Abstract
The sterol regulatory element–binding protein-2 (SREBP-2) is produced as a large precursor molecule attached to the endoplasmic reticulum membrane. In response to the sterol depletion, the N-terminal segment of the precursor, which contains a basic helix-loop-helix–leucine zipper domain, is released by two sequential cleavages and is translocated to the nucleus, where it activates the transcription of target genes. The data herein show that released SREBP-2 uses a distinct nuclear transport pathway, which is mediated by importin β. The mature form of SREBP-2 is actively transported into the nucleus when injected into the cell cytoplasm. SREBP-2 binds directly to importin β in the absence of importin α. Ran-GTP but not Ran-GDP causes the dissociation of the SREBP-2–importin β complex. G19VRan-GTP inhibits the nuclear import of SREBP-2 in living cells. In the permeabilized cell in vitro transport system, nuclear import of SREBP-2 is reconstituted only by importin β in conjunction with Ran and its interacting protein p10/NTF2. We further demonstrate that the helix-loop-helix–leucine zipper motif of SREBP-2 contains a novel type of nuclear localization signal, which binds directly to importin β.
INTRODUCTION
Eukaryotic cells can be subdivided into various membrane-bound compartments, each of which provides an optimal environment for specific biochemical reactions. As a result, the specialized systems have evolved, which permit the transport of macromolecules from one compartment to another. Because the nucleus is the central apparatus that coordinates all cellular activity, via gene expression, DNA replication, and ribosome assembly, proteins that are involved in these nuclear events must be selectively transported into the nucleus. At the same time, tRNAs and mRNAs are synthesized in the nucleus and are subsequently exported to the cytoplasm. The nuclear pore complex (NPC), which provides the gateway for this nucleocytoplasmic traffic, has recently been reviewed (Davis, 1995; Fabre and Hurt, 1997). Small molecules up to ∼9 nm diameter, which corresponds to a globular protein of ∼60 kDa, are able to pass through the aqueous pore by passive diffusion, whereas larger molecules are selectively transported via an energy- and signal-dependent mechanism. Proteins that are actively transported between nucleus and cytoplasm have specific signals for import, termed nuclear localization signals (NLSs), or for export, termed nuclear export signals. Previous studies have concluded that multiple transport pathways specified by distinct signals exist in cells, and this area has been reviewed by several groups (Corbett and Silver, 1997; Nigg, 1997; Yoneda, 1997; Mattaj and Englmeier, 1998; Ohno et al., 1998). Among the enormous amount of nucleocytoplasmic traffic, the nuclear import pathway, which depends on a classical NLS that consists of one or two clusters of basic amino acids, is best characterized.
Nuclear import of classical NLS-containing proteins is initiated by binding to an importin α/β heterodimer (Imamoto et al., 1995c). Importin α directly interacts with the NLS as well as with importin β, and the resultant heterotrimer docks at the nuclear pore through the interaction of importin β with the NPC components (Adam and Adam, 1994; Görlich et al., 1994, 1995; Chi et al., 1995; Imamoto et al., 1995a,b; Moroianu et al., 1995; Radu et al., 1995; Weis et al., 1995). Subsequently, the docked complex is translocated through the NPC, via a pathway that is dependent of a small GTPase Ran and several Ran-interacting proteins (Melchior et al., 1993; Moore and Blobel, 1993, 1994; Paschal and Gerace, 1995; Paschal et al., 1996; Nehrbass and Blobel, 1996). The import reaction is terminated at the nucleoplasmic side of the NPC, where the binding of Ran-GTP to importin β causes the dissociation of the importin heterodimer (Rexach and Blobel, 1995; Chi et al., 1996; Görlich et al., 1996). Thus, importin β acts as a receptor molecule for targeting to NPC and NPC translocation, whereas importin α acts as an adapter for karyophiles. Ran represents a key regulator of nucleocytoplasmic transport.
A number of studies have demonstrated that several other importin β-related proteins function as import or export receptors for distinct cargoes (for reviews see Pemberton et al., 1998; Wozniak et al., 1998). Aside from differences in their cargo specificity, they share the following two properties: 1) binding to the NPC components and 2) binding to Ran-GTP (Görlich et al., 1997). Transport receptors are able to shuttle between the nucleus and the cytoplasm via direct interaction with NPC components (Kose et al., 1997, 1999; Kutay et al., 1998). Ran controls the assembly and disassembly of transport complexes. The binding of Ran-GTP to the import receptors releases their cargoes or adapter molecules (Chi et al., 1996; Izaurralde et al., 1997). In contrast, binding of Ran-GTP to the export receptors stabilizes the complex, along with their export cargoes (Fornerod et al., 1997; Kutay et al., 1997, 1998; Arts et al., 1998). Because the only known nucleotide exchange factor for Ran, RCC1, is located in the nucleus (Ohtsubo et al., 1989; Bischoff and Ponstingl, 1991), and the only known Ran-GTPase–activating protein, RanGAP1, is exclusively cytoplasmic (Bischoff et al., 1995; Matunis et al., 1996; Mahajan et al., 1997), it would be predicted that nuclear Ran is predominantly the GTP-bound form and that cytoplasmic Ran is mainly the GDP-bound form. Therefore, a low level of Ran-GTP in the cytoplasm allows the import receptors to form a complex, whereas a high level of Ran-GTP in the nucleus favors the dissociation of the import complex. In contrast, the export complexes are formed in the nucleus and dissociate in the cytoplasm. Thus, nuclear import or export, as mediated by importin β-family receptors, is believed to depend on a steep Ran-GTP gradient across the nuclear envelope (Izaurralde et al., 1997).
Among the currently identified importin β-family members, only importin β uses an adapter, whereas the others directly bind to their cargoes. However, recent reports have suggested that importin β is capable of binding directly to some nuclear proteins and to mediate their import. The ability of importin β to function without an adapter was first demonstrated for a fusion protein containing the importin β binding domain of importin α (IBB domain) (Görlich et al., 1996; Weis et al., 1996). It has been also shown that the yeast mRNA binding protein Nab2p interacts directly with human importin β and is imported into the nucleus of human cells, whereas the yeast homologue of importin β (Kap95p) is not able to mediate the import in human cells (Truant et al., 1998). In another example, Jäkel and Görlich (1998) reported that ribosomal proteins are directly imported by at least four importin β-family import receptors, namely importin β, transportin, RanBP5, and RanBP7. Interestingly, the importin β binding domains of these substrates share no obvious sequence similarities. This raises questions about how diverse substrates are imported by importin β, and what is the underlying mechanism by which a single receptor is able to recognize and carry distinct cargoes.
Import into the nucleus occurs not only as a continuous flux but also as temporally controlled events. For many signal transduction pathways, specific proteins have been identified, which are translocated to the nucleus in response to particular signals (Vandromme et al., 1996). Nuclear import of these signaling molecules appears to be controlled in two ways, with one involving the masking and unmasking of its NLS. Masking is ordinarily regulated by phosphorylation. For example, a transcription factor, NF-κB, is transported into the nucleus after its cytoplasmic masking protein, IkB, is phosphorylated and then undergoes degradation (Beg et al., 1992). The nuclear accumulation of the NF-AT family of proteins is triggered by the dephosphorylation of critical serine residues, allowing the two basic NLSs to be exposed on the molecular surface (Shibasaki et al., 1996; Beals et al., 1997). STAT1 is a transcription factor, which is translocated from the cytoplasm to the nucleus when cells are stimulated by interferon-γ. Interferon-γ stimulation leads to the tyrosine phosphorylation of STAT1, which enables it to form a nuclear pore-targeting complex with NPI-1 (a family of importin α) and importin β (Sekimoto et al., 1996, 1997).
