densities determined by culture and histology. The mean grades of the densities estimated by histology, culture, and LC-PCR were 1.09 ± 0.13, 0.91 ± 0.11, and 1.59 ± 0.16, respectively. There was significant correlation between the grade of bacterial density estimated by LC-PCR and those estimated by histology (P < 0.001) or culture (P < 0.001) (Fig. 1).
FIG. 1.
Correlations between the grade of bacterial density estimated by real-time PCR and that determined by histology (P < 0.001) (A) or culture (P < 0.001) (B). The results of real-time PCR are expressed in decimal logarithmic values.
Comparison of clarithromycin susceptibility testing by E-test and PCR.
Comparison between PCR and E-test could be performed for 89.4% (59 of 66) of the H. pylori-positive biopsy specimens. There was 98.3% concordance (58 of 59) between the two tests (Table 2). The prevalence of clarithromycin resistance in this study was 18.5% (12 of 65). MICs of clarithromycin for susceptible and resistant strains were ≤0.016 to 0.12 and 24 to ≥256 μg/ml, respectively. There was only one discrepant result, concerning a culture which was in fact a mixture of mutant and wild type, with a susceptible-to-resistant ratio of 11.5. The resistant population was detected by E-test but was missed by PCR. Comparison was impossible in five cases because of negativity of culture, in one because of negativity of PCR and in another because of negativities of both techniques. Concerning the five cases of negativity of culture, clarithromycin susceptibility testing by PCR could be performed for all of them, with susceptibility occurring in four cases and resistance occurring in one case. On the other hand, the strain detected by culture but not by PCR was clarithromycin susceptible. To summarize the data, clarithromycin susceptibility testing could be performed for 59, 64, and 65 of the 66 H. pylori-positive biopsy specimens by culture, PCR, and a combination of the two techniques, respectively.
TABLE 2.
Comparison of clarithromycin susceptibility testinga by E-test and by LC-PCR with melting analysis
| E-test result | LC-PCR result | No. of biopsy specimens |
|---|---|---|
| S | S | 48 |
| R | R | 10 |
| Culture negative | S | 4 |
| Culture negative | R | 1 |
| S | Negative | 1 |
| Heterogeneously R | S | 1b |
S, susceptible; R, resistant.
The susceptible-to-resistant ratio was 11.5.
DISCUSSION
We have developed a real-time PCR technique based on the amplification of a fragment of the 23S rRNA gene with the Light Cycler apparatus aimed at detecting H. pylori directly from gastric biopsy specimens, quantifying the bacterial density, and testing the susceptibility of the strain to clarithromycin. The sensitivity and the negative predictive value of PCR were better than those of culture and of histology performed by a trained physician, although positivity of PCR alone was not a criterion standard in our study. Indeed, an indirect test (serology or breath test) had to be concomitantly positive to consider isolated PCR positivity as a true positive, but indirect tests were rarely performed in our study. Thus, only four of the nine cases of isolated PCR positivity benefited from an indirect diagnostic test which was positive for two of them. The five other cases were classified as false positives, but given that the bacterial densities determined by quantitative PCR for these cases were low, it may be that at least some of them corresponded, in fact, to true infections with bacterial loads too low to be detected by other techniques. Thus, the performances of our LC-PCR assay could, in fact, be higher than those reported. This point could be clarified in another study with the systematic use of more diagnostic tests. Nevertheless, even with the criteria used, the performances of our PCR assay are still very good.
The quantitative sensitivity of our assay was 300 bacteria per reaction tube, i.e., 600 copies of the 23S rRNA gene and as few as 30 bacteria (60 copies of the gene) could be detected but not accurately quantified. These thresholds are similar to those of published reports (7, 12), and our work confirms that real-time PCR can accurately quantify the gastric mucosal density of H. pylori. We have shown that this can be done by the amplification of the 23S rRNA gene, whereas precedent works were based on amplification of either the ureC or 16S rRNA gene (12, 15). Other techniques are usable for quantification; however, each technique has weaknesses. Culture is only semiquantitative and is time consuming. Histology is also semiquantitative, but its accurateness is relatively weak because of great interobserver variation (2, 6, 9). Urea breath test has been shown to be uncorrelated to culture-determined bacterial density (3), but on the other hand, Kobayashi et al. found significant correlation between the results of quantification obtained by urea breath test and TaqMan real-time PCR (15). The accurateness of quantification by breath test would need to be further examined.
