Abstract
Multilocus sequence typing (MLST) has been proven useful for the study of the global population structure of Campylobacter jejuni; however, its usefulness for the investigation of outbreaks of disease caused by C. jejuni has not been proven. In this study, MLST plus sequencing of the flaA short variable region (SVR) were applied to 47 isolates from 12 outbreaks of C. jejuni infection whose relatedness has been determined previously, and the results were compared to those of serotyping and pulsed-field gel electrophoresis (PFGE). Isolates implicated in an outbreak were indistinguishable by all four subtyping methods, with sporadic isolates being distinguished from outbreak isolates. Two sporadic isolates from one outbreak were resistant to SmaI digestion and therefore nontypeable by PFGE but were differentiated from the outbreak strain by the other methods. PFGE and flaA SVR typing were the most discriminatory methods, with discriminatory indices (DI) of 0.930 and 0.923, respectively. However, an epidemic strain from one outbreak was distinguished from the other outbreak isolates by flaA SVR typing; its flaA allele was different at five nucleotides, suggesting that this change was possibly mediated by recombination. MLST was less discriminatory than PFGE and flaA SVR typing (DI = 0.859), and many of the epidemic strains possessed common sequence types (STs) including ST-8, -21, -22, and -42. However, further discrimination within STs was achieved by flaA SVR typing or PFGE. The results from this study demonstrate that a combined approach of MLST plus flaA SVR typing provides a level of discrimination equivalent to PFGE for outbreak investigations.
Campylobacter jejuni is a pathogen that primarily causes gastrointestinal disease in humans. Gastrointestinal infection with C. jejuni causes significant morbidity and mortality, with the estimated number of cases a year in the United States exceeding 2 million and an estimated 2,000 deaths attributed to the infection (21). Human gastrointestinal infection by C. jejuni is thought to occur through the zoonotic transmission of the organism from natural reservoirs via food or water. C. jejuni forms part of the normal gastrointestinal flora of many farm animals, including poultry, pigs, and cattle (2), and is also found in environmental surface waters (3). Most human infections are sporadic (31); however, outbreaks do occur occasionally.
Contaminated milk has been responsible for large outbreaks of C. jejuni enteritis (24). Contamination of milk can be caused by direct excretion from an asymptomatic cow with mastitis (17) or, more commonly, by fecal contamination during milking (43). The majority of outbreaks have occurred after the consumption of raw milk (44), although improperly pasteurized milk (1) and postpasteurization contamination have been implicated as sources of outbreaks. Consumption of untreated surface water and cross-contamination from raw or undercooked products to other foods have also been demonstrated to be responsible for outbreaks of infection (34).
The detection of outbreaks of gastrointestinal disease caused by C. jejuni is important for the identification of the source or vehicle of infection and for limiting further infection in the community. Accurate and sensitive subtyping methods are required if we are to recognize outbreaks of infection, to match case isolates with those from potential vehicles of infection, and to discriminate these from unrelated strains.
A multitude of subtyping methods have been described for C. jejuni. Phenotypic methods include serotyping (32), phage typing (36), and biotyping (4). A number of molecular subtyping methods have been developed for C. jejuni, including ribotyping (10), flaA restriction fragment length polymorphism typing (25), amplified fragment length polymorphism typing (5), random amplified polymorphic DNA (19), and pulsed-field gel electrophoresis (PFGE) (28). PFGE has proven to be useful and discriminatory for the investigation of outbreaks of gastroenteritis caused by C. jejuni (11) and is considered to be the current “gold standard.” Ribot and colleagues (33) described a rapid standardized 1-day protocol for PFGE typing of C. jejuni that may facilitate timely investigation of outbreaks. Standardized protocols for PFGE and computer-assisted comparison of the PFGE patterns facilitates interlaboratory comparisons and has enabled the development of a nationwide surveillance network (PulseNet) within the United States, which has also aided in the detection of outbreaks of food-borne disease (39). However, interlaboratory comparisons of PFGE patterns require strict adherence to complex, standardized protocols, thereby limiting these comparisons to laboratories that have access to the appropriate equipment and software.
DNA sequencing-based methods for subtyping virtually eliminate experimental variation, facilitating interlaboratory comparisons. The text data generated by these methods can be easily transmitted electronically via the internet and compared to DNA sequences of other strains, allowing the development of global databases. The development of automated DNA sequencers and significant reductions in the price of reagents has led to an increasing use of DNA sequence data for routine clinical applications. DNA sequencing-based subtyping methods have been reported for C. jejuni. Sequencing a 267-bp segment of the flagellin (flaA) gene of C. jejuni, including the short variable region (SVR) may be useful for the investigation of C. jejuni outbreaks (22). Dingle and colleagues sequenced a 321-bp segment of the flaA gene that included the SVR in a study investigating the genetic diversity of C. jejuni strains associated with Guillain-Barré syndrome (7). This study identified 20 different SVR nucleotide sequences in 25 strains, illustrating the variability present in the sequence.