The other manner in which nuclear transport is regulated is based on an anchoring–releasing mechanism. Membranous organelles as well as the plasma membrane are involved in the cytoplasmic anchoring of certain signaling molecules. One extreme example of this type of regulation is provided by a transcription factor, referred to as the sterol regulatory element–binding protein (SREBP) (for review, see Brown and Goldstein, 1997). Three SREBPs are known to exist in animal cells. Two of these, designated SREBP-1a and -1c, are synthesized from a single gene through the use of alternate promoters and first exons, and SREBP-2 is synthesized from a different gene (Hua et al., 1993; Tontonoz et al., 1993; Yokoyama et al., 1993). Each SREBP is synthesized as a large precursor molecule of ∼1150 amino acids, which consists of three domains. The N-terminal domain (∼480 amino acids) contains a basic-helix-loop-helix–leucine zipper (bHLH-Zip) motif, which is followed by a membrane attachment domain of ∼80 amino acids with two transmembrane segments, and a C-terminal regulatory domain of ∼590 amino acids. The precursor SREBPs are attached to the endoplasmic reticulum membrane and the outer nuclear envelope in a hairpin manner with their N- and C-terminal domains projecting into the cytoplasm. The middle attachment domain projects into the endoplasmic reticulum lumen. When the cholesterol content of cells is reduced, the N-terminal domain of SREBP is released from the membranes by sequential proteolytic cleavages at two sites, designated Site-1 and Site-2 (Rawson et al., 1997; Sakai et al., 1998). The cleaved N-terminal fragment, referred to as the mature form of SREBP, travels to the nucleus, where it activates the transcription of genes involved in cholesterol and fatty acid metabolism. When cells accumulate cholesterol, the activity of the Site-1 protease is reduced, and the SREBP remains bound to the membranes. As a result, the transcription of the target genes is decreased. This regulation assures a steady supply of cholesterol and fatty acids by preventing their overaccumulation. It has been reported that when the cDNAs that terminate before the first transmembrane domain of SREBPs are transfected into cells, the SREBPs constitutively accumulate in the nuclei independently of the intracellular sterol content (Sato et al., 1994; Wang et al., 1994; Yang et al., 1994, 1995). Therefore, it is probable that nuclear import of the mature form occurs via a sterol-independent mechanism. However, SREBPs possess no consensus sequence with previously identified NLSs, and significantly less is known about the overall nuclear import mechanism.
This paper reports a study of the molecular mechanism of the nuclear import of the mature form SREBP-2. A variety of in vivo and in vitro experiments show that importin β interacts directly with SREBP-2 and mediates import in a Ran-dependent manner. In addition, we show that the HLH-Zip motif of the SREBP-2 contains a novel type of NLS, which directly binds to importin β.
MATERIALS AND METHODS
Construction of Plasmids
The XhoI–NotI fragment encoding an active form of human SREBP-2 (amino acids 1–481) was obtained from pSREBP2(1–481) (Sato et al., 1996) and subcloned into the SalI–NotI sites of pGEX-6P-3 (Pharmacia, Piscataway, NJ). A FLAG tag with BglII and BamHI sites at the ends was generated by annealing two synthetic oligonucleotides (5′-GATCTGACTACAAGGACGACGATGACAAGG-3′ and 5′-GATCCCTTGTCATCGTCGTCCTTGTAGTCA-3′) and inserting them into the BamHI site of the above construct. The resultant construct is referred to as pGEX FL-SREBP2. To generate a His-tagged SREBP-2(1–481) expression vector, pRSETA-SREBP2, the BamHI–NotI fragment from the pGEX FL-SREBP2 was inserted into the BamHI–PvuII sites of pRSETA (Invitrogen, San Diego, CA) after blunting the NotI site. To construct the expression vector for the His-tagged SREBP-2(1–370) mutant, the fragment encoding amino acids 1–370 of SREBP-2 was amplified using specific primer pairs with BamHI and EcoRI at the ends and subcloned into the BamHI–EcoRI sites of the pRSETA. To generate the plasmids which encode for the His-tagged green fluorescent protein (GFP) chimera, the expression vector pRSETA-GFP* was engineered from the pRSETA by inserting an amplified GFP into the BamHI site and introducing NotI and EcoRV cutting sequences in the multicloning site using two oligonucleotides (5′-CGCGGCGGCAGATCTGATATCG-3′ and 5′-AATTCGATATCAGATCTGCGGCCGGGAGCT-3′). To construct pRSETA GFP-SREBP2, the expression vector encoding for the His-tagged SREBP-2 protein fused with GFP at the N terminus, the BamHI-NotI fragment from pGEX FL-SREBP2 was inserted into the same restriction sites of pRSETA-GFP*. To produce the expression vectors encoding the His-tagged GFP chimera of SREBP-2 deletion mutants, pRSETA GFP-SREBP2(1–403), pRSETA GFP-SREBP2(1–370), pRSETA GFP-SREBP2(1–317), and pRSETA GFP-SREBP2(343–403), each appropriate fragment from SREBP-2 was amplified using the specific primer pairs with BamHI and EcoRI at the ends and subcloned into the BamHI–EcoRI sites of the pRSETA-GFP*. The expression vector encoding GST-GFP-SREBP-2(343–460), pGEX GFP-SREBP2(343–460), was constructed by inserting an amplified fragment into the BamHI–EcoRI sites of the pGEX-6P-2-hGFP, which carries the S65A/Y145F humanized GFP gene at the N terminus of the multicloning site (kindly provided by Dr. S. Kuroda, Institute of Scientific and Industrial Research, Osaka University). The expression vector encoding His-tagged mouse importin α (PTAC 58), pRSETA-PTAC58, was constructed by ligating the encoding full-length m-importin α (Imamoto et al., 1995b) to pRSETA in frame.
Expression and Purification of Recombinant Proteins
To express GST-FLAG-SREBP-2, Escherichia coli strain BL21, which had been transformed with pGEX FL-SREBP2, was grown in Luria–Bertani medium containing 100 μg/ml ampicillin at 37°C to a density of 1.2 (OD550). Expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside and incubated for 14 h at 20°C. Cells were harvested by centrifugation and resuspended in high-salt buffer (50 mM Tris-HCl, pH 8.0, and 500 mM NaCl) containing 1 mM PMSF, 1 mM DTT, and protease inhibitor mixture (1 μg/ml each aprotinin, leupeptin, and pepstatin), using 1/25 vol of the original cell culture. After two freeze–thaw cycles, PMSF was again added to the cell suspension to a final concentration of 1 mM, and cells were lysed by sonication. After the extract was clarified by centrifugation, glycerol was added to the supernatant to a final concentration of 10%, and the extract was incubated with glutathione-Sepharose (Pharmacia) at 4°C. The recombinant protein-bound Sepharose was washed extensively with cleavage buffer (50 mM Tris-HCl, pH 7.0, 100 mM NaCl, and 1 mM DTT) containing protease inhibitor mixture and incubated with Prescission Protease (Pharmacia) at 5°C for 4 h. Partially purified recombinant FLAG-SREBP-2, which is cleaved from the GST moiety but associated with an E. coli DnaK protein, was collected from the flow-through of the glutathione-Sepharose column and then desalted with a PD10 column (Pharmacia) equilibrated with transport buffer (20 mM HEPES, pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 5 mM sodium acetate, and 0.5 mM EGTA) containing 2 mM DTT and protease inhibitor mixture, followed by concentration by ultrafiltration using Centricon 30 (Amicon, Beverly, MA).
