Abstract
Sla2p, also known as End4p and Mop2p, is the founding member of a widely conserved family of actin-binding proteins, a distinguishing feature of which is a C-terminal region homologous to the C terminus of talin. These proteins may function in actin cytoskeleton-mediated plasma membrane remodeling. A human homologue of Sla2p binds to huntingtin, the protein whose mutation results in Huntington’s disease. Here we establish by immunolocalization that Sla2p is a component of the yeast cortical actin cytoskeleton. Deletion analysis showed that Sla2p contains two separable regions, which can mediate association with the cortical actin cytoskeleton, and which can provide Sla2p function. One localization signal is actin based, whereas the other signal is independent of filamentous actin. Biochemical analysis showed that Sla2p exists as a dimer in vivo. Two-hybrid analysis revealed two intramolecular interactions mediated by coiled-coil domains. One of these interactions appears to underlie dimer formation. The other appears to contribute to the regulation of Sla2p distribution between the cytoplasm and plasma membrane. The data presented are used to develop a model for Sla2p regulation and interactions.
INTRODUCTION
SLA2 (Synthetic Lethal with ABP1), also known as END4 and MOP2, is essential in yeast for correct organization of the cortical actin cytoskeleton (Holtzman et al., 1993). It is also required for endocytosis and for accumulation or maintenance of a plasma membrane ATPase at the cell surface (Raths et al., 1993; Na et al., 1995). Sla2p might therefore facilitate actin-mediated plasma membrane remodeling.
Cells lacking Sla2p are temperature sensitive for growth. In addition, they exhibit a disorganized actin cytoskeleton, and their cell surface growth is depolarized (Holtzman et al., 1993). Instead of the characteristic ellipsoid cell shape displayed by wild-type diploid cells, sla2 mutants are spherical. The cortical actin patches, normally spatially restricted to growing domains within the cell cortex, are no longer concentrated at the surface of the bud and instead are distributed evenly across the surface of the mother cell and the bud. Finally, sla2 mutant cells accumulate post-Golgi vesicles, possess an abnormally thick cell wall, and do not undergo the wild-type bipolar budding pattern (Mulholland et al., 1997; Yang et al., 1997). Several of these defects are characteristic of mutants affecting the cortical actin cytoskeleton.
A deletion mutation of SLA2 is synthetically lethal with null alleles of at least three genes encoding components of the cortical actin cytoskeleton. These genes are ABP1 (Holtzman et al., 1993), which encodes a protein containing a C-terminal Src homology 3 domain and an N-terminal ADF homology domain (Drubin et al., 1990; Lappalainen et al., 1998), SRV2 (Lila and Drubin, 1997), which encodes a protein that binds to actin monomers and to adenylyl cyclase, a component of the Ras signaling pathway in yeast (Freeman et al., 1995, 1996), and SAC6 (Holtzman et al., 1993), which encodes the actin filament-bundling protein fimbrin. A deletion mutant of SLA2 is also synthetic lethal with a deletion mutant of GCS1. Gcs1p is a GTPase-activating protein for Arf proteins in yeast that also appears to have a direct effect on actin dynamics (Blader et al., 1999). These genetic interactions suggest that Sla2p, Sac6p, Srv2p, Abp1p, and Gcs1p contribute to common properties of the cytoskeleton.
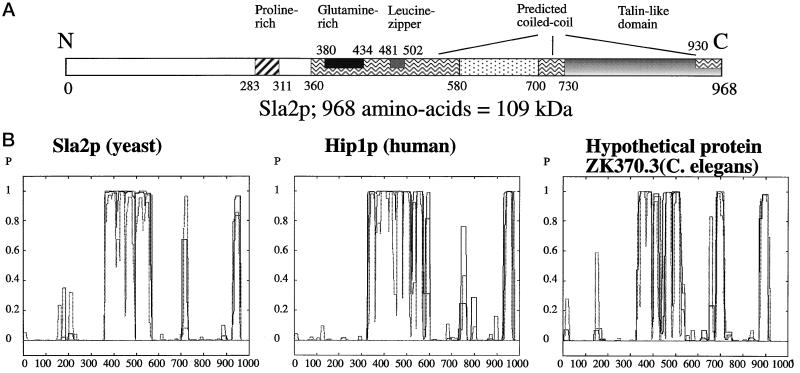
SLA2 encodes a protein of 968 amino acids with a predicted molecular mass of 109 kDa. Homologues of Sla2p have been identified in nematodes, fruit flies, mice, and humans. Hip1p, a human homologue of Sla2p, binds to huntingtin, the protein product of the Huntington’s disease (HD) gene (Kalchman et al., 1997; Wanker et al., 1997). Thus, studies of Sla2p might provide insights into the etiology of HD. Sla2p and related proteins all share a similar arrangement of three predicted coiled-coil regions (amino acids 360–580, 700–730, and 930–960 of Sla2p), as well as a C-terminal domain similar to the C terminus of talin, a protein found in focal adhesion plaques (Figure 1A). Talin contains at least three actin-binding sites, one of which, amino acids 2269–2541, spans the region that shows homology to Sla2p (Hemmings et al., 1996). Indeed, this region of Sla2 has been shown by in vitro biochemical studies to bind to actin (McCann and Craig, 1997).
Figure 1.
Domain arrangement of Sla2p. (A) Schematic diagram of identifiable domains in Sla2p. The three predicted coiled-coil forming domains are indicated, as well as glutamine- and proline-rich regions and the C-terminal talin-like domain. (B) The COILS program (Lupas et al., 1991) was used to predict the potential coiled-coil–forming domains of Sla2p and its homologues in humans (Hip1p) and nematodes (GenBank accession number 1353119). The probability of coiled coil formation is plotted as a function of the amino acid position in the protein. All three proteins are ∼950 amino acids in length, and, in addition to sharing secondary structure, they show ∼20% identity over their lengths.