Multilocus sequence typing (MLST) is a subtyping method that is analogous to multilocus enzyme electrophoresis. Similarly to multilocus enzyme electrophoresis, MLST indexes the neutral genetic variation in housekeeping genes, which evolve slowly and are not under selective pressure. This technique was originally described for Neisseria meningitidis (20) but has been applied to other organisms, including C. jejuni (6). The sequences of 400- to 500-bp segments of seven housekeeping genes are determined by using automated sequencing, and the alleles from the seven genes are assigned a number based upon those already described in the Campylobacter database accessible through the internet at http://www.mlst.net. The 7-digit number, or allele profile, is assigned a sequence type (ST) based on those already present in the database, with novel allele profiles being assigned new STs arbitrarily.
MLST has been useful for investigating the DNA sequence diversity within C. jejuni and estimating the rates of intraspecies recombination (6, 38). MLST has also been proven very useful for longitudinal studies of population structure. However, the utility of MLST as an epidemiological tool for the investigation of outbreaks of disease caused by C. jejuni has not been established. The sequencing of more variable genes in conjunction with MLST has proven useful in investigations of outbreaks of N. meningitidis infection and may increase the discriminatory power of the method (9). The aim of the present study was to compare the subtyping of 47 isolates from 12 C. jejuni outbreaks by using PFGE, serotyping, MLST, sequencing of the 321-bp region of the flaA SVR, and a combined approach of MLST and SVR sequencing in an attempt to increase the discriminatory power of the DNA sequencing-based method.
MATERIALS AND METHODS
Bacterial isolates and culture conditions.
C. jejuni isolates were obtained from the National Campylobacter and Helicobacter Reference Laboratory, Centers for Disease Control and Prevention. Forty-seven isolates were included in the study from 12 separate outbreaks (Table 1). Prior to testing, we recovered C. jejuni isolates from storage at −70°C and cultured the isolates on heart infusion agar containing 5% (vol/vol) defibrinated rabbit blood (Becton Dickinson Biosciences, Sparks, Md.). We incubated all cultures under microaerobic conditions at 37°C for 48 h.
TABLE 1.
Demographic information for the 12 outbreaks included in this study
| Outbreak no. | Location | Date (yr) | Source | No. of isolates | Comment(s) | Reference |
|---|---|---|---|---|---|---|
| 1 | Kansas | 1998 | Food handler | 6 | 161 cases at a school luncheon | 27 |
| 2 | Florida | 1983 | City water supply | 5 | 865 cases | 35 |
| 3 | Kansas | 1981 | Raw milk (dairy A) | 6 | 18 | |
| 4 | Vermont | 1983 | Raw milk | 4 | 15 | |
| 5 | Vermont | 1982 | Raw milk | 2 | 15 cases | 40 |
| 6 | Vermont | 1986 | Inadequately pasteurized milk | 4 | 104 cases at a boarding school | 1 |
| 7 | Oklahoma | 1988 | Fecal contamination of milk | 5 | 104 cases in 4 prisons | Not previously reported |
| 8 | New York | 1986 | Tuna salad | 4 | Prison outbreak | Not previously reported |
| 9 | Kansas | 1988 | Raw milk | 4 | 87 cases at a bible school | Not previously reported |
| 10 | Wisconsin | 1995 | Tuna salad | 2 | 79 cases at a summer camp | 34 |
| 11 | Maine | 1989 | Food handler | 3 | Restaurant associated | Not previously reported |
| 12 | Louisiana | 1995 | Contaminated garlic bread | 2 | 30 cases, restaurant associated | Not previously reported |
HS serotyping.
The isolates had been previously serotyped at the National Campylobacter and Helicobacter Reference Laboratory, Centers for Disease Control and Prevention, by using the heat-stable (HS) serotyping scheme of Penner and Hennessy. The serotyping results of seven of the outbreaks were previously reported (1, 11, 29, 32).
PFGE.
We carried out PFGE typing by using SmaI to macrorestrict DNA and electrophoretic conditions as previously described by Ribot and colleagues (33). Electrophoresis was carried out by using a CHEF mapper (Bio-Rad Laboratories, Hercules, Calif.). Following electrophoresis, we visualized the fragments by ethidium bromide staining and UV transillumination. We captured gel images with a Gel Doc 2000 image analysis system (Bio-Rad Laboratories). PFGE gel images were normalized, and PFGE profiles were grouped together by using the Dice coefficient and clustering with the unweighted pair group method with arithmetic averages by using BioNumerics (version 3.0; Applied Maths, Austin, Tex.). Isolates were assigned to the same PFGE macrorestriction profile (mrp) when they clustered at greater than 95% similarity (0.41% optimization and 1.5% position tolerance), and we considered isolates differing at one band as different, with types being numbered arbitrarily (e.g., mrp 1 and 2, etc.). We confirmed the results of the clustering visually.
flaA SVR and MLST sequencing.