Further purification was performed as follows. The flow-through was dialyzed against 20 mM HEPES-NaOH, pH 7.5, 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, and protease inhibitor mixture and then incubated in the presence of 2 mM ATP for 10 min at room temperature. After clarification by ultracentrifugation, the recombinant protein solution was subjected to chromatography on a Mono Q column (1 ml) on an FPLC system (Pharmacia) at a flow rate of 0.25 ml/min using a linear gradient from 0.05 to 1.0 M NaCl in 20 mM HEPES-NaOH, pH 7.5, 1 mM MgCl2, 1 mM DTT, 2 μM ATP, and protease inhibitor mixture. Peak fractions containing FLAG-SREBP-2 (eluted between 300 and 350 mM NaCl) were pooled and desalted with a PD10 column (Pharmacia) equilibrated with transport buffer containing 2 mM DTT and protease inhibitor mixture and then concentrated. Note that the partially purified FLAG-SREBP-2 had the same activity relative to nuclear import as the purified FLAG-SREBP-2 when examined in digitonin-permeabilized cell transport assays, as well as by microinjection.
GST-FLAG-SREBP-2 protein was eluted from the protein-bound glutathione beads with elution buffer (100 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 20 mM glutathione) containing 1 mM DTT and protease inhibitor mixture and purified on a Mono S column (1 ml; Pharmacia) at a flow rate of 0.5 ml/min with a linear gradient of 0.02–1.0 M KCl in 10 mM potassium phosphate, pH 6.7, 1 mM DTT, and protease inhibitor mixture. Pooled fractions containing GST-FLAG-SREBP-2 were dialyzed against transport buffer containing 2 mM DTT and protease inhibitor mixture.
GFP-SREBP-2(343–460) protein was purified from E. coli strain BL21 transformed with pGEX GFP-SREBP2(343–460) using glutathione-Sepharose in the same manner as for partially purified FLAG-SREBP-2.
His-SREBP-2(1–370) protein was expressed in E. coli strain BL21(DE3)pLysS transformed with pRSETA SREBP2(1–370) and purified by nickel nitrilo-triacetic acid agarose (Qiagen, Chatsworth, CA) affinity chromatography, followed by the chromatography on a Mono Q column in the same manner as for FLAG-SREBP-2.
Expression and purification of mouse importin β (PTAC 97) was performed as described previously (Kose et al., 1997), as were the purification of GST-mouse importin α (GST-PTAC 58), and GST-importin β (GST-PTAC 97) (Imamoto et al., 1995b). Recombinant human p10/NTF2 protein was expressed and purified as described previously (Tachibana et al., 1996). E. coli strains expressing wild-type and G19V Ran were obtained as described previously (Sekimoto et al., 1996), and recombinant wild-type and G19V Ran were expressed, purified, and charged with GDP and GTP, respectively, as described previously (Hieda et al., 1999). To generate the GST-NLS-GFP expression vector, pGST-NLS-GFP, the oligonucleotide encoding SV40 large T-antigen NLS (PKKKRKVEDP) was ligated into the BamHI–SmaI sites of pGST-GFP (Tachibana et al., 1996). GST-NLS-GFP fusion protein was expressed and purified to homogeneity using glutathione-Sepharose following the manufacturer’s recommendations. Aliquots of each recombinant protein were frozen in liquid nitrogen and stored at −80°C.
Antibodies
Rabbit anti-importin β polyclonal antibodies were prepared as described previously (Kose et al., 1997), as were rabbit anti-importin α (PTAC58, mouse Rch1) polyclonal antibodies (Sekimoto et al., 1997). A polyclonal antibody (RS004) against human SREBP-2 was produced by immunizing rabbits with a fusion protein encoding six histidines followed by amino acids 1–481 of human SREBP-2. The fusion protein constructs were cloned into a pET28(a) vector (Novagen, Madison, WI), expressed in E. coli, and purified by Ni2+-Sepharose affinity chromatography. Murine IgG1 monoclonal anti-FLAG M2 antidody was purchased from Kodak (Rochester, NY). Monoclonal anti-penta His antibody was purchased from Qiagen. Monoclonal anti-human transportin antidody was purchased from Transduction Laboratories (Lexington, KY).
Microinjection
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (Life Technologies, Gaithersburg, MD) supplemented with 5% FBS and plated on coverslips 36–48 h before use. Proteins were injected through a glass capillary into the cytoplasm of HeLa cells, which were grown on coverslips. After incubation for 30 min at 37°C or on ice, the cells were fixed with 3.7% formaldehyde in PBS. To examine the localization of injected FLAG-SREBP-2, fixed cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature, incubated with 3% skim milk in PBS for 20 min, and then incubated with 30 μg/ml monoclonal anti-FLAG M2 antibody for 1 h at room temperature. The mouse antibody was detected with Cy3-labeled goat anti-mouse IgG (Amersham, Arlington Heights, IL). To examine the localization of injected His-SREBP-2(1–370), fixed and permeabilized cells were subjected to indirect immunofluorescence using rabbit anti-human SREBP-2 antisera at 1:200 dilution and Cy3-labeled goat anti-rabbit IgG (Amersham). All samples were examined by Axiophot microscopy (Carl Zeiss, Thornwood, NY).
In Vitro Transport Assay
Transport assays were performed essentially as described previously (Adam et al., 1990; Imamoto et al., 1995c). Briefly, HeLa cells were plated at a density of 1 × 105 cells/ml on an eight-well multitest slide (6040805; ICN, Costa Mesa, CA) 36–48 h before use. The cells grown on slides were rinsed twice in an ice-cold transport buffer and permeabilized for 5 min in ice-cold transport buffer containing 40 μg/ml digitonin (nacalai tesque, 123-50; diluted from a 20 mg/ml stock solution in DMSO), 2 mM DTT, and protease inhibitor mixture. After removing the digitonin-containing buffer, the slides were washed twice and immersed in an ice-cold transport buffer containing 2 mM DTT and protease inhibitor mixture for 5 min. The slides were then blotted to remove excess buffer, and 10 μl of reaction mixture per single well were applied to the cells. Import reactions were performed by incubating the slides for 20 min at 30°C unless otherwise indicated. After incubation, the cells were rinsed twice in transport buffer and fixed with 3.7% formaldehyde in transport buffer for 15 min at room temperature. For wheat germ agglutinin (WGA) treatment, permeabilized cells were incubated with 0.5 mg/ml WGA (E.Y. Laboratories, San Mateo, CA) in transport buffer containing 2 mM DTT and protease inhibitor mixture for 5 min on ice before the import reaction. All reaction mixtures contained 2% BSA, 2 mM DTT, and protease inhibitor mixture in transport buffer. As described in the respective figure legends, each reaction mixture contained an import substrate combined with cytosol or a combination of recombinant transport factors at the indicated amounts in the presence or absence of a ATP regeneration system (1 mM ATP, 5 mM creatine phosphate, and 20 U/ml creatine phosphokinase) and 0.5 mM GTP. Total cytosol from Ehrlich ascites tumor cells was prepared as described previously (Imamoto et al., 1995c). For the competition experiments, biotinylated BSA, which was chemically coupled to a synthetic peptide containing the SV40 T-antigen NLS (T-bBSA) was prepared as described previously (Imamoto et al., 1995c) and added to the reaction mixtures as an unlabeled competitor. For the deprivation of ATP, 1.8 U/μl hexokinase (Toyobo, Osaka, Japan) and 5 mM glucose were added to a reaction mixture in the absence of the ATP regeneration system and GTP.