Previous studies have shown that Sla2p is a peripheral membrane protein (Na et al., 1995; Wesp et al., 1997) and is likely to be part of a protein complex in vivo. In addition, Sla2p sediments as a single peak at ∼10.6 S, which correlates to 220 kDa, or about twice its molecular mass (Wesp et al., 1997). This behavior required the presence of the large, central coiled-coil domain. Wesp and colleagues (1997) performed a domain analysis in an attempt to correlate Sla2p’s function with its various regions. A Sla2p N-terminal deletion protein lacking amino acids 114–284 was unable to restore endocytosis, actin organization, and growth at high temperature in sla2Δ mutant cells. Because none of the other deletion mutants of Sla2p exhibited this severe lack of activity, they concluded that the N-terminal region of Sla2p is indispensable for its cellular activity. Somewhat surprisingly, the talin-like domain was shown to be dispensable for endocytosis and growth at high temperature. Finally, as well as appearing to be involved in the formation of either a dimer or a multiprotein complex, the central coiled-coil domain of Sla2p was found to be required for endocytosis in genetic backgrounds mutant for ABP1 or SRV2 (Wesp et al., 1997).
In this study, we report on the cellular localization of Sla2p, demonstrating that it is a component of actin cortical patches. We also provide further analysis showing that different domains of the protein interact with each other and thereby regulate the localization and activity of Sla2p.
MATERIALS AND METHODS
Strains and Growth Conditions
Yeast strains used in this study are listed in Table 1. Standard yeast genetic procedures and media were used (Rose et al., 1990). Except where noted, yeast strains were grown at 25°C in rich YPD media.
Table 1.
Yeast strains used in this study
| Name | Genotype | Source |
|---|---|---|
| DDY131 | MATa his3-Δ200 ura3-52 leu2-3,112 lys2-801 ade2-1 | 1 |
| DDY288 | MATa//MATα ura3-52/ura3-52 leu2-3,112/+ his4-619/+ | 1 |
| DDY426 | MATa/MATα ura3-52/ura3-52 leu2-3,112/leu2-3,112 lys2-801/lys2-801 his3-Δ200/his3-Δ200 ade2-1/+ | 1 |
| DDY540 | MATa/MATα sla2Δ::URA3/sla2Δ::URA3 ura3/ura3 his4/+ leu2/+a | 2 |
| DDY1076 | MATa/MATα sla2Δ::URA3/sla2Δ::URA3 his3/his3 ura3/ura3 leu2/leu2 lys2/lys2 ade2/+b | 2 |
| DDY1166 | MATa sla2Δ::HIS3 his3 ura3 leu2 lys2 ade2 | 2 |
| DDY1168 | MATa/MATα sla2Δ::HIS3/sla2Δ::HIS3 his3/his3 ura3/ura3 leu2/leu2 lys2/lys2 ade2/+b | 2 |
| DDY1194 | MATa/MATα sla2Δ768-968::URA3 leu2/leu2 lys2/lys2 ura3/ura3 his3/his3 ade2/+b | 1 |
| DDY1195 | MATa/MATα sla2Δ501-968::URA3/sla2Δ501-968::URA3 leu2/leu2 lys2/lys2 ura3/ura3 his3/his3 ade2/+b | 1 |
| DDY1196 | MATa/MATα sla2Δ360-968::URA3/sla2Δ360-968::URA3 leu2/leu2 lys2/lys2 ura3/ura3 his3/his3 ade2/+b | 1 |
| DDY1550 | MATa 10His-SLA2::URA3 his3 ura3 leu2 lys2 ade2c | 2 |
| DDY1551 | MATa 6myc-SLA2::URA3 his3 ura3 leu2 lys2 ade2c | 2 |
| DDY1554 | MATα 6myc-SLA2::URA3 his3 ura3 leu2 lys2 ade2c | 2 |
| DDY1556 | MATa/α 10His -SLA2::URA3/6myc-SLA2::URA3 his3/his3 ura3/ura3 leu2/leu2 lys2/lys2 ade2/ade2c | 2 |
| DDY1557 | MATa/α 6myc-SLA2::URA3/6myc-SLA2::URA3 his3/his3 ura3/ura3 leu2/leu2 lys2/lys2 ade2/ade2c | 2 |
| Y190 | MATa gal4 gal80 his3 leu2 trp1 ura3 ade2 cyhr, URA3::GALp–lacZ, LYS2::GALp–HIS3 | 3 |
For each strain, the strain collection number, genotype, and source are indicated. Sources: 1, Drubin lab; 2, this study; 3, Elledge lab.
Derived from DDY288.
Derived from DDY426.
Derived from DDY131.
Plasmid Constructions and Other DNA Manipulations
All DNA manipulations were performed by standard techniques (Maniatis et al., 1982; Ausubel et al., 1989). The PCR was used to create restriction enzyme sites for cloning purposes or to build gene disruption constructs (Lorenz et al., 1995). Plasmids used in this study are listed in Table 2.
Table 2.