We carried out MLST as previously described by Dingle and colleagues (6). Briefly, we suspended a 1-μl loop of cells from a fresh overnight plate culture of the isolate in 0.5 ml of reagent grade water. We then heated the cell mixture in a heating block at 100°C for 15 min to lyse the cells. The mixture was centrifuged at 19,000 × g for 10 min, and we carefully removed 10 μl of the supernatant and added it to 90 μl of reagent grade water in a 0.5-ml microtube. We used PCR to amplify fragments of the seven housekeeping gene loci by using the reaction conditions and primers of Dingle and colleagues (6) and then amplified the entire flaA gene with consensus primers (42). The presence of the correct size PCR product was confirmed by gel electrophoresis. The PCR products were purified with a QiaAmp PCR purification kit according to the manufacturer's instructions (Qiagen, Valencia, Calif.). The purified products were diluted (1:9, vol/vol) in reagent grade water. Sequencing reaction mixtures were prepared with 3 μl of BigDye reaction mix (Applied Biosystems, Foster City, Calif.), 11 μl of the diluted PCR product, and 1 μl of primer to give a final concentration of 3.2 pmol per reaction mixture. Sequencing of the 7 MLST loci (aspA, glnA, gltA, glyA, pgm, tkt, and uncA) was carried out by using the previously described primers (6), and the flaA SVR was sequenced by using the following novel primers: flaA 130, GAT GCT TCA GGG ATG GCG ATA; flaA 624, CAA GTC CTG TTC CAA C(TA)G A(AG)G; flaA 612, GTT CCA ACT GAA GTT GAA ATC ACA. The reaction products were purified by column purification (Centri Sep 96; Princeton Separations, Adelphia, N.J.). We then separated and detected these reaction products with an ABI Prism 377 automated DNA sequencer (Applied Biosystems). Chromatograms were exported into SeqMan II (DNAStar Inc., Madison, Wis.), and the forward and reverse sequences were assembled. We cut the sequences to previously described allele lengths (between 402 and 507 bp) for each of the 7 MLST loci and then assigned allele numbers based on those already described in the MLST database at http://www.MLST.net. STs were assigned based upon those already present in the MLST database. The flaA SVR sequences were cut to 321 nucleotides, corresponding to nucleotides 283 to 603 of the coding sequence as previously described (7) and assigned allele numbers or flaA SVR types arbitrarily in order of description based on their nucleotide sequences.
DI.
Simpson's index of diversity (DI) was calculated for each of the subtyping methods by using the formula of Hunter and Gaston (16).
Nucleotide sequence accession numbers.
We submitted the partial flaA sequences to GenBank (accession numbers AY226587 to AY226590). Sequences that were identical to other sequences in GenBank were not submitted to the database.
RESULTS
Strain discrimination within outbreaks.
A summary of the results of the subtyping methods is presented in Table 2. In general, isolates within an outbreak were indistinguishable from each other by all four subtyping methods. Further, geographically and temporally related isolates that had been shown not to be part of the outbreak were differentiated from outbreak isolates, with the following exceptions. (i) Two isolates from wild birds associated with the second outbreak were resistant to SmaI digestion and therefore nontypeable by PFGE, but they were differentiated from the outbreak strains by MLST, serotyping, and flaA SVR typing. (ii) One isolate (Table 2, EDL2) that was not epidemiologically implicated in outbreak 3 was indistinguishable from the outbreak strains by all methods except for the detection of an additional HS antigen, HS:22. This antigen can be variably detected, and the sporadic strain was considered the same serotype as the outbreak strains (29). (iii) In outbreak 4, a fecal isolate from a cow implicated in the raw milk outbreak was found to be distinct from the outbreak strain by all four methods; however, a milk isolate from the same cow matched the outbreak strains. (iv) In outbreak 9, an epidemiologically implicated milk isolate was differentiated from other outbreak isolates by flaA SVR typing but was found to be indistinguishable by other typing methods. The flaA allele from the milk isolate contained five single-nucleotide polymorphisms (SNPs) relative to the outbreak strain in a 150-nucleotide-long segment located at the 3′ end of the 329 bases sequenced.
TABLE 2.