To examine the import of FLAG-SREBP-2, the fixed cells were rinsed in PBS, permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature, and subjected to indirect immunofluorescence using a monoclonal anti-FLAG M2 antibody (see Microinjection). The import of GST-NLS-GFP and GFP-SREBP-2(343–460) was detected by Axiophot microscopy after fixation.
Immunoprecipitation
Total cytosol from Ehrlich ascite tumor cells was prepared in transport buffer containing 2 mM DTT and protease inhibitor mixture as described above and preclarified by incubation with protein G-Sepharose (Pharmacia). Five-microliter aliquots of protein G-Sepharose beads were incubated with 6 μg of monoclonal anti-FLAG M2 antibody for 1 h at room temperature; the unbound antibody was then removed. The resultant anti-FLAG antibody-coupled beads were incubated on a turntable for 2 h at 4°C with 90 μl of clarified cytosol in the presence or absence of 10 μg of partially purified recombinant FLAG-SREBP-2. After incubation, the beads were pelleted and washed three times in transport buffer containing 200 mM NaCl. The bound fraction was eluted by boiling in SDS-PAGE sample buffer and analyzed by 12.5% SDS-PAGE followed by Coomassie blue staining and immunoblotting using the antibodies described in the figure legends. The protein bands were visualized by an ECL Western blotting detection kit (Amersham).
Solution Binding Assay Using Cytosolic Extract
Binding assays were performed in transport buffer containing 2 mM DTT and protease inhibitor mixture. To preclear the cytosol, purified GST and glutathione-Sepharose were incubated with the cytosol for 1 h at 4°C, and the resin was removed by centrifugation. Ten micrograms of purified GST-SREBP-2, GST-NLS-GFP, or GST were incubated with 90 μl of clarified cytosol for 30 min on ice and centrifuged at 15,000 rpm for 20 min to remove any insoluble material. The supernatants were then incubated with 10 μl of glutathione-Sepharose on a turntable for 1 h at 4°C. After incubation, the beads were washed five times with transport buffer containing 2 mM DTT, after which the bound fractions were eluted by boiling in SDS-PAGE sample buffer, separated on 10% SDS-PAGE, and then analyzed by immunoblotting using the antibodies described in the figure legends.
Solution Binding Assay Using Recombinant Proteins
His-tagged fusion proteins were expressed in E. coli strain BL21(DE3)pLysS. The cells were suspended in binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 1 mM DTT) containing 1 mM PMSF and protease inhibitor mixture and lysed. The lysates were clarified by ultracentrifugation (150,000 × g, 30 min). The expression level of the fusion protein in each lysate was examined by immunoblotting using the monoclonal anti-penta His antibody, and the lysates were appropriately diluted with binding buffer to adjust the concentration of expressed recombinant proteins. Diluted lysates were precleared by purified GST and glutathione Sepharose as described above (see Solution Binding Assay Using Cytosolic Extract). Thirty micrograms of purified GST or GST-importin β were incubated with 270 μl of clarified lysates for 30 min on ice and centrifuged at 15,000 rpm for 20 min to remove any insoluble material. The supernatants were then incubated with 15 μl of glutathione-Sepharose on a turntable for 1 h at 4°C. After incubation, the beads were washed five times with binding buffer. The bound fractions were analyzed by 10% SDS-PAGE and immunoblotting using the monoclonal anti-penta His antibody.
Immunoblotting
Proteins were separated on 10 or 12.5% SDS-PAGE and transferred electrophoretically to nitrocellulose membranes. After blocking with 3% skim milk in TBS-T (20 mM Tris-HCl, pH 7.6, 150 mM NaCl, and 0.05% Tween 20), the blots were probed with rabbit polyclonal antibodies: anti-importin β (diluted 1:1000) and anti-PTAC58 (diluted 1:5000), or mouse monoclonal antibodies: anti-transportin (diluted 1:100) and anti-penta His antibody (diluted 1:1000). The probed antibodies were detected by standard methodology using alkaline phosphatase–coupled secondary antibodies unless otherwise indicated.
RESULTS
SREBP-2 Is Actively Imported into the Nucleus by Soluble Mediator(s)
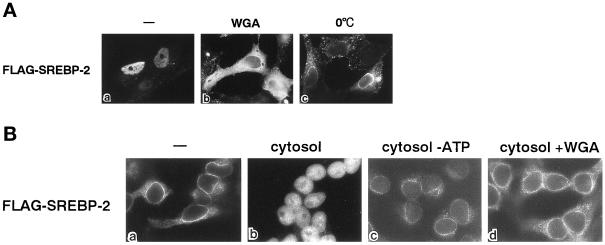
The mature form SREBP-2 is an N-terminal fragment (∼480 amino acids) with an apparent molecular mass of ∼60 kDa, which is referred to herein as SREBP-2. This molecule lacks the classical basic-type NLSs. To understand the mechanism of the nuclear import of SREBP-2, we prepared various epitope-tagged recombinant N-terminal 481-amino-acid fragments. As shown in Figure 1A, when FLAG-tagged SREBP-2 was injected into the cell cytoplasm, the SREBP-2 accumulated in the nucleus 30 min after injection. The import was found to be temperature dependent and sensitive to the coinjection of WGA (Finlay et al., 1987; Yoneda et al., 1987).
Figure 1.
(A) SREBP-2 is actively imported into the nuclei of living cells. Purified recombinant FLAG-SREBP-2 (0.5 mg/ml) was injected into the cytoplasm of HeLa cells with (b) or without (a and c) 2 mg/ml WGA. After incubation for 30 min at 37°C (a and b) or on ice (c), cells were fixed, and the localization of FLAG-SREBP-2 was detected by indirect immunofluorescence with an anti-FLAG M2 antibody. (B) Nuclear import of SREBP-2 requires soluble factors. Digitonin-permeabilized HeLa cells were incubated with 10 μl of a reaction mixture containing 4 pmol of FLAG-SREBP-2 with ATP regeneration system only (a), 4 mg/ml cytosol and an ATP regeneration system (b and d), or 4 mg/ml cytosol and 18 U of hexokinase (c). WGA pretreatment was performed by incubating the cells with 0.5 mg/ml WGA for 5 min on ice before incubating with reaction mixture (d). Import reactions were performed for 20 min at 30°C, and FLAG-SREBP-2 was detected by indirect immunofluorescence.
Next, to examine factor requirements for the import, we subjected the FLAG-tagged SREBP-2 to an in vitro cell-free transport assay. Nuclear accumulation of FLAG-SREBP-2 was reconstituted in the presence of cytosol and an ATP regenerating system and inhibited by WGA and energy depletion (Figure 1B). Note that sterol depletion had no effect on either the in vivo or in vitro assays (our unpublished results). These results indicate that the nuclear import of SREBP-2 is an active process, which is mediated by soluble factor(s), and that this import is not dependent on cellular sterol levels, which is consistent with the previous findings obtained by transfection experiments (Sato et al., 1994; Wang et al., 1994; Yang et al., 1994, 1995).