Plasmids used in this study
| Name | Features | Use |
|---|---|---|
| GST-SLA2 fusion protein plasmids | ||
| pDD356 | Plac GST-SLA2 | Contains Sla2p amino acids 664-968 |
| pDD358 | Plac GST-SLA2 | Contains Sla2p amino acids 503-968 |
| pDD361 | Plac GST-SLA2 | Contains Sla2p amino acids 90-547 |
| SLA2 plasmids | ||
| pDD353 | CEN, HIS3, SLA2 | pRS313 with 4.4-kb EcoRI SLA2 insert |
| pDD354 | 2μ, HIS3, SLA2 | YEP430 with 4.4-kb EcoRI SLA2 insert |
| pDD355 | CEN, URA3, GAL1p-SLA2 | Sla2p inducible overexpression (in pYES-R) |
| pDD362 | CEN, HIS3, sla2Δ33-750 | Sla2Δ33-750 deletion protein |
| pDD363 | 2μ, HIS3, sla2Δ33-750 | Sla2Δ33-750 deletion protein |
| pDD364 | CEN, HIS3, sla2Δ33-501 | Sla2Δ33-501 deletion protein |
| pDD365 | 2μ, HIS3, sla2Δ33-501 | Sla2Δ33-501 deletion protein |
| pDD366 | CEN, URA3, GAL1p-sla2Δ33-750 | Sla2Δ33-750 inducible overexpression |
| pDD367 | CEN, HIS3, sla2Δ33-359 | Sla2Δ33-359 deletion protein |
| pDD368 | 2μ, HIS3, sla2Δ33-359 | Sla2Δ33-359 deletion protein |
| pDD369 | CEN, HIS3, sla2Δ33-359, 576stop | Sla2Δ33-359,576stop deletion protein |
| pDD370 | 2μ, HIS3, sla2Δ33-359, 576stop | Sla2Δ33-359,576stop deletion protein |
| pDD371 | CEN, HIS3, sla2Δ360-575 | Sla2Δ360-575 deletion protein |
| pDD372 | 2μ, HIS3, sla2Δ360-575 | Sla2Δ360-575 deletion protein |
| Two-hybrid plasmids | ||
| pDD73 (pSE1111) | CEN, LEU2 | Snf4p fused to activation domain of Gal4p in pACT |
| pDD74 (pSE1112) | CEN, TRP1 | Snf1p fused to the DNA-binding domain of Gal4p in pAS1 |
| pDD373 | CEN, TRP1 | Sla2p aa 503-968 fused to the DNA-binding domain of Gal4p in pAS1-CYH2 |
| pDD374 | CEN, TRP1 | Sla2p aa 768-968 fused to the DNA-binding domain of Gal4p in pAS1-CYH2 |
| pDD375 | CEN, LEU2 | Sla2p aa 502-767 fused to the activation domain of Gal14p in pACTII |
| pDD376 | CEN, LEU2 | Sla2p aa 575-767 fused to the activation domain of Gal4p in pACTII |
| pDD377 | CEN, TRP1 | Sla2p aa 751-930 fused to the DNA-binding domain of Gal4p in pAS1-CYH2 |
| pDD378 | CEN, TRP1 | Sla2p aa 5-576 fused to the DNA-binding domain of Gal14p in pAS1-CYH2 |
| pDD379 | CEN, LEU2 | Sla2p aa 5-576 fused to the activation domain of Gal4p in pACTII |
| pDD381 | CEN, LEU2 | Sla2p aa 5-358 fused to the activation domain of Gal4p in pACTII |
| pAIP70a | CEN, LEU2 | Actin fused to the activation domain of Gal4p in pACTII |
| pRB1516a | CEN, TRP1 | Actin fused to the DNA-binding domain of Gal4p in pAS1-CYH2 |
GST-Sla2p Fusion Proteins
Different portions of SLA2 were cloned into pGEX-2T or pGEX-3X (Amersham Pharmacia Biotech, Piscataway, NJ) to create GST fusion proteins. The GST moiety could be cleaved from the fusion protein because pGEX-2T contains a thrombin cleavage site, and pGEX-3X contains a Factor Xa site. To construct a plasmid encoding a GST-Sla2p fusion containing amino acids 664–968 of Sla2p (pDD356), BamHI linkers were added to a 1.1-kb RsaI SLA2 fragment, which was then was cloned into the BamHI site of pGEX-2T. For the GST-Sla2p fusion protein expressing amino acids 503–968 of Sla2p (pDD358), a plasmid containing SLA2 was digested with PstI and AatII. The ends of the 1.4-kb fragment were filled in using T4 DNA polymerase. BamHI linkers were then added, and the fragment was cloned into the BamHI site of pGEX-3X. To construct a plasmid encoding the GST-Sla2p fusion expressing amino acids 90–547 of Sla2p (pDD361), a plasmid containing SLA2 was digested with BstYI. The resulting 1.4-kb fragment was subcloned into the BamHI site of pGEX-2T. The resulting plasmids were screened by restriction analysis for those containing the insert in the correct orientation.
pDD356 and pDD358 produce soluble fusion proteins, which were induced and purified as described (Ausubel, 1990). Fusion proteins were eluted from the glutathione–agarose beads with an excess of free glutathione (5 mM, pH 8.0; Sigma, St. Louis, MO). Alternatively, thrombin (Sigma) or Factor Xa (Boehringer Mannheim, Indianapolis, IN) was used to cleave and, in doing so, solubilize the Sla2p portion of the fusion protein. pDD361 produces mainly insoluble fusion protein. However, an “inclusion body” preparation from a strain expressing this plasmid provided partially purified protein that was used for generating antibodies (see below). Cells were prepared in the same manner as above, except that here the majority of the fusion protein is insoluble and present in the pellet. The fusion protein comprised most (90%) of the protein in the pellet. To purify the protein, the pellet was solubilized using sample buffer containing 2% SDS and then run on an SDS-PAGE gel. Protein was eluted from the gel as described in (Drubin et al., 1988).
Antibodies
Antibodies against Sla2p were raised in New Zealand White rabbits, essentially as described in (Harlow and Lane, 1988). To generate antibodies against the Sla2p C terminus, Sla2p amino acids 664–968 were fused to GST (pDD356). A GST fusion protein containing Sla2p amino acids 90–547 (pDD361) was used to generate the Sla2p N-terminal antibodies. The GST-Sla2 fusion from pDD356 was not further purified after glutathione elution from Sepharose beads, whereas the GST-Sla2 fusion protein from pDD361 was purified by excision of Coomassie blue–stained bands from SDS-PAGE gels. Approximately 100 μg of fusion protein were used for each injection. Freund’s complete adjuvant was used for the first immunization, and Freund’s incomplete adjuvant was used for subsequent injections (days 21, 36, and 78).