Comparison of MLST, PFGE, and flaA SVR sequencing for the discrimination of outbreak-associated C. jejuni isolates
| Isolate | HS type(s) | PFGE mrp | ST | flaA SVR type | Outbreak no. | Source | EIa |
|---|---|---|---|---|---|---|---|
| D5482 | 19 | 1 | 22 | 6 | 1 | Human | Y |
| D5479 | 19 | 1 | 22 | 6 | 1 | Human | Y |
| D5475 | 19 | 1 | 22 | 6 | 1 | Human | Y |
| D5476 | 19 | 1 | 22 | 6 | 1 | Human | Y |
| D5480 | 19 | 1 | 22 | 6 | 1 | Human | Y |
| D5478 | 4 | 2 | 48 | 13 | 1 | Human | N |
| D0445 | 19 | 3 | 22 | 6 | 2 | Human | Y |
| D0450 | 19 | 3 | 22 | 6 | 2 | Human | Y |
| D0452 | 19 | 3 | 22 | 6 | 2 | Human | Y |
| D0462 | 19, 23, 34 | NTb | 470 | 15 | 2 | Wild bird | N |
| D0467 | 19, 23, 34 | NT | 470 | 15 | 2 | Wild bird | N |
| EDL18 | 36, 23, 22 | 4 | 42 | 9 | 3 | Human | Y |
| EDL22 | 36, 23, 22 | 4 | 42 | 9 | 3 | Human | Y |
| SSU9892 | 36, 23, 22 | 4 | 42 | 9 | 3 | Cow, dairy A | Y |
| SSU9894 | 36, 23, 22 | 4 | 42 | 9 | 3 | Cow, dairy A | Y |
| SSU9896 | 2 | 5 | 21 | 3 | 3 | Cow, dairy A | N |
| EDL2 | 36, 23 | 4 | 42 | 9 | 3 | Cow, dairy B | N |
| D1117 | 2 | 6 | 21 | 4 | 4 | Child | Y |
| D1118 | 2 | 6 | 21 | 4 | 4 | Milk from cow A | Y |
| D1114 | 2 | 6 | 21 | 4 | 4 | Calf (drank cow A's milk) | Y |
| D1108 | 4 | 7 | 38 | 7 | 4 | Feces from cow A | Y |
| D0224 | 2 | 6 | 21 | 4 | 5 | Human | Y |
| D0226 | 2 | 6 | 21 | 4 | 5 | Cow feces | Y |
| D2286 | 2 | 8 | 21 | 3 | 6 | Human | Y |
| D2287 | 2 | 8 | 21 | 3 | 6 | Human | Y |
| D2290 | 2 | 8 | 21 | 3 | 6 | Cow feces | Y |
| D2289 | 36, 23, 22 | 9 | 42 | 11 | 6 | Cow feces | N |
| D2643 | 1, 8 | 10 | 8 | 1 | 7 | Human | Y |
| D2642 | 1, 8 | 10 | 8 | 1 | 7 | Human | Y |
| D2651 | 1, 8 | 10 | 8 | 1 | 7 | Human | Y |
| D2641 | 1, 8 | 10 | 8 | 1 | 7 | Cow feces (feed barn II) | Y |
| D2640 | 23, 36 | 4 | 42 | 10 | 7 | Cow feces (feed barn I) | N |
| D2248 | 55 | 11 | 45 | 12 | 8 | Human | Y |
| D2256 | 55 | 11 | 45 | 12 | 8 | Human | Y |
| D2253 | 4, 13 | 12 | 122 | 14 | 8 | Human | N |
| D2261 | 2 | 8 | 21 | 3 | 8 | Human | N |
| D2669 | 2 | 8 | 21 | 3 | 9 | Human | Y |
| D2678 | 2 | 8 | 21 | 3 | 9 | Human | Y |
| D2692 | 2 | 8 | 21 | 5 | 9 | Milk sample | Y |
| D2677 | 15, 23, 36 | 13 | 459 | 11 | 9 | Cow feces | N |
| D5157 | 33 | 11 | 679 | 12 | 10 | Human | Y |
| D5161 | 33 | 11 | 679 | 12 | 10 | Human | Y |
| D2763 | 2 | 14 | 8 | 2 | 11 | Human | Y |
| D2770 | 2 | 14 | 8 | 2 | 11 | Human | Y |
| D2769 | 2 | 14 | 8 | 2 | 11 | Human | Y |
| D5163 | 41 | 15 | 41 | 8 | 12 | Human | Y |
| D5166 | 41 | 15 | 41 | 8 | 12 | Human | Y |
EI, epidemiological implication; Y, yes; N, no.
NT, not typeable, did not restrict with SmaI.
Strain discrimination between outbreaks by each method.
The DI of the subtyping methods varied between 0.839 for serotyping and 0.930 for PFGE (Table 3). We identified 12 HS serotypes among the 47 isolates. The most common serotype was HS:2, which was the serotype of the epidemic strains in outbreaks 4, 5, 6, 9, and 11 and was also the serotype of temporally related strains from two other outbreaks. Serotype HS:19 was the second most common serotype and was the serotype of the epidemic strains in outbreaks 1 and 2. Serotype HS:36,23,22 was the serotype of the epidemic strain in outbreak 3.
TABLE 3.