The Nuclear Import of SREBP-2 Is Not Identical to the Basic NLS-mediated Import
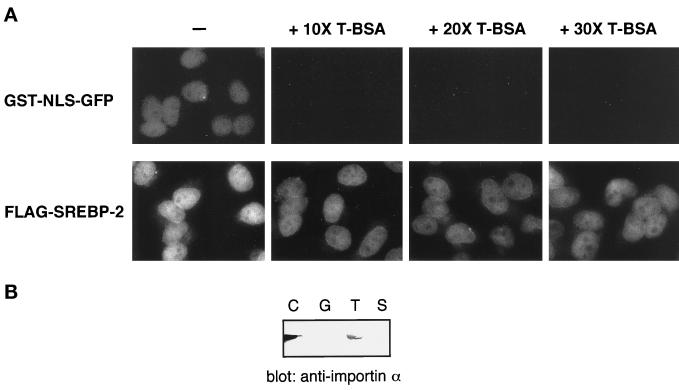
To characterize the nuclear transport pathway of SREBP-2, we tested whether the excess amount of the basic-type NLS conjugates are able to compete with the SREBP-2 import in vitro. We used a chimeric protein consisting of GST fused with SV40 T antigen NLS and GFP (GST-NLS-GFP) as a typical basic-type NLS-bearing transport substrate. BSA conjugated with peptides containing the NLS of SV40 T antigen (T-BSA) was used as an unlabeled competitor. As expected, the import of GST-NLS-GFP was completely inhibited in the presence of a 10-fold excess of unlabeled competitor. In contrast, the nuclear import of FLAG-SREBP-2 was only slightly inhibited even in the presence of a 30-fold excess of T-BSA (Figure 2A). This result suggests that the nuclear import machinery of SREBP-2 is not identical to that of the basic NLS-mediated import. However, the possibility remains that both SREBP-2 and basic NLS-bearing proteins are imported by the importin α/β heterodimer but independently bind to different sites of importin α.
Figure 2.
(A) Basic NLS-containing substrate cannot compete with the nuclear import of SREBP-2 in permeabilized cells. Digitonin-permeabilized cells were incubated with 10 μl of a reaction mixture containing 4 pmol of GST-NLS-GFP or FLAG-SREBP-2 in the absence (−) or presence of 40 pmol (+ 10× T-BSA), 80 pmol (+ 20× T-BSA), or 120 pmol (+ 30× T-BSA) of T-bBSA. All reaction mixtures contained 4 mg/ml cytosol and an ATP regeneration system. After incubation for 20 min at 30°C, the cells were fixed, and the localization of GST-NLS-GFP and FLAG-SREBP-2 was examined by fluorescent microscopy. All photographs for the same substrate were taken with the same exposure time, and the images were obtained by scanning photographic negatives using Adobe (Mountain View, CA) Photoshop version 5.0 under the same conditions. (B) SREBP-2 does not interact with importin α. Cytosol (90 μl) was incubated with GST (G), GST-NLS-GFP (T), or GST-SREBP-2 (S). GST fusion proteins were then captured on glutachione-Sepharose beads. The beads were then washed, and the bound proteins were eluted by boiling in SDS sample buffer. Eluates were separated on 10% SDS-PAGE and analyzed by immunoblotting using anti-mouse importin α polyclonal antibodies. Cytosol (2 μl) was loaded directly onto SDS-PAGE (C).
To exclude the possibility, we examined whether SREBP-2 is capable of interacting with importin α in the cytosol using immobilized recombinant GST-SREBP-2 followed by immunoblotting with antibodies that are specific for importin α family members. GST-NLS-GFP was used as a positive control. As shown in Figure 2B, no interaction of GST-SREBP-2 with the Rch1 family of importin α was observed. Neither of the two other families of importin α, NPI-1 and Qip1, interacted with SREBP-2 (our unpublished results). These collective findings indicate that the nuclear import of SREBP-2 occurs in a manner that is independent of importin α.
SREBP-2 Directly Binds to Importin β
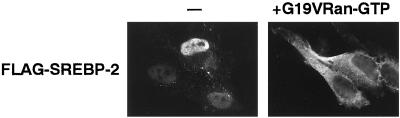
We next studied the involvement of small GTPase Ran in the nuclear import of SREBP-2 by using a G19VRan mutant, which is deficient in GTPase activity and remains in the GTP-bound state, even in the presence of cytoplasmic RanGAP1 (Carey et al., 1996). Because the GTP-bound Ran triggers the release of cargoes from the importin β family import factors, the addition of G19VRan-GTP has been shown to block several nuclear import pathways that are mediated by the importin β family import factors (Sekimoto et al., 1996; Kose et al., 1997). By coinjecting G19VRan-GTP into the cell cytoplasm, the nuclear import of SREBP-2 was strongly inhibited (Figure 3). This predicts that the nuclear import of SREBP-2 would be mediated by the importin β family molecule and dependent on the Ran GTPase cycle.
Figure 3.
Nuclear import of SREBP-2 is inhibited by G19VRan-GTP in living cells. FLAG-SREBP-2 (0.3 mg/ml) was injected into the cytoplasm of HeLa cells with (+G19VRan-GTP) or without (−) 3 mg/ml G19VRan-GTP. After incubation for 30 min at 37°C, cells were fixed, and the localization of FLAG-SREBP-2 was examined as in Figure 1A.
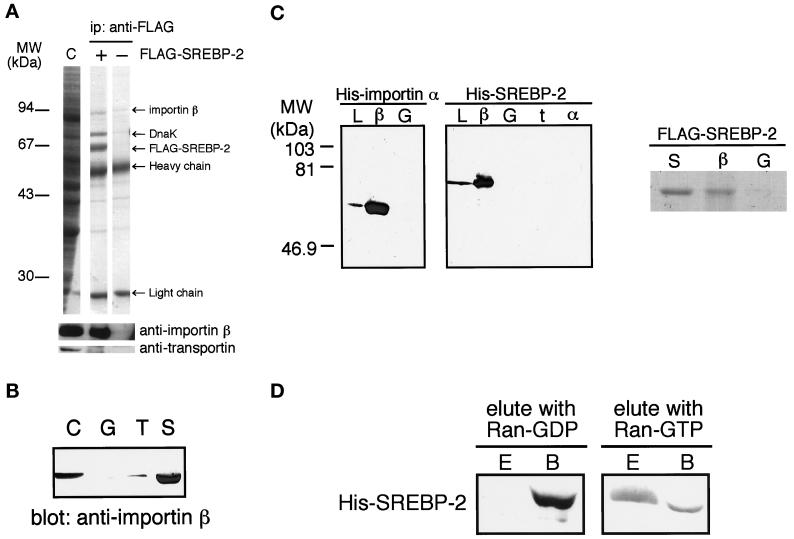
In this regard, we attempted to examine the issue of whether the importin β family transport factors interact with SREBP-2. As shown in Figure 4A, one major protein of ∼94 kDa was coprecipitated with FLAG-SREBP-2 from the Ehrlich ascites tumor cell cytosol by the anti-FLAG antibody. Immunoblotting in conjunction with importin β-specific antibodies showed that the coimmunoprecipitated fraction actually contained importin β. No significant interaction of transportin with SREBP-2 was detected, as evidenced by the transportin-specific antibody. By using the solution binding assay with GST-SREBP-2 as in Figure 2B, we were able to confirm that SREBP-2 efficiently interacts with importin β (Figure 4B).
Figure 4.