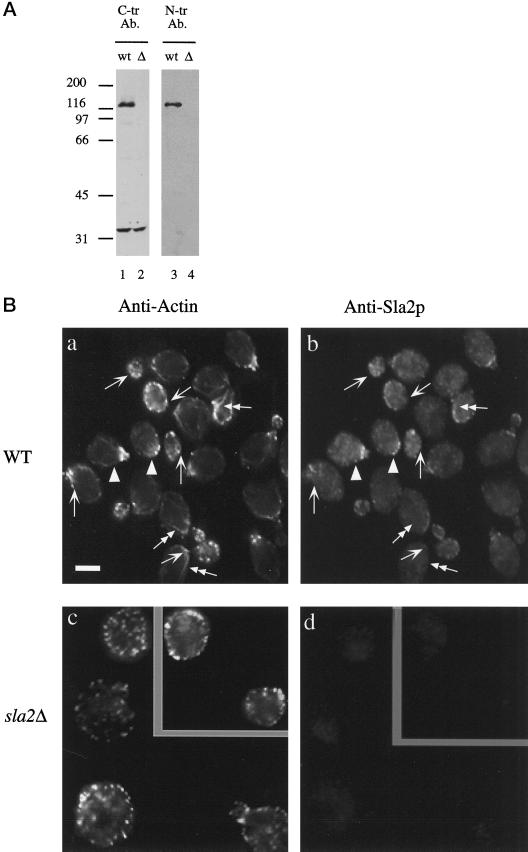
Antisera were affinity purified against Sla2p as follows. The GST-Sla2 fusion protein from pDD358 (amino acids 503–968 of Sla2p) was cleaved with Factor Xa and then purified further by excision of Coomassie blue–stained bands from SDS-PAGE gels. The protein was coupled to cyanogen bromide (CNBr)–activated beads, generating a column containing only Sla2p amino acids 503–968. This column was then used for affinity purification of the C-terminal antibody. The resulting affinity-purified antiserum recognized a protein with apparent molecular mass of 116 kDa in immunoblots of whole-cell extracts (1:5000 dilution) (Figure 2A). This band was absent in sla2Δ whole-cell extract. However, the antiserum also recognized another protein with apparent molecular mass of 35 kDa in whole-cell extract from wild-type or sla2Δ cells. Cross-reactivity of the antibody with this protein did not effect the localization of Sla2p by indirect immunofluorescence, however, because sla2Δ cells showed only faint background staining using this antibody. For the N-terminal antibody, antisera were first adsorbed to GST-coupled CNBr beads to deplete the anti-GST antibodies. The GST-Sla2 fusion protein from pDD361 was purified in the same manner as that used for the rabbit injections and coupled to CNBr beads. Additionally, a smaller amount of soluble GST-Sla2 fusion protein was purified by excision of Coomassie blue–stained bands from SDS-PAGE gels and then coupled to CNBr-activated beads to generate an affinity column containing only the GST-Sla2 fusion protein. Antisera affinity-purified on either column recognized similar proteins in immunoblots, although lower-titer antiserum was obtained using the latter column. These antisera (1:5000) recognized a protein with apparent molecular mass of 116 kDa that is absent in sla2Δ whole-cell extract. They also recognized less strongly a protein with apparent molecular mass of 70 kDa that is present in sla2Δ whole-cell extract. However, the reactivity against the 70-kDa protein was variable and not always observed.
Figure 2.
Subcellular localization of Sla2p. (A) Affinity-purified anti-Sla2p antibodies recognize a 116-kDa protein species in wild-type yeast whole-cell extracts. Lanes 1 and 2, immunoblot using the affinity-purified antibody against the C terminus of Sla2p. Lanes 3 and 4, immunoblot using affinity-purified antibody against the N terminus of Sla2p. Lanes 1 and 3 contain whole-cell extract from wild-type cells. Lanes 2 and 4 contain whole-cell extract from sla2Δ cells. (B) Indirect immunolocalization shows that Sla2p localizes to cortical structures the yeast cell. This localization largely, but not completely, overlaps with that of cortical actin. Arrows indicate patches that contain actin but no detectable Sla2p. Arrowheads show patches that contain Sla2p but no detectable actin. Double-headed arrows indicate actin cables, which contain no Sla2p. Diploid wild-type (DDY288; a and b) and sla2Δ cells (DDY540; c and d) were processed for immunofluorescence using affinity-purified antibody against the C terminus of Sla2p and anti-yeast actin antibodies, as described in MATERIALS AND METHODS. (a and c) Fluorescein staining of actin structures. (b and d) CY3 staining of Sla2p structures.
Polyclonal rabbit anti-myc antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Immunofluorescence Microscopy
Yeast cells grown to early exponential phase in either YPD or supplemented synthetic media were prepared for immunofluorescence as described previously (Pringle et al., 1991). The cold methanol–acetone fixation–permeabilization step, which is required for anti-actin reactivity, is not required for anti-Sla2p reactivity but results in better staining (probably because of the flattening of the cells allowing easier visualization of the staining pattern). Both anti-N- and anti-C-terminal affinity-purified rabbit anti-Sla2p antisera were used for immunofluorescence at a dilution of 1:50. Anti-actin guinea pig antiserum (Mulholland et al., 1994) was used at a 1:1000 dilution. Detection was accomplished by applying either FITC-conjugated goat anti-rabbit or FITC-conjugated goat anti-guinea pig antibodies (Cappel/Organon Teknika, Malvern, PA) at a dilution of 1:1000 or CY3-conjugated sheep anti-rabbit antibodies (Sigma) at a dilution of 1:200. Cells were visualized with a Zeiss Axioscop fluorescence microscope with an HB100 W/Z high-pressure mercury lamp and a Zeiss 100× Plan-Neofluar oil immersion objective (Carl Zeiss, Thornwood, NY). Images were captured electronically using a 200-E charge-coupled device camera (Sony Electronics, San Jose, CA) using Northern Exposure software (Phase 3 Imaging Systems, Milford, MA).
Endocytosis Assays
Endocytosis was assayed using the internalization of Lucifer yellow (LY; Sigma-Aldrich-Fluka, Milwaukee, WI) as a marker, according to the method described by Dulic et al. (1991).
Creation of a Diploid Strain Expressing myc Epitope-tagged and Histidine-tagged Sla2p
A 10-histidine tag flanked by BamHI sites was placed at the N terminus of Sla2p using a PCR-stitching strategy. All PCR reactions used high-fidelity DNA polymerases, and all relevant constructs were sequenced for accuracy. Two first-round PCR reactions were performed between 1) primers Sla2Htag5 (CAGGATGATGATTCGGA-TCCTCATCATCATCATCATCATCATCATCATCATGGGGATCCA -AACTGATCATGAATTGTAC) and M13Universal (GTAAAACGAC-GGCCAGTG) and 2) primers Sla2Htag6 (GTACAATTCATGATCAGTTTGGATCCCCATGATGATGATGATGATGATGATGATG-ATGAGGATCCGAATCATCATCCTG) and Sla2Htag7 (GATGTTAC-CGATGCATGC) using pDD550 as template, which is an EcoRI genomic fragment containing SLA2 inserted into pBluescript II SK+ (Stratagene, La Jolla, CA). The products of the first two PCR reactions were mixed and reamplified using the outside primers M13Universal and Sla2Htag7. The resulting amplified DNA was digested with HindIII and PflMI and used to replace a HindIII–PflMI fragment from pDD550, generating an SLA2 gene encoding Sla2p with a C-terminal 10-His tag (… DDDSDPHHHHHHHHHHGDPNstop), flanked by BamHI sites.