Susceptibility testing by PCR could be performed for all PCR-positive biopsy specimens. In comparison with phenotypic clarithromycin resistance measurement by the E-test, the detection of resistance mutations by PCR revealed 98.2% correlation. The only discrepancy concerned a mixture of susceptible and resistant cells which was classified as susceptible by PCR. However, susceptibility was predominant in this strain. Our observation is in agreement with the results obtained by Matsumura et al. (16). These authors reported that, in a mixture of susceptible and resistant cells, resistance was missed by LC-PCR when the susceptible-to-resistant ratio became higher than 10. Nonetheless, to our knowledge there is no proof that the identification of clarithromycin-resistant cells in a mixture of susceptible and resistant organisms has clinical relevance when susceptible cells largely predominate. Furthermore, it is evident that when bacterial densities are high, resistant mutants preexist even in apparently homogeneous susceptible bacterial populations. This issue would need to be clarified by further studies.
Our LC-PCR technique was fast. The PCR for detection could be performed in 2 to 3 h, including DNA extraction. The PCR for clarithromycin susceptibility testing required only one additional hour. Thus, our technique allows easily for same-day diagnosis of H. pylori infection with clarithromycin susceptibility testing, which is much more rapid than culture, which requires at least 5 days. This new method can thus permit us to obtain the information regarding clarithromycin susceptibility or resistance before treating the patient and so choose the appropriate regimen more efficiently. In addition, this method appears very well suited for large, multicentric, epidemiological studies on the prevalence of infection and of clarithromycin resistance. Indeed, a great advantage of PCR is that it does not require viable bacteria. The transport conditions are thus not as critical as they are for culture, and shipment costs are cheaper. The cost of the reagents necessary for our technique is reasonable: $6.20 for H. pylori detection only and $8.60 for detection and clarithromycin susceptibility testing. By microbiological techniques, detection is cheaper, $1.70, but the price of detection with clarithromycin susceptibility testing by E-test, $6.90, is comparable to that of PCR.
In conclusion, we have developed an LC-PCR assay that permits accurate, fast, and cost-effective detection and quantification of H. pylori directly from gastric biopsy specimens as well as determination of the susceptibility of the strain to clarithromycin. The specificity of our technique was excellent, and its sensitivity was higher than those of culture or histology despite the fact that, in this study, isolated PCR positivity was classified as false positivity. Thus, we think that this PCR technique is able to replace culture in routine as well as in epidemiological studies.
Acknowledgments
This work was supported by grant TBI 97017 from the Direction de la Recherche Clinique, Assistance Publique-Hôpitaux de Paris.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noohan, B. C. Guild, B. L. DeJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Atherton, J. C., K. T. Tham, R. M. Peek, Jr., T. L. Cover, and M. J. Blaser. 1996. Density of Helicobacter pylori infection in vivo as assessed by quantitative culture and histology. J. Infect. Dis. 174:552-556. [DOI] [PubMed] [Google Scholar]
- 3.Auroux, J., D. Lamarque, J. Tankovic, R. Benamouzig, S. Mahé, M.-T. Chaumette, and J.-C. Delchier. 1998. Comparaison de la quantification de l'infection gastrique à Helicobacter pylori par culture, histologie, et test respiratoire à l'urée radiomarquée au 13C. Gastroenterol. Clin. Biol. 22:407-412. [PubMed] [Google Scholar]
- 4.Bravos, E. D., and R. H. Gilman. 2000. Accurate diagnosis of Helicobacter pylori. Other tests. Gastroenterol. Clin. N. Am. 29:925-929. [DOI] [PubMed] [Google Scholar]
- 5.Chisholm, S. A., R. J. Owen, E. L. Teare, and S. Saverymuttu. 2001. PCR-based diagnosis of Helicobacter pylori infection and real-time determination of clarithromycin resistance directly from human gastric biopsy samples. J. Clin. Microbiol. 39:1217-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, A. H., T. Gjorup, J. Hilden, C. Fenger, B. Henriksen, M. Vyberg, K. Ostergaard, and B. F. Hansen. 1992. Observer homogeneity in the histologic diagnosis of Helicobacter pylori. Latent class analysis, kappa coefficient, and repeat frequency. Scand. J. Gastroenterol. 27:933-939. [DOI] [PubMed] [Google Scholar]