DIs for C. jejuni subtyping methods
| Method | No. of types | No. of isolates with the most common type/total no. of isolates (%) | DI |
|---|---|---|---|
| Serotyping | 12 | 16/47 (34) | 0.839 |
| PFGE | 15 | 7/47 (15) | 0.930 |
| MLST | 12 | 13/47 (28) | 0.859 |
| flaA SVR | 15 | 8/47 (17) | 0.923 |
| MLST and flaA SVR | 17 | 8/47 (17) | 0.924 |
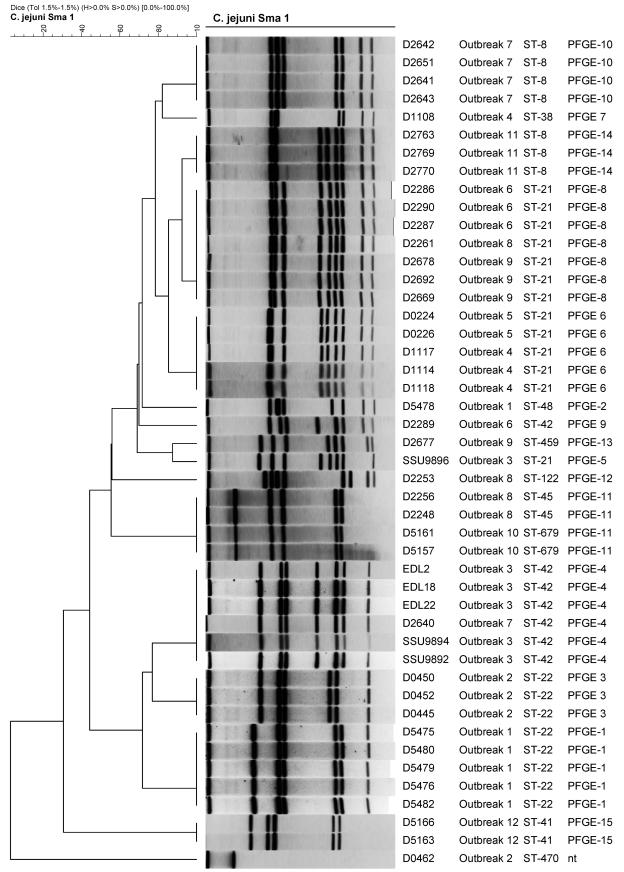
PFGE identified 15 mrp's with SmaI, with some outbreak patterns associated with more than one outbreak; the number of bands per mrp varied between four and nine (Fig. 1). The most common type was mrp 8, which was the type of the epidemic strains in outbreaks 6 and 9 and a temporally related strain in outbreak 8. The epidemic strain in outbreak 3 was indistinguishable from a temporally related strain from outbreak 7 (mrp 4). The epidemic strains in outbreaks 4 and 5 (mrp 6) and 8 and 10 (mrp 11) were indistinguishable.
FIG. 1.
C. jejuni outbreak isolates clustered by using the unweighted pair group method with arithmetic averages of PFGE profiles. The PFGE profile from strain D0467 was not included due to lack of restriction by SmaI (nt).
MLST discriminated the isolates into 12 STs, with some STs associated with more than one outbreak. The epidemic strains from outbreaks 1 and 2 were both ST-22. The epidemic strain from outbreak 3, plus temporally related strains from outbreaks 3, 6, and 7 were all ST-42. The epidemic strain from outbreaks 4, 5, 6, and 9 plus temporally related strains from outbreaks 3 and 8 were all ST-21. The epidemic strain from outbreaks 7 and 11 was ST-8. The DI was 0.859 for MLST; however, if the flaA SVR results were included as an additional allele, the DI was increased to 0.924.
We discriminated the isolates into 15 subtypes by using flaA SVR sequencing. Some subtypes were unique to each outbreak set; however, some were common to more than one outbreak: SVR type 6 was the subtype identified in the epidemic strains of outbreaks 1 and 2; SVR type 4 was the epidemic subtype in the strains from outbreaks 4 and 5; SVR type 3 was the subtype of the outbreak 6 epidemic strains and also a temporally related strain from outbreak 3; SVR type 12 was identified in the epidemic strains of outbreaks 8 and 10.
Discrimination within ST by PFGE and flaA SVR sequencing.
Some of the isolates indistinguishable by MLST were further discriminated by using PFGE or flaA SVR sequencing (Table 4). Four STs, ST-8, -21, -22, and -42, were separated into two or more PFGE types and into between two and four types by flaA SVR sequencing. The differences between the SVR sequences within each of the STs varied by between 1 and 24 SNPs.
TABLE 4.
Discrimination within STs by PFGE and flaA SVR sequencing
| ST | PFGE mrp | flaA SVR type | No. of isolates | No. of nucleotide differences (SNPs) between SVR types |
|---|---|---|---|---|
| ST-8 | 10 | 1 | 4 | 11 |
| 14 | 2 | 3 | ||
| ST-21 | 5 | 3 | 1 | 4, 5, or 7 |
| 6 | 4 | 5 | ||
| 8 | 3 | 6 | ||
| 8 | 5 | 1 | ||
| ST-22 | 1 | 6 | 5 | NAa |
| 3 | 6 | 3 | ||
| ST-42 | 2 | 9 | 1 | 1, 11, 23, or 24 |
| 4 | 9 | 4 | ||
| 4 | 10 | 1 | ||
| 9 | 11 | 1 |
NA, not applicable.
DISCUSSION
Serotyping with the Penner HS scheme has been successfully used to investigate outbreaks of C. jejuni disease for many years (29); however, this method has a number of limitations that restrict its use mainly to reference laboratories. Serotyping requires the production of a large panel of antisera, which is labor intensive and expensive. Furthermore, this method has relatively poor discriminatory power and suffers from problems with cross-reactivity between antigens. Approximately 20% of strains are nontypeable by this method (42). The strains in this study were previously serotyped by using the Penner serotyping scheme, and none were found to be nontypeable. The serotypes of the strains in this study included HS:1, HS:2, HS:4, HS:19, and HS:55, all common serotypes frequently found among C. jejuni strains from sporadic infections in the United States (30). Serotyping correctly distinguished sporadic isolates from outbreak isolates in 11 of the 12 outbreaks; it failed to differentiate the sporadic strain EDL2 in outbreak 3 from the epidemic strain, as did the other three subtyping methods. In this study, Penner serotyping was the least discriminatory subtyping method (DI = 0.839) for strain discrimination among all of the 47 strains from the 12 outbreaks. Previous studies have found that using Penner serotyping in conjunction with another subtyping method such as phagetyping or Lior (heat-labile) serotyping can increase the discriminatory power of subtyping (36, 45).