The 10-His tag was replaced with a 6-myc tag (6xMEQKLISEEDLNE) flanked by BamHI sites. A NotI site was placed after the predicted polyadenylation signals 3′ of SLA2 using the mutagenic primer Sla2NotIins (CATATCACATCTCTGGCGGCCGCTACCATTTTGCTTC). The URA3 gene, flanked by NotI sites (from pJC56, which is pBluescript II SK+ containing URA3 flanked by SmaI/XbaI and NotI sites; our unpublished data) was inserted to generate auxotrophically marked constructs. During these manipulations, pUC118 and pGEM-9Z were used as intermediate vectors.
The EcoRI-digested inserts from pDD551 (10-His C-terminal tagged Sla2 with URA3 marker in pUC118) and pDD552 (6-myc C-terminal tagged Sla2 with URA3 marker in pGEM-9Z) were used to transform DDY1166, which contains the HIS3 gene in place of the deleted SLA2 gene. Colonies that were Ura+ and His− were selected, and the insertion of tagged SLA2 was confirmed by PCR using JCYNL243F (CTGGATCCTTGACAATGTTAC) and JCYNL243R (CTACTACTGTCTGAAGAGC) as primers. Both the 10-His and 6-myc constructs (in DDY1550 and DDY1551, respectively) complemented various sla2 null phenotypes fully (our unpublished results). DDY1551 was crossed with DDY131 and sporulated, and haploid, Ura+, and Matα progeny were identified (DDY1554). DDY1550 and DDY1554 were crossed to create diploid yeast (DDY1556) containing one copy of SLA2 tagged with 6-His and one copy tagged with 6-myc. DDY1551 and DDY1554 were crossed to create diploid yeast (DDY1557) with both copies of SLA2 tagged with 6-myc.
Fractionation of Yeast Cell Extracts
Cell extracts for sucrose gradient fractionation were prepared using a liquid nitrogen grinding method. Exponentially growing cells were harvested by centrifugation and washed once with cold double-distilled H2O and once with buffer C (50 mM HEPES, pH 7.4, 50 mM KCl, 2 mM MgCl2, 1 mM EDTA, and 1 mM DTT). Cells were then resuspended into an equal-weight volume of buffer C with the addition of protease inhibitors (antipain, leupeptin, pepstatin A, chymostatin, and aprotinin at 1 μg/ml and 10 μM PMSF and 1 mM benzamidine). The cell suspension was then drop frozen in liquid nitrogen. Depending on the amount of material, cells were lysed either manually using a mortar and pestle, or in a 1-l Waring blender. Liquid nitrogen was used liberally to ensure that cells remained frozen at all times. The cell lysate was then thawed at room temperature to obtain crude extract. All subsequent steps were performed at 4°C. At this point, if detergent-solubilized extract was desired, Nonidet P-40 was added to 0.2%. The lysate was then spun at 1000 × g to clear unlysed cells and cell debris. The extract was then spun at 100,000 × g to obtain the high-speed supernatant fraction for loading onto the sucrose gradient. The pellet and supernatant fractions were assayed by immunoblotting to determine the amount of Sla2p present; usually, Sla2p was present at approximately equal levels in both fractions.
For isolation of 6-histidine–tagged Sla2p, the above procedure was modified as follows: no EDTA was present in the extraction buffer, and 15 mM β-mercaptoethanol was used instead of DTT. Also, a medium-speed centrifugation was introduced (15,000 × g for 10 min), after which the pellet was resuspended in modified buffer C plus 100 mM NaCl and 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid. The resulting solutions were spun at 250,000 × g for 30 min. The supernatants were preincubated with Sepharose CL-6B beads (Amersham-Pharmacia Biotech) to remove proteins that bind to this matrix nonspecifically, and then applied to an Ni-NTA column (Qiagen, Chatsworth, CA). The columns were washed, and the bound protein was eluted according to the manufacturer’s instructions. The high-speed supernatant derived from the medium-speed pellet (i.e., the material that was extractable only in the presence of high salt and detergent) was termed the membrane extract.
Sucrose Gradients
Linear sucrose gradients (11 ml of 3–30%) in buffer C were prepared in Ultra-Clear centrifuge tubes (Beckman, Palo Alto, CA). Samples (400 μl) of yeast extract or protein molecular mass standards were layered on top. The gradients were spun at 34,000 rpm in an SW41 rotor for 15 h at 4°C. Fractions (400 μl) were removed manually and then assayed on SDS-PAGE. Immunoblotting was used to determine the position of various proteins. Native high-molecular-mass proteins (thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; aldolase, 158 kDa; and BSA, 66 kDa) (Pharmacia, Piscataway, NJ) were used to calibrate the gradients. For immunoblots, anti-Srv2p antibody (Freeman et al., 1996) was used at a dilution of 1:5000, anti-Sac6p antibody (Drubin et al., 1988) at 1:500, and anti-Abp1p antibody at 1:5000 (Drubin et al., 1988). HRP-conjugated secondary antibody was used at 1:5000, and detected using the ECL chemiluminescence kit (Amersham, Arlington Heights, IL).
Sla2p Deletions and Truncation
Sla2p C-terminal truncation mutants were generated using a PCR gene deletion strategy (Lorenz et al., 1995). Oligonucleotides containing 54 or 55 nucleotides (nt) homologous to portions of SLA2 and 20 or 21 nt homologous to pRS vector sequences flanking URA3 were used as primers in PCR reactions amplifying the URA3 gene. All PCR reactions used primer SY16 (5′-AAATATATTTATATTAACGTTTATCTTTAT-ATATAAAAAGTACAATTCATGATCACTGTGCGGTATTTCACA-CCGC-3′), which contains 55 nt homologous to the 3′ region of SLA2 immediately following its stop codon as well as 21 nt homologous to a region 3′ of the URA3 gene. The other primer used in the PCR reaction contains 54 nt homologous to various regions of the SLA2 (depending on the desired truncation), a stop codon, and 20 nt homologous to a region 5′ of the URA3 gene (∼100 nt before the URA3 start codon). Primer SY18 (5′-GTTGCCACTGCTGACAAAATTGTCAAATCCTCAGAACACTTACGT -GTTGACGTTTGAGATTGTACTGAGAGTGCACC-3′) contains homology until amino acid 768, primer SY20 (5′-CTATACTCCCAG-TTGCGTCAAGAGCATCTAAATCTTTTACCTCGTTTTAAAAAAT -GAGATTGTACTGAGAGTGCACC-3′) contains homology until amino acid 501, and primer SY21 (5′-GGTGCAGCTAACGCCATTTTTCCACAGGCGACGGCACAAATGCAGCCGGACTTCTGAGATTGTAC TG-AGAGTGCACC-3′) contains homology until amino acid 359.