- 7.Costa, J.-M., C. Pautas, P. Ernault, F. Foulet, C. Cordonnier, and S. Bretagne. 2000. Real-time PCR for diagnosis and follow-up of toxoplasma reactivation after allogeneic stem cell transplantation using fluorescence resonance energy transfer hybridization probes. J. Clin. Microbiol. 38:2929-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Zimaity, H. M. T., D. Y. Graham, M. T. Al-Assi, H. Malaty, T. J. Karttunen, D. P. Graham, R. M. Huberman, and R. M. Genta. 1996. Interobserver variation in the histopathological assessment of Helicobacter pylori gastritis. Hum. Pathol. 27:35-41. [DOI] [PubMed] [Google Scholar]
- 10.Fontana, C., M. Faverole, S. Minelli, A. A. Criscuolo, A. Pietroiusti, A. Galante, and C. Favalli. 2002. New site of modification of 23S rRNA associated with clarithromycin resistance of Helicobacter pylori clinical isolates. Antimicrob. Agents Chemother. 46:3765-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grignon, B., J. Tankovic, F. Mégraud, Y. Glupczynski, M.-O. Husson, M.-C. Conroy, J.-P. Emond, J. Loulergue, J. Raymond, and J.-L. Fauchère. 2002. Validation of diffusion methods for macrolide susceptibility testing of Helicobacter pylori. Microb. Drug Resist. 8:61-66. [DOI] [PubMed] [Google Scholar]
- 12.He, Q., J.-P. Wang, M. Osato, and L. B. Lachman. 2002. Real-time quantitative PCR for detection of Helicobacter pylori. J. Clin. Microbiol. 40:3720-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houben, M. H., D. van der Beek, E. F. Hensen, A. J. Craen, E. A. J. Rauws, and G. N. J. Tytgat. 1999. A systematic review of Helicobacter pylori eradication therapy-the impact of resistance on eradication rates. Aliment. Pharmacol. Ther. 13:1047-1055. [DOI] [PubMed] [Google Scholar]
- 14.Hulten, K., A. Gibreel, O. Sköld, and L. Engstrand. 1997. Macrolide resistance in Helicobacter pylori: mechanism and stability in strains from clarithromycin-treated patients. Antimicrob. Agents Chemother. 41:2550-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi, D., Y. Eishi, T. Ohkusa, I. Ishige, T. Suzuki, J. Minami, T. Yamada, T. Takizawa, and M. Koike. 2002. Gastric mucosal density of Helicobacter pylori estimated by real-time PCR compared with results of urea breath test and histological grading. J. Med. Microbiol. 51:305-311. [DOI] [PubMed] [Google Scholar]
- 16.Matsumura, M., Y. Hikiba, K. Ogura, G. Togo, I. Tsukuda, K. Ushikawa, Y. Shiratori, and M. Omata. 2001. Rapid detection of mutations in the 23S rRNA gene of Helicobacter pylori that confers resistance to clarithromycin treatment to the bacterium. J. Clin. Microbiol. 39:691-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Occhialini, A., M. Urdaci, F. Doucet-Populaire, C. M. Bébéar, H. Lamouliatte, and F. Mégraud. 1997. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob. Agents Chemother. 41:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oleastro, M., A. Ménard, A. Santos, H. Lamouliatte, L. Monteiro, P. Barthélémy, and F. Mégraud. 2003. Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J. Clin. Microbiol. 41:397-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owen, R. J. 2002. Molecular testing for antibiotic resistance in Helicobacter pylori. Gut 50:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Perez, G. I. 2000. Accurate diagnosis of Helicobacter pylori. Culture, including transport. Gastroenterol. Clin. N. Am. 29:879-884. [DOI] [PubMed] [Google Scholar]
- 21.Price, A. B. 1991. The Sydney system: histological division. J. Gastroenterol. Hepatol. 6:209-222. [DOI] [PubMed] [Google Scholar]
- 22.Tankovic, J., D. Lamarque, C. Lascols, C.-J. Soussy, and J.-C. Delchier. 2001. Impact of Helicobacter pylori resistance to clarithromycin on the efficacy of the omeprazole-amoxicillin-clarithromycin therapy. Aliment. Pharmacol. Ther. 15:707-713. [DOI] [PubMed] [Google Scholar]
- 23.Tomb, J. F., O. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzgerald, M. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete gene sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 24.Vaira, D., L. Gatta, C. Ricci, and M. Miglioli. 2002. Review article: diagnosis of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 16(Suppl. 1):16-23. [DOI] [PubMed] [Google Scholar]
- 25.van Doorn, L.-J., Y. Glupczynski, J. G. Kusters, F. Mégraud, P. Midolo, N. Maggi-Solca, D. M. M. Queiroz, N. Nouhan, E. Stet, and W. G. V. Quint. 2001. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob. Agents Chemother. 45:1500-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittwer, C. T., K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry, and U. J. Balis. 1997. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques 22:176-181. [DOI] [PubMed] [Google Scholar]