(A) Importin β is coprecipitated with FLAG-SREBP-2 from cytosol. Cytosol was incubated with (+) or without (−) recombinant partially purified FLAG-SREBP-2 and precipitated with an anti-FLAG M2 antibody. Coprecipitated proteins were separated on 12.5% SDS-PAGE and analyzed by Coomassie blue staining followed by immunoblotting using polyclonal anti-importin β antibodies and monoclonal anti-transportin antibody. Importin β (∼94 kDa) was coprecipitated by an anti-FLAG antibody with FLAG-SREBP-2 (+) but not by antibody alone (−). Note that although E. coli protein DnaK was contaminated with partially purified FLAG-SREBP-2, we verified that it does not affect the results of immunoprecipitation by using purified FLAG-SREBP-2 (our unpublished results). (B) Importin β is pulled down by GST-SREBP-2 from cytosol. Cytosol (90 μl) was pulled down with GST (G), GST-NLS-GFP (T), or GST-SREBP-2 (S) and analyzed as in Figure 2B except for using polyclonal anti-importin β antibodies. Cytosol (2 μl) was loaded directly onto SDS-PAGE (C). (C) SREBP-2 binds directly to importin β. Left, E. coli lysate expressing His-SREBP-2(1–481) or His-importin α was incubated with GST (G), GST-importin β (β), GST-transportin (t), or GST-importin α (α), and the GST-fusion proteins were captured on glutathione-Sepharose beads. The beads were then washed, and the bound proteins were analyzed by 12.5% SDS-PAGE followed by immunoblotting using anti-penta His antibody. E. coli lysate expressing each His-tagged protein was directly loaded onto the gel (L). Right, purified FLAG-SREBP-2 (5 μg) was incubated with 8 μg of GST-importin β (β) or GST (G) in transport buffer for 1.5 h at 4°C, and the GST fusion proteins were then captured on glutathione-Sepharose beads. The beads were then washed with transport buffer, and the bound proteins were eluted by boiling in SDS sample buffer. Half of the eluted proteins were separated on 10% SDS-PAGE and visualized by Coomassie blue staining. Purified FLAG-SREBP-2 (1 μg) was directly loaded onto the gel (S). (D) SREBP-2 is released from importin β by Ran-GTP but not by Ran-GDP. His-SREBP-2(1–481) was bound to GST-importin β and absorbed on glutathione beads as in C. After extensive washing, the beads were incubated with the transport buffer containing either 10 μM Ran-GDP or G19VRan-GTP for 30 min at 4°C and pelleted by centrifugation. Eluates (E) and bound fractions (B) were separated on 10% SDS-PAGE and analyzed by immunoblotting using anti-penta His antibody.
The finding that SREBP-2 interacts with importin β but not with importin α raises two possibilities: first, that SREBP-2 binds directly to importin β, and second, that SREBP-2 requires an adapter molecule other than importin α to form a complex with importin β. To address these issues, we tested whether recombinant GST-importin β is able to bind directly to His-tagged recombinant SREBP-2. The E. coli lysate, which contained a His-tagged importin α, was used as a positive control material. Each lysate was incubated with immobilized GST-importin β, and the bound proteins were analyzed by immunoblotting with anti-His tag antibody. The results clearly show that His-SREBP-2 is able to bind directly to importin β, and that His-importin α binds to GST-importin β (Figure 4C, left panel). By using the purified recombinant FLAG-SREBP-2 and GST-importin β, we confirmed that SREBP-2 directly binds to importin β independently of an adapter protein (Figure 4C, right panel). Moreover, as shown in Figure 4C, left panel, it was confirmed that neither importin α nor transportin binds to SREBP-2.
Importin β Mediates the Nuclear Import of SREBP-2 in a Ran-dependent Manner
To test whether the GTP-bound state of Ran causes the dissociation of the SREBP-2–importin β complex, immobilized GST-importin β, which had been prebound to His-SREBP-2, was incubated with buffer containing Ran-GDP or Ran-GTP (G19VRan-GTP), and the eluate and bound fractions were examined by immunoblotting using anti-His tag antibody. As shown in Figure 4D, SREBP-2 was released after incubation with Ran-GTP but not with Ran-GDP.
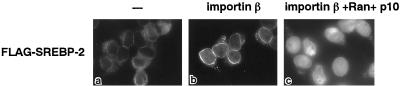
We examined the issue of whether importin β mediates the nuclear import of SREBP-2 by using the in vitro transport assay. As shown in Figure 5, in the presence of importin β alone, FLAG-SREBP-2 was targeted to the nuclear rim of the permeabilized cells. With the further addition of Ran and p10/NTF2, the efficient nuclear accumulation was completely reconstituted. These results clearly indicate that the nuclear import of SREBP-2 is mediated by importin β in conjunction with Ran and p10/NTF2.
Figure 5.
Nuclear import of SREBP-2 is reconstituted in the presence of importin β, Ran, and p10/NTF2 in permeabilized cells. Digitonin-permeabilized HeLa cells were incubated with 10 μl of reaction mixture containing 4 pmol of FLAG-SREBP-2 in the absence (a) or presence of recombinant transport factors: (b) 4 pmol of importin β alone; (c) 4 pmol of importin β, 4 pmol of Ran, and 3 pmol of p10/NTF2. All reactions contained an ATP regeneration system and 0.5 mM GTP and were performed as in Figure 1B.
An HLH-Zip Domain Is Required for the Nuclear Import of SREBP-2
To determine the regions of SREBP-2 that are required for the binding to importin β, we constructed the subsets of SREBP-2 deletion mutants (shown in Figure 6A) and performed binding assays using E. coli lysate expressing each SREBP-2 deletion mutant and GST-importin β as described in Figure 4C. As shown in Figure 6B, residues 1–403 of SREBP-2 bound as efficiently as the full-length protein (1–481). Further C-terminal deletion of 33 amino acids (1–370) severely abolished binding activity. The N-terminal deletion of 342 residues (343–403) did not reduce but, rather, increased the binding activity. These results indicate that residues 371–403 of SREBP-2 are necessary and, at most, residues 343–403 are sufficient for binding to importin β.
Figure 6.
Mapping of the importin β binding domain of SREBP-2. (A) Deletion mutants of SREBP-2 used in this study. All mutants were expressed as His-tagged GFP fusion protein. (B) Binding activity of SREBP-2 deletion mutants to importin β. E. coli lysate expressing each His-GFP-SREBP-2 deletion mutant was incubated with GST (G) or GST-importin β (β), and proteins bound to glutathione beads were analyzed by an anti-penta His antibody as in Figure 4C. E. coli lysate (L) expressing the indicated deletion mutant was directly loaded onto the gel.
The issue of whether the importin β binding domain (343–403) confers import was examined by means of in vivo and in vitro assays. For this, we produced GFP fused with SREBP-2(343–460) mutant protein instead of the (343–403) mutant, because GFP-SREBP-2(343–403) protein is sufficiently small (30 kDa) to diffuse through the NPC. When injected into the cytoplasm, purified GFP-SREBP-2(343–460) accumulated efficiently in the nucleus (Figure 7A). The import was completely blocked when WGA was coinjected (our unpublished results). In the permeabilized cells, GFP-SREBP-2(343–460) docked at the nuclear rim as the result of the exogenous addition of importin β alone and was imported into the nucleus with further addition of Ran and p10/NTF2 (Figure 7B). These results indicate that GFP-SREBP-2(343–460) was sufficient to reproduce the nuclear import of the mature form SREBP-2(1–481) in vivo and in vitro. On the other hand, recombinant SREBP-2(1–370) was not imported into the nucleus (Figure 7A). Collectively, these results suggest that the minimum domain that is sufficient for SREBP-2 import lies within residues 343–460, probably residues 343–403 of SREBP-2, which contains an HLH-Zip motif but not the preceding basic domain, which consists of the cluster of basic amino acids (see Figure 6A and DISCUSSION). This is the first evidence that the HLH-Zip region functions as an NLS.
Figure 7.