The resulting PCR products consist of the URA3 gene flanked by ∼50 nt of SLA2 sequence. They were purified using the Qiaquick PCR purification kit (Qiagen) and then used to transform wild-type diploid yeast (DDY426). If integrated at the SLA2 locus, the PCR construct should delete a specific amount SLA2 (depending on the construct) and insert the URA3 gene. PCR was performed on genomic DNA from Ura+ transformants to determine whether the PCR product integrated at the SLA2 locus. The presence of a truncated gene product of the predicted length was further confirmed by immunoblotting using an Sla2p antibody. The heterozygous diploids were then sporulated to generate haploids expressing a truncated Sla2p. Homozygous diploids were obtained by mating the appropriate haploids.
Two NsiI sites are naturally present in SLA2, one before codon 33 and one after codon 896. Additional NsiI sites were inserted before codon 751, codon 502, or codon 360 through PCR using primers SY5 (5′-GCATGCATAGTTGCCACTGCTGAC-3′) and SY7 (5′-GTTGCTGCTGTTGCGAAG-3′), SY6 (5′-GCATGCATACTGCAGTTAAAGGT-C-3′) and SY7, or SY26 (5′-GCATGCATATGGGCCAATCAACAAG-CC-3′) and SY7 respectively. Primers SY26 and SY34 (5′-CGATGCA-TACTAGGCATCCAGAATAGGGTTC-3′) were used to insert an NsiI site before codon 360 and to insert an NsiI site and a stop codon after codon 576. The PCR-generated NsiI fragments were then used to replace the normal NsiI fragment of pDD353 or pDD354 (SLA2 in pRS313; Sikorski and Hieter, 1989) or YEP430 (Ma et al., 1987). All PCR-generated DNA that was not later replaced with plasmid DNA was sequenced, and no PCR-induced base pair changes were found. Thus, eight plasmids were obtained (Table 2): pDD362 (sla2Δ33–750 in pRS313), pDD363 (sla2Δ33–750 in YEP430), pDD364 (sla2Δ33–501 in pRS313), pDD365 (sla2Δ33–501 in YEP430), pDD367 (sla2Δ33–359 in pRS313), pDD368 (sla2Δ33–359 in YEP430), pDD369 (sla2Δ33–359,576stop in pRS313), and pDD370 (sla2Δ33–359,576stop in YEP430). To construct a GAL1 promoter-driven sla2Δ33–750 (pDD366), the NsiI fragment of pDD362 was used to replace the NsiI fragment of pDD355, a plasmid containing GAL1-SLA2. In the above cloning procedures, when necessary, pBluescript, which contains no NsiI sites, was used as the shuttle vector. Primer SY23 (5′-GCACAAATGCAGCCGGACTTCGATGCCATATTGGAAAGCGGTATC-3′) was used with the Clontech Transformer Mutagenesis kit (Clontech, Palo Alto, CA) to delete Sla2p codons 360–576, generating pDD371 (sla2Δ360–575 in pRS313) and pDD372 (sla2Δ360–575 in YEP430).
Two-Hybrid Interactions
Residues 503–968 of Sla2p were fused to the Gal4p DNA-binding domain by cloning the BamHI fragment of pDD356 into pAS1-CYH2 (Durfee et al., 1993) (to generate pDD373). Residues 768–968 of Sla2p were fused to the Gal4p DNA-binding domain by cloning the HincII–RsaI SLA2 fragment into pAS1-CYH2 (to generate pDD374). SY11 (5′-CGGGATCCTGCAGTTAAAGGTG-3′) and SY12 (5′-GCGGATCCTCAGTCAACACGTAAGTG-3′) were used as primers in a PCR reaction to create BamHI sites flanking Sla2p residues 502–767, and the resulting fragment was cloned into pACTII (Durfee et al., 1993) (which contains the Gal4p activation domain) to generate pDD375. SY13 (5′-CGCCATGGATGCCATATTGGAAAGCGG-3′) and SY12 were used as primers in a PCR reaction to create NcoI and BamHI sites flanking Sla2p residues 575–767. The resulting fragment was cloned into pACTII to generate pDD376. SY14 (5′-CGCCATGGTTGCCACTGCTGAC-3′) and SY15 (5′-GCGGATCCTTATTCGGATGTGAAGTCCAA-3′) were used as primers in a PCR reaction to create NcoI and BamHI sites flanking Sla2p residues 751–930. The resulting fragment was cloned into pAS1-CYH2 to generate pDD377. SY29 (5′-CGCCATGGATTCAGATCTGCAGAAAGCG-3′) and SY31 (5′-GCGGATCCTCAGGCATCCAGAATAGGG-3′) were used as primers in a PCR reaction to create NcoI and BamHI sites flanking Sla2p residues 5–576. The resulting fragment was subcloned into pACTII to generate pDD379 or into pAS1-CYH2 to generate pDD378. All PCR products were sequenced to confirm the absence of any Taq polymerase-generated errors or were replaced with plasmid DNA. The plasmids were transformed into yeast strain Y190 (Durfee et al., 1993). Immunoblotting of whole-cell extract from these strains, using either anti-Sla2p antibody or anti-hemagglutinin antibody (both pACTII and pAS1-CYH2 contain a hemagglutinin tag), confirmed the expression of a fusion protein of the expected size. To determine whether the different proteins can interact with each other, pairwise combinations of the plasmids were transformed into Y190. This strain contains the Escherichia coli lacZ gene and the HIS3 gene under control of the GAL1 promotor. If the proteins bind to each other, the cells can grow in media (His−/3-AT) lacking histidine and containing 25 mM 3-aminotriazole, and β-galactosidase is expressed. The plasmids pAS1-CYH2, pACTII, pSE1111, and pSE1112, and the yeast strain Y190 were kindly provided by Steve Elledge (Baylor College of Medicine, Houston, TX). The actin 2-hybrid plasmids were kindly provided by David Amberg (State University of New York, Health Science Center, Syracuse, NY).