In this study, PFGE correctly differentiated temporally related isolates from outbreak strains in 10 of the 12 outbreaks. It did not distinguish isolate EDL2 in outbreak 3, as described above. DNA from two bird isolates in outbreak 2 did not restrict with the SmaI enzyme, and therefore, these strains could not be typed by PFGE. The inability of restriction enzymes to cut DNA from some isolates may reduce the sensitivity of PFGE among some strains (26). PFGE was the most discriminatory subtyping method in this study (DI = 0.930); however, some strains from separate outbreaks had indistinguishable PFGE profiles. The PFGE profiles reported in this study with SmaI contained between 4 and 9 bands per profile. PFGE subtyping of C. jejuni by using SmaI with profiles based upon <10 bands may limit the discriminatory power of the method. Further studies of the prevalence and diversity of C. jejuni SmaI PFGE types of isolates from sporadic human infections in the United States are required to validate the usefulness of this method. In a recent study, the restriction enzyme KpnI was found to be more discriminatory than SmaI (23). In addition, the KpnI clustering of strains related to ingestion of raw milk and specific water sources correlated better, leading the authors to conclude KpnI to be the enzyme of choice for molecular epidemiological studies of C. jejuni (23).
In our study, we used a rapid PFGE protocol that can be completed in 24 to 30 h, (33) unlike previous methods that require 4 to 5 days to complete the analysis. However, even rapid protocols are labor intensive, and PFGE requires considerable financial investment, both in analysis equipment to generate the profiles and in computer software to normalize the data for interlaboratory comparisons. PFGE profiles may not provide a stable fingerprint; chromosomal rearrangements may lead to minor or major changes in profiles, making this method unsuitable for long-term or global epidemiological studies of C. jejuni (12, 41).
Sequencing the SVR of the flaA gene of C. jejuni has been useful both for investigating outbreaks of human disease (22) and for investigating the genetic diversity of C. jejuni strains within poultry operations (14). The variation in flaA indexed by SVR typing is thought be under diversifying selection, thereby making it a suitable target for short-term epidemic investigations (8). However, studies have demonstrated that flaA can undergo intra- and intergenomic recombination (13); thus, the instability in this locus may make flaA SVR typing unsuitable either for epidemic or for longitudinal epidemiological studies. In this study, flaA SVR typing correctly differentiated sporadic isolates from outbreak isolates in 10 of the 12 outbreaks. In outbreak 9, one of the outbreak strains was distinguished from the others by flaA SVR typing. The isolate had several nucleotide substitutions distributed throughout a 150-nucleotide region of the SVR, suggesting that the change was due to recombination rather than spontaneous mutation. Thus, this outbreak may be an example of why flaA typing alone may not be adequate for epidemiologic investigation of outbreaks.
MLST indexes the variation in housekeeping genes, which accumulates slowly since the genes are under stabilizing selection to conserve metabolic function. This method has been found to be very useful for investigating the genetic diversity of C. jejuni and for estimating the role of recombination in shaping its population structure (6, 8, 38); however, its utility for short-term epidemiological or outbreak investigations has not been established. In a recent study, 12 isolates of C. jejuni representing three outbreaks were typed using amplified fragment length polymorphism, MLST, and clustered regularly interspersed short palindromic repeat typing (37). All three methods were reported to be equally useful for identifying strains from outbreaks; however, the epidemic strains from each outbreak possessed very common STs (ST-21, -45, and -53), suggesting that recognition of outbreaks caused by strains with common STs may be problematic. In this study, MLST was as discriminatory as PFGE for distinguishing temporally related isolates from the epidemic strain in all of the outbreaks. However, MLST was not as discriminatory as PFGE for strain discrimination between outbreaks (DI = 0.859). Incorporating the flaA SVR sequencing results as an additional locus provided increased discrimination within the more common STs and raised the DI to 0.923, a DI very similar to that provided by PFGE.
Theoretically, MLST and other DNA sequencing-based approaches have the potential for high intra- and interlaboratory reproducibility. MLST, with its standardized nomenclature, the unambiguous nature of sequence data, and the potential for long-term global comparisons via a centralized database represents a significant advantage over PFGE and other gel-based methods. However, the method in its current form is rather unwieldy and labor intensive, requiring the sequencing of >3,000 bp per isolate. Generation of an ST requires the combination of 14 separate sequencing reactions or experiments; therefore, sequencing errors may lead to the generation of incorrect data. A single nucleotide sequencing error can lead to the designation of a new allele and ST; therefore, great care must be exercised when setting up reactions and loading samples on sequencing gels. Sequencing the allele in both directions can reduce the likelihood of such errors, and strict controls must be maintained to allow potential new alleles to be deposited in global databases only after the original sequencing results are reconfirmed and the original sequencing traces are sent to the database curator.