The importin β binding domain acts as an NLS. (A) SREBP-2 (343–460) is sufficient for nuclear import in living cells. Purified recombinant His-SREBP-2(1–370) (a) or GFP-SREBP-2(343–460) (b) was injected into the cytoplasm of HeLa cells. After incubating for 30 min at 37°C, the cells were fixed, and the localization of each protein was examined by indirect immunofluorescence using anti-human SREBP-2 antibodies (a) or directly by fluorescent microscopy (b). (B) SREBP-2 (343–460) exactly reproduces the nuclear import of full-length SREBP-2 in vitro. Permeabilized HeLa cells were incubated with 10 μl of reaction mixture containing 6.5 pmol of GFP-SREBP-2(343–460) in the absence (a) or the presence (b and c) of recombinant transport factors: (b) 4 pmol of importin β; (c) 4 pmol of importin β, 4 pmol of Ran, and 3 pmol of p10/NTF2. All reactions contained an ATP regeneration system and 0.5 mM GTP. Import reactions were performed as in Figure 1B and examined by fluorescent microscopy.
DISCUSSION
The present study describes the characterization of the nuclear import of the mature form of SREBP-2 by using in vivo microinjection experiments and in vitro transport assay. The results show that SREBP-2 is actively imported via SREBP-2–importin β complex formation in a Ran-dependent manner. Furthermore, it was found that the HLH-Zip is responsible for the binding to importin β and hence acts as a novel NLS. These findings extend our understanding relative to the nuclear import mechanisms of bHLH-Zip–containing transcription factors and the versatility of importin β.
We have shown that a saturable amount of basic NLS-containing substrate does not completely prevent the import of SREBP-2 in permeabilized cells (Figure 2A). It is noteworthy, however, that a slight (∼30%) decrease in SREBP-2 import was observed in the presence of a 10-fold excess of the competitor but that no more decrease was observed on increasing dose of the competitor. This observation is somewhat surprising, because basic NLS-bearing substrates also use importin β via importin α-family adapters. Several possible explanations exist for explaining this phenomenon. First, the recycling of importin β may occur more efficiently than importin α, and, as a result, a significant amount of free importin β may remain unoccupied by importin α even in the presence of a large amount of cargoes for importin α. Because it has been clearly shown that importin β alone can shuttle between the nucleus and the cytoplasm (Kose et al., 1997, 1999), whereas importin α is exported via a specific export receptor, CAS (Kutay et al., 1997), it is reasonable to speculate that importin β returns to the cytoplasm more efficiently than importin α without limitation by export carrier molecules. Alternatively, considering the fact that this experiment was performed using the total cytosol as a source of transport factors, unknown modifying factor(s), which may regulate importin β-mediated import, are present.
In the immunoprecipitation experiment using the cell extract, SREBP-2 significantly bound to importin β, whereas no significant binding to other importin β family members was observed (Figure 4A). Even transportin, a closely related homologue of importin β, failed to interact with SREBP-2 (Figure 4, A and C). Therefore, although the possibility that other members of importins also mediate the SREBP-2 import cannot be completely excluded, it is probable that importin β is the principal import receptor for SREBP-2. As mentioned in INTRODUCTION, importin β has been shown to carry nuclear proteins in two ways: 1) via the importin α-family adapters and 2) via direct interaction with the nuclear protein. Cargoes that are carried by importin β appear to be divided into two groups by virtue of receptor selectivity: 1) karyophiles, which can be imported by several importin β-related receptors, and 2) molecules, which are transported exclusively by importin β. The former includes ribosomal proteins, whereas the latter includes SREBP-2 as well as importin α family proteins. It has been suggested that, during evolution, an importin α-independent common ancestor gave rise to the importin α-dependent importin β molecule together with importin α, and, at some point afterward, importin α was divided into several groups (Malik et al., 1997). Therefore, the former group might consist of evolutionary old karyophiles, which were already in existence before importin β diverged, whereas the latter might be newer, having appeared at the stage of evolution of importin β. Homologues of SREBP are consistently observed only in higher eukaryotes, ranging from the fly (Drosophila melanogaster) (Rosenfeld and Osborne, 1998) to mammals (human, mouse, rat, and hamster).
The bHLH-Zip motifs are found in a large number of eukaryotic transcription factors. The basic region of bHLH-Zip proteins binds to specific sequences in DNA, and the adjacent HLH-Zip region mediates homo- and heterodimerization (for review, see Ferré-D’Amaré and Burley, 1995). We have demonstrated herein that the HLH-Zip domain is responsible for binding to importin β and the nuclear import of SREBP-2 (Figures 6 and 7). Three SREBPs, designated SREBP-1a, -1c, and -2, are known to exist in the nucleus (Hua et al., 1993; Tontonoz et al., 1993; Yokoyama et al., 1993). The mature forms of these SREBPs are most highly conserved (∼71% identical) in the bHLH-Zip region, whereas they are varied in other regions. Therefore, it is possible that SREBP-1a and -1c are also recognized and imported by importin β by virtue of the conserved HLH-Zip domain. Furthermore, it would be worthwhile to determine whether importin β binds to dimerized HLH-Zip of SREBP-2. If this is the case, homo- or heterodimerization with the proper partner would be required, not only for the productive binding to target DNA sequences but also for efficient nuclear import, leading to the highly precise regulation of lipid metabolism.
SREBP-2 represents the first example of a protein that contains an NLS within the HLH-Zip motif. c-Myc and USF2, both of which contain bHLH-Zip motifs that are structurally very similar to SREBP-2, have two NLSs, one within the basic region and the other upstream of the basic region (Dang and Lee, 1988; Luo and Sawadogo, 1996). Either of the two NLSs consists of basic amino acids, implying that the nuclear import of these proteins is importin α/β dependent. In fact, Saphire et al. (1998) have demonstrated that the importin α/β heterodimer mediates the nuclear import of c-Myc basic NLS-containing substrate in vitro. Conversely, both the basic region and the upstream domain of SREBP-2 are dispensable for its nuclear localization. One possible explanation for the difference in the importance of the basic region arises from structural aspects. All bHLH-Zip–containing proteins including SREBPs bind to palindromic sequences containing a so-called E box (CANNTG). Unlike other E box–binding proteins, SREBPs have an atypical tyrosine, instead of a conserved arginine in the basic regions, allowing them to recognize an asymmetric target sequence called the sterol regulatory element (SRE; 5′-ATCACCCCAC-3′) as well as an E box sequence (Párraga et al., 1998). Therefore, it is possible that such an unusual structure in the basic region might affect not only the DNA-binding properties but also the affinity for importin α.
Recent studies have revealed that each member of the import receptor can import a variety of cargoes rather than a single specific class of karyophiles. To date, three classes of importin β-binding sequences have been identified: the IBB domain of importin α (Görlich et al., 1996; Weis et al., 1996), the beta-like import receptor binding domain on ribosomal proteins (Jäkel and Görlich, 1998), and the RGG repeat of yeast Nab2p (Truant et al., 1998). This study points to the fact that importin β recognizes a wider variety of signals than has previously been expected. These importin β-binding signals, including HLH-Zip of SREBP-2, share little homology with one another except for some basic residues. How does importin β carry these different classes of cargoes into the nucleus? The binding site in importin β for ribosomal proteins has been shown to be distinct from that for importin α (Jäkel and Görlich, 1998). We also have collected some preliminary data that indicate that the SREBP-2 binding site and the importin α binding site in importin β are not identical (our unpublished results). Thus, does importin β sort out various cargoes via the use of its own different regions? Or is there a regulatory mechanism in cells for cargo selectivity? Further studies, including structural analysis, will be required to address these questions.