RESULTS
Immunolocalization of Sla2p
The genetic interactions and morphological defects of the sla2 mutants show that Sla2p is necessary for the correct functioning of the cortical actin cytoskeleton. To determine the cellular localization of Sla2p, we raised polyclonal antisera against its N terminus (aa 90–547) and its C terminus (aa 664–968). Both antisera recognize a 116-kDa protein that is absent in sla2Δ extracts (Figure 2A), and both produce similar immunofluorescence staining patterns (below).
In wild-type cells, Sla2p localizes to cortical patches at the growing surfaces of the cell (Figure 2). The Sla2p staining pattern, however, is distinct from that of actin. First, although staining is detectable in medium- and large-budded cells, the staining is strongest in unbudded and small budded cells. Second, Sla2p colocalizes with the actin cortical patches, but it appears that not every actin cortical patch contains Sla2p (Figure 2B). Finally, Sla2p appears to be present in some cortical patches independent of actin. This is most apparent in medium- and large-budded cells, in which only ∼45% of Sla2p patches overlap with actin patches, compared with an 85% overlap in unbudded and small-budded cells. Similarly, only ∼40% of actin patches in medium- and large-budded cells also contain Sla2p, compared with 85% in unbudded and small-budded cells. Thus, a subset of the actin patches contains Sla2p, and a subset of the Sla2p patches contains actin. The latter observation is consistent with the ability of Sla2p to achieve a partial polarized cortical localization in cells lacking filamentous actin (Ayscough et al., 1997).
Furthermore, we also observed that known actin cortical patch components (i.e., Abp1p, Sac6p, and cofilin) colocalize 100% with cortical actin, and their localization is unchanged in sla2Δ mutants (our unpublished results).
Effects of Sla2p Deletion and Truncation Mutations on Cell Growth Properties
Sla2p shares with its homologues in other organisms an extensive, internal, predicted coiled-coil region. In addition, its C terminus is similar to that of talin and contains an actin-binding site (McCann and Craig, 1997). Previously, Wesp et al. (1997) determined that only the N terminus of Sla2p is absolutely essential for growth at high temperatures, for endocytosis, and for correct actin cytoskeleton organization. In strains carrying mutations in ABP1 or SRV2, the large central coiled-coil domain of Sla2p becomes essential for the above-listed functions as well.
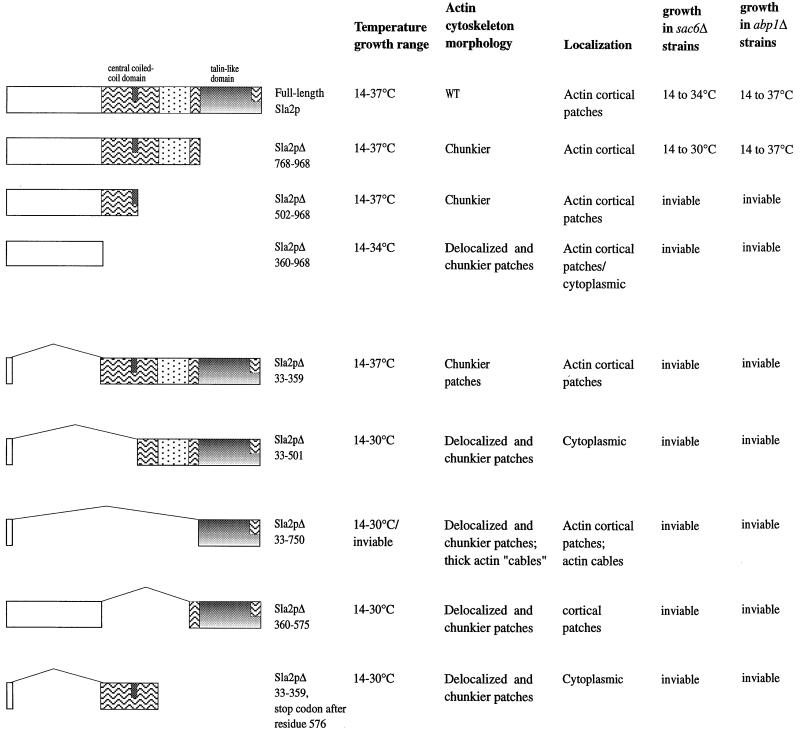
The in vivo roles of the various Sla2p domains, however, have yet to be fully explored. To further identify and clarify the roles of Sla2p domains, we have produced eight new truncation and deletion mutants of SLA2 (summarized in Figure 3) and have tested these for their ability to complement the various phenotypes caused by a complete deletion of SLA2. Our analysis supports conclusions reached by Wesp et al. (1997) but also provides novel insights, which lead us to propose a revised model for Sla2p function.
Figure 3.
Expression and temperature growth profile of Sla2p deletion proteins. (A) C-terminal deletions. Crude extracts were prepared from wild-type (full-length Sla2p; DDY426) and mutant cells containing the Sla2p C-terminal deletions Sla2Δ768–968 (DDY1194), Sla2Δ501–968 (DDY1195), and Sla2Δ360–968 (DDY1196), grown at 25°C, and then shifted to 37°C for 3 h. Equal cell equivalents (based on A600 absorbance) were immunoblotted using the N-terminal Sla2p antibody. Cells expressing wild-type or Sla2p C-terminal deletion mutants were then replica plated and grown for 36 h (37 and 34°C), 48 h (30 and 25°C), 72 h (20°C), or 5 d (14°C) on YPD plates before being photographed. (B) N-terminal and central coiled-coil domain deletions. Crude extracts were prepared from wild-type (DDY426) cells containing pRS313 (empty vector) and sla2Δ (DDY540) cells containing pDD367 (Sla2Δ33–359), pDD364 (Sla2Δ33–501), pDD362 (Sla2Δ33–750), pDD371(Sla2Δ360–575), pDD369(Sla2Δ33–359, 576stop), or vector alone, grown at 25°C, and then shifted to 37°C for 3 h. Equal cell equivalents from the 25°C cultures or the 37°C-shifted cultures were immunoblotted using the C-terminal Sla2p antibody, except for extracts containing pDD369, in which case the N-terminal Sla2p antibody was used. Wild-type and sla2Δ cells containing vector alone or expressing these Sla2p deletion mutants were then replica plated and grown for 2 d (37 and 34°C), 3 d (30 and 25°C), 5 d (20°C), or 8 d (14°C) on SC-his plates before being photographed as shown. The first spot of each panel represents growth with a low-copy-number plasmid, and the second spot represents growth with a high-copy-number plasmid. (C) Sla2Δ33–750 overexpression is recessive-lethal. Wild-type and sla2Δ cells expressing either Sla2Δ33–750 under the control of the galactose-inducible GAL1 promotor or containing vector alone (pYES-R) were grown on glucose- or galactose-containing media at 25 or 37°C for 72 h before being photographed. (1) Wild-type cells expressing vector only (pYES-R). (2) Wild-type cells containing galactose-inducible Sla2Δ33–750. (3) sla2Δ cells containing vector only. (4) sla2Δ cells expressing galactose-inducible Sla2Δ33–750.