The results from this study demonstrate that a combined approach of MLST plus sequencing of the flaA SVR provides a level of discrimination equivalent to that of PFGE for outbreak investigations. Determining the ST of strains will also facilitate global comparisons via the centralized database, providing long-term epidemiological data for future comparisons. Although PFGE may be less laborious and costly when compared to MLST, reductions in cost and improvements in sequencing technology, including further automation of the technique, will make MLST more accessible for routine clinical microbiology laboratories in the future. Incorporation of additional variable loci such as the flaA SVR will provide a level of discrimination suitable for outbreak investigations while providing data suitable for longitudinal comparisons. Further application of DNA sequencing-based subtyping methods to the surveillance of C. jejuni infection will lead to significant improvements in our understanding of the epidemiology of this important pathogen.
Acknowledgments
We thank Mabel Ann Nicholson, National Campylobacter and Helicobacter Reference Laboratory, Centers for Disease Control and Prevention, for serotyping the isolates.
The use of trade names is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention or by the U.S. Department of Health and Human Services.
REFERENCES
- 1.Birkhead, G., R. L. Vogt, E. Heun, C. M. Evelti, and C. M. Patton. 1988. A multiple-strain outbreak of Campylobacter enteritis due to consumption of inadequately pasteurized milk. J. Infect. Dis. 157:1095-1097. [DOI] [PubMed] [Google Scholar]
- 2.Blaser, M. J., D. N. Taylor, and R. A. Feldman. 1983. Epidemiology of Campylobacter jejuni infections. Epidemiol. Rev. 5:157-176. [DOI] [PubMed] [Google Scholar]
- 3.Bolton, F. J., D. Coates, D. N. Hutchinson, and A. F. Godfree. 1987. A study of thermophilic campylobacters in a river system. J. Appl. Bacteriol. 62:167-176. [DOI] [PubMed] [Google Scholar]
- 4.Bolton, F. J., D. R. A. Wareing, M. B. Skirrow, and D. N. Hutchinson. 1992. Identification and biotyping of campylobacters, p. 151-162. In R. G. Board, D. Jones, and F. A. Skinner (ed.), Identification methods in applied and environmental microbiology. Blackwell Scientific Publications, Oxford, United Kingdom.
- 5.Desai, M., J. M. Logan, J. A. Frost, and J. Stanley. 2001. Genome sequence-based fluorescent amplified fragment length polymorphism of Campylobacter jejuni, its relationship to serotyping, and its implications for epidemiological analysis. J. Clin. Microbiol. 39:3823-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. J. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dingle, K. E., N. Van Den Braak, F. M. Colles, L. J. Price, D. L. Woodward, F. G. Rodgers, H. P. Endtz, A. Van Belkum, and M. C. Maiden. 2001. Sequence typing confirms that Campylobacter jejuni strains associated with Guillain-Barré and Miller-Fisher syndromes are of diverse genetic lineage, serotype, and flagella type. J. Clin. Microbiol. 39:3346-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. A. Wareing, and M. C. J. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feavers, I. M., S. J. Gray, R. Urwin, J. E. Russell, J. A. Bygraves, E. B. Kaczmarski, and M. C. Maiden. 1999. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J. Clin. Microbiol. 37:3883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald, C., R. J. Owen, and J. Stanley. 1996. Comprehensive ribotyping scheme for heat-stable serotypes of Campylobacter jejuni. J. Clin. Microbiol. 34:265-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgerald, C., L. O. Helsel, M. A. Nicholson, S. J. Olsen, D. L. Swerdlow, R. Flahart, J. Sexton, and P. I. Fields. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanninen, M. L., M. Hakkinen, and H. Rautelin. 1999. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrington, C. S., F. M. Thomson-Carter, and P. E. Carter. 1997. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J. Clin. Microbiol. 35:2386-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiett, K. L., N. J. Stern, P. Fedorka-Cray, N. A. Cox, M. T. Musgrove, and S. Ladely. 2002. Molecular subtype analyses of Campylobacter spp. from Arkansas and California poultry operations. Appl. Environ. Microbiol. 68:6220-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson, P. J., R. L. Vogt, J. Brondum, and C. M. Patton. 1984. Isolation of Campylobacter jejuni from milk during an outbreak of campylobacteriosis. J. Infect. Dis. 150:789. [DOI] [PubMed] [Google Scholar]
- 16.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchinson, D. N., F. J. Bolton, P. M. Hinchliffe, H. C. Dawkins, S. D. Horsley, P. A. Robertshaw, and D. E. Counter. 1985. Evidence of udder excretion of Campylobacter jejuni as the cause of milkborne Campylobacter outbreak. J. Hyg. 94:205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornblatt, A. N., T. Barrett, G. K. Morris, and F. E. Tosh. 1985. Epidemiologic and laboratory investigation of an outbreak of Campylobacter enteritis associated with raw milk. Am. J. Epidemiol. 122:884-889. [DOI] [PubMed] [Google Scholar]