ACKNOWLEDGMENTS
We thank Dr. S. Kuroda (Institute of Scientific and Industrial Research, Osaka University) for the gift of pGEX-6P-2 hGFP. This work was supported by grant-in-aid for scientific research on priority areas 07282103, grant-in-aid for scientific research (B) 08458229, grant-in-aid for scientific research (C) 09680692, and grant-in-aid for Center-of-Excellence research 07CE2006 from the Japanese Ministry of Education, Science, Sports and Culture.
REFERENCES
- Adam EJ, Adam SA. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Beals CR, Clipstone NA, Ho SN, Crabtree GR. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin- sensitive intramolecular interaction. Genes & Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- Beg AA, Ruben SM, Scheinman RI, Haskill S, Rosen CA, Baldwin AS., Jr IkB interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes & Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc Natl Acad Sci USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Carey KL, Richards SA, Lounsbury KM, Macara IG. Evidence using a green fluorescent protein-glucocorticoid receptor chimera that the Ran/TC4 GTPase mediates an essential function independent of nuclear protein import. J Cell Biol. 1996;133:985–996. doi: 10.1083/jcb.133.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Visser GD, Adam SA. RanBP1 stabilizes the interaction of Ran with p97 nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Lee WM. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988;8:4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- Fabre E, Hurt E. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu Rev Genet. 1997;31:277–313. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- Ferré-D’Amaré AR, Burley SK. DNA recognition by helix-loop-helix proteins. Nucleic Acids Mol Biol. 1995;9:285–298. [Google Scholar]
- Finlay DR, Newmeyer DD, Price TM, Forbes DJ. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J Cell Biol. 1987;104:189–200. doi: 10.1083/jcb.104.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Henklein P, Laskey RA, Hartmann E. A 41 amino acid motif in importin α confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Görlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Hieda M, Tachibana T, Yokoya F, Kose S, Imamoto N, Yoneda Y. A monoclonal antibody to the COOH-terminal acidic portion of Ran inhibits both the recycling of Ran and nuclear protein import in living cells. J Cell Biol. 1999;144:645–655. doi: 10.1083/jcb.144.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, Goldstein JL, Wang X. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci USA. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Kose S, Takao T, Tachibana T, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 1995a;368:415–419. doi: 10.1016/0014-5793(95)00699-a. [DOI] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore- targeting complex in nuclear protein import. EMBO J. 1995b;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto N, Tachibana T, Matsubae M, Yoneda Y. A karyophilic protein forms a stable complex with cytoplasmic components before nuclear pore binding. J Biol Chem. 1995c;270:8559–8565. doi: 10.1074/jbc.270.15.8559. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S, Görlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose S, Imamoto N, Tachibana T, Shimamoto T, Yoneda Y. Ran-unassisted nuclear migration of a 97-kDa component of nuclear pore-targeting complex. J Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose S, Imamoto N, Tachibana T, Yoshida M, Yoneda Y. β-Subunit of nuclear pore-targeting complex (Importin β) can be exported from the nucleus in a Ran-independent manner. J Biol Chem. 1999;274:3946–3952. doi: 10.1074/jbc.274.7.3946. [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Luo X, Sawadogo M. Functional domains of the transcription factor USF2: atypical nuclear localization signals and context-dependent transcriptional activation domains. Mol Cell Biol. 1996;16:1367–1375. doi: 10.1128/mcb.16.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Malik HS, Eickbush TH, Goldfarb DS. Evolutionary specialization of the nuclear targeting apparatus. Proc Natl Acad Sci USA. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Párraga A, Bellsolell LR, Ferré-D’Amaré AR, Burley SK. Cocrystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure. 1998;6:661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Delphin C, Gerace L. Nucleotide-specific interaction of Ran/TC4 with nuclear transport factors NTF2 and p97. Proc Natl Acad Sci USA. 1996;93:7679–7683. doi: 10.1073/pnas.93.15.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Blobel G, Rosenblum JS. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- Radu A, Blobel G, Moore MS. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JM, Osborne TF. HLH106, a Drosophila sterol regulatory element-binding protein in a natural cholesterol auxotroph. J Biol Chem. 1998;273:16112–16121. doi: 10.1074/jbc.273.26.16112. [DOI] [PubMed] [Google Scholar]
- Sakai J, Rawson RB, Espenshade PJ, Cheng D, Seegmiller AC, Goldstein JL, Brown MS. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- Saphire AC, Bark SJ, Gerace L. All four homochiral enantiomers of a nuclear localization sequence derived from c-Myc serve as functional import signals. J Biol Chem. 1998;273:29764–29769. doi: 10.1074/jbc.273.45.29764. [DOI] [PubMed] [Google Scholar]
- Sato R, Inoue J, Kawabe Y, Kodama T, Takano T, Maeda M. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J Biol Chem. 1996;271:26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- Sato R, Yang J, Wang X, Evans MJ, Ho YK, Goldstein JL, Brown MS. Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1) J Biol Chem. 1994;269:17267–17273. [PubMed] [Google Scholar]
- Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto T, Nakajima K, Tachibana T, Hirano T, Yoneda Y. Interferon-γ-dependent nuclear import of Stat1 is mediated by the GTPase activity of Ran/TC4. J Biol Chem. 1996;271:31017–31020. doi: 10.1074/jbc.271.49.31017. [DOI] [PubMed] [Google Scholar]
- Shibasaki F, Price ER, Milan D, McKeon F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature. 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- Tachibana T, Hieda M, Sekimoto T, Yoneda Y. Exogenously injected nuclear import factor p10/NTF2 inhibits signal-mediated nuclear import and export of proteins in living cells. FEBS Lett. 1996;397:177–182. doi: 10.1016/s0014-5793(96)01180-5. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Kim JB, Graves RA, Spiegelman BM. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant R, Fridell RA, Benson RE, Bogerd H, Cullen BR. Identification and functional characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+ RNA binding protein. Mol Cell Biol. 1998;18:1449–1458. doi: 10.1128/mcb.18.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandromme M, Gauthier-Rouviere C, Lamb N, Fernandez A. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem Sci. 1996;21:59–64. [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Weis K, Mattaj IW, Lamond AI. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- Weis K, Ryder U, Lamond AI. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- Wozniak RW, Rout MP, Aitchison JD. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- Yang J, Brown MS, Ho YK, Goldstein JL. Three different rearrangements in a single intron truncate sterol regulatory element binding protein-2 and produce sterol-resistant phenotype in three cell lines. Role of introns in protein evolution. J Biol Chem. 1995;270:12152–12161. doi: 10.1074/jbc.270.20.12152. [DOI] [PubMed] [Google Scholar]
- Yang J, Sato R, Goldstein JL, Brown MS. Sterol-resistant transcription in CHO cells caused by gene rearrangement that truncates SREBP-2. Genes & Dev. 1994;8:1910–1919. doi: 10.1101/gad.8.16.1910. [DOI] [PubMed] [Google Scholar]
- Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein JL, Brown MS. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- Yoneda Y. How proteins are transported from cytoplasm to the nucleus. J Biochem. 1997;121:811–817. doi: 10.1093/oxfordjournals.jbchem.a021657. [DOI] [PubMed] [Google Scholar]
- Yoneda Y, Imamoto-Sonobe N, Yamaizumi M, Uchida T. Reversible inhibition of protein import into the nucleus by wheat germ agglutinin injected into cultured cells. Exp Cell Res. 1987;173:586–595. doi: 10.1016/0014-4827(87)90297-7. [DOI] [PubMed] [Google Scholar]