- 19.Madden, R. H., L. Moran, and P. Scates. 1996. Sub-typing of animal and human Campylobacter spp. using RAPD. Lett. Appl. Microbiol. 23:167-170. [DOI] [PubMed] [Google Scholar]
- 20.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinersmann, R. J., L. O. Helsel, P. I. Fields, and K. L. Hiett. 1997. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35:2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaud, S., S. Menard, C. Gaudreau, and R. D. Arbeit. 2001. Comparison of SmaI-defined genotypes of Campylobacter jejuni examined by KpnI: a population-based study. J. Med. Microbiol. 50:1075-1081. [DOI] [PubMed] [Google Scholar]
- 24.Morgan, D., C. Gunneberg, D. Gunnell, T. D. Healing, S. Lamerton, N. Soltanpoor, D. A. Lewis, and D. G. White. 1994. An outbreak of Campylobacter infection associated with the consumption of unpasteurised milk at a large festival in England. Eur. J. Epidemiol. 10:581-585. [DOI] [PubMed] [Google Scholar]
- 25.Nachamkin, I., K. Bohachick, and C. M. Patton. 1993. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J. Clin. Microbiol. 31:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newell, D. G., J. A. Frost, B. Duim, J. Wagenaar, R. H. Madden, J. van der Plas, and S. L. W. On. 2000. New developments in the subtyping of Campylobacter species, p. 27-44. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 27.Olsen, S. J., G. R. Hansen, L. Bartlett, C. Fitzgerald, A. Sonder, R. Manjrekar, T. Riggs, J. Kim, R. Flahart, G. Pezzino, and D. L. Swerdlow. 2001. An outbreak of Campylobacter jejuni infections associated with food handler contamination: the use of pulsed-field gel electrophoresis. J. Infect. Dis. 183:164-167. [DOI] [PubMed] [Google Scholar]
- 28.On, S. L., E. M. Nielsen, J. Engberg, and M. Madsen. 1998. Validity of SmaI-defined genotypes of Campylobacter jejuni examined by SalI, KpnI, and BamHI polymorphisms: evidence of identical clones infecting humans, poultry, and cattle. Epidemiol. Infect. 120:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patton, C. M., I. K. Wachsmuth, G. M. Evins, J. A. Kiehlbauch, B. D. Plikaytis, N. Troup, L. Tompkins, and H. Lior. 1991. Evaluation of 10 methods to distinguish epidemic-associated Campylobacter strains. J. Clin. Microbiol. 29:680-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patton, C. M., M. A. Nicholson, S. M. Ostroff, A. A. Ries, I. K. Wachsmuth, and R. V. Tauxe. 1993. Common somatic O and heat-labile serotypes among Campylobacter strains from sporadic infections in the United States. J. Clin. Microbiol. 31:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pebody, R. G., M. J. Ryan, and P. G. Wall. 1997. Outbreaks of campylobacter infection: rare events for a common pathogen. Commun. Dis. Rep. CDR Rev. 7:R33-R37. [PubMed]
- 32.Penner, J. L., and J. N. Hennessy. 1980. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J. Clin. Microbiol. 12:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roels, T. H., B. Wickus, H. H. Bostrom, J. J. Kazmierczak, M. A. Nicholson, T. Kurzynski, and J. P. Davis. 1998. A foodborne outbreak of Campylobacter jejuni (O:33) infection associated with tuna salad: a rare strain in an unusual vehicle. Epidmiol. Infect. 121:281-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sacks, J. J., S. Lieb, L. M. Baldy, S. Berta, C. M. Patton, M. C. White, W. J. Bigler, and J. J. Witte. 1986. Epidemic campylobacteriosis associated with a community water supply. Am. J. Public Health 76:424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salama, S. M., F. J. Bolton, and D. N. Hutchinson. 1990. Application of a new phagetyping scheme to campylobacters isolated during outbreaks. Epidemiol. Infect. 104:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schouls, L. M., S. Reulen, B. Duim, J. A. Wagenaar, R. J. Willems, K. E. Dingle, F. M. Colles, and J. D. Van Embden. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swaminathan, B., T. J. Barrett, S. B. Hunter, R. V. Tauxe, and The CDC PulseNet Task Force. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogt, R. L., A. A. Little, C. M. Patton, T. J. Barrett, and L. A. Orciari. 1984. Serotyping and serology studies of campylobacteriosis associated with consumption of raw milk. J. Clin. Microbiol. 20:998-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wassenaar, T. M., B. Geilhausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterman, S. C., R. W. Park, and A. J. Bramley. 1984. A search for the source of Campylobacter jejuni in milk. J. Hyg. 93:333-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood, R. C., K. L. MacDonald, and M. T. Osterholm. 1992. Campylobacter enteritis outbreaks associated with drinking raw milk during youth activities. A 10-year review of outbreaks in the United States. JAMA 268:3228-3230. [PubMed] [Google Scholar]
- 45.Woodward, D. L., and F. G. Rodgers. 2002. Identification of Campylobacter heat-stable and heat-labile antigens by combining the Penner and Lior serotyping schemes. J. Clin. Microbiol. 40:741-745. [DOI] [PMC free article] [PubMed] [Google Scholar]