First, the importance of Sla2p’s C terminus was explored by creating three Sla2p C-terminal truncation mutants (Figure 3A), each of which was integrated into the genome. Sla2pΔ768–968 expresses residues 1–767 and lacks the entire talin homology region. This mutant is almost identical to the sla2Δtalin mutant described by Wesp et al. (1997), which expresses residues 1–766. Sla2pΔ501–968 expresses residues 1–500, a region that includes two-thirds of the predicted large coiled-coil region and terminates at the end of a predicted leucine zipper motif. Sla2pΔ360–968 expresses residues 1–359 and lacks this predicted coiled-coil region altogether. Immunoblots performed on yeast extracts generated from strains expressing the different C-terminal truncation mutants showed that each expressed a protein of approximately the predicted size (Figure 3A).
Although sla2Δ mutant cells are unable to grow at 34°C or above (Figure 3B, vector alone) all of the C-terminal truncation mutants were able to grow at 34°C. In addition, only cells carrying the largest truncation, sla2Δ360–968, failed to grow at 37°C (Figure 3A). These results suggest that the N-terminal third of Sla2p is sufficient to fulfill a critical role of Sla2p and perhaps represents the essential functional region of Sla2p, consistent with the observations of Wesp et al. (1997).
To delineate further the functionality of the N-terminal region of Sla2p, three Sla2p N-terminal deletion mutations were generated (Figure 3B). Sla2pΔ33–750 comprises almost solely the talin-like domain. Sla2pΔ33–501 contains the talin-like domain and the 250 amino acids preceding it. Sla2pΔ33–359 contains both the extensive internal coiled-coil region and the talin-like domain but lacks the N-terminal third of the protein. These Sla2p deletion mutants were introduced into sla2Δ mutant cells on either low- or high-copy plasmids and were tested for expression (Figure 3B).
One of the three Sla2p N-terminal deletion mutants, Sla2pΔ33–359, rescued the temperature sensitivity of the sla2Δ cells (Figure 3B). Thus, the N terminus is not absolutely required for Sla2p functionality. A less severe deletion (of residues 114–284) did not support growth in the hands of Wesp et al. (1997). The reason for this discrepancy is unknown (possibilities discussed below). Overexpression of Sla2pΔ33–750, the talin-like domain, not only failed to support growth at the higher temperatures, but it seemed to have a deleterious effect on growth, in contrast to the overexpression of full-length Sla2p, which has no effect on cell growth (our unpublished results). To characterize this deleterious effect further, the Sla2pΔ33–750 deletion protein was expressed from the powerful GAL1 promoter (pDD366). Overexpression of the talin-like domain in sla2Δ cells was lethal, even at 25°C (Figure 3C). Wild-type cells transformed with pDD366 or plasmid vector alone, however, showed no obvious difference in their ability to grow on galactose, showing that this effect is recessive. Additional effects of overexpression of the talin-like domain are discussed below.
In the experiments described above, only a portion of the large predicted coiled-coil domain (residues 360–500), as well as the first 32 Sla2p residues, were found to be indispensable for growth at high temperatures. To test the importance of the central coiled-coil domain in Sla2p function directly, two additional deletion mutants were generated. Sla2pΔ360–575 lacks the entire predicted coiled-coil domain, whereas Sla2pΔ33–359,576stop comprises almost solely the predicted coiled-coil domain. These mutants were again expressed in sla2Δ cells on either low- or high-copy plasmids and were tested for expression (Figure 3B). We found that sla2Δ cells expressing the various constructs had growth characteristics similar to those of cells carrying vector alone (Figure 3B). Intriguingly, the corresponding coiled-coil deletion of Wesp et al. (1997), which removes residues 376–573, was able to complement most of the sla2 mutant phenotype. This result is further discussed below.
Effects of Sla2 Deletion Mutations on Cell Morphology and Actin Organization
The relative ability of different Sla2p mutants to rescue the temperature-sensitive growth of sla2Δ mutant cells is a sensitive measure of function. However, the evaluation of growth properties alone does not provide insight into the nature of the underlying defects in cells expressing the Sla2p mutants. Therefore, we next examined the effects of sla2 mutations on actin cytoskeleton organization. The data presented below indicate that Sla2p is capable of mediating the polarization of actin patches in the absence of either the C-terminal 468 residues or the N-terminal 360 residues (or, more specifically, residues 33–359). Effects on the actin cytoskeleton thus parallel effects on growth at high temperatures, suggesting that the ability to develop polarized actin cytoskeleton organization is required for growth at high temperatures.
The actin cytoskeletons of cells expressing the different Sla2p deletion mutants were visualized after growth at 25 and 37°C. The phenotypes were generally more severe at 37°C (our unpublished results). Cells grown at 25°C are shown in Figure 4, and results are summarized in Figure 5. sla2Δ mutant cells expressing Sla2pΔ360–968 (Figure 4D), Sla2pΔ360–575 (Figure 4H), Sla2pΔ33–501 (Figure 4F), or Sla2pΔ33–359,576stop (Figure 4I) all exhibited morphological and actin phenotypes similar to those observed for the null mutant. These cells were round, they exhibited an increased number of cortical actin patches, and their patches were not restricted to the bud.
Figure 4.
Actin cytoskeleton organization in sla2Δ cells and cellular localization of the Sla2 deletion mutant proteins. The yeast strains are those described in Figure 3, A and B, with all plasmids being low copy number (CEN), except for pDD368, which is a high-copy-number (2 μ) plasmid expressing Sla2Δ33–359. Cells were grown at 25°C and were then fixed and stained with either anti-actin antibodies, or anti-Sla2p antibodies, as indicated. Cells expressing Sla2pΔ360–968, Sla2pΔ501–968, and Sla2pΔ33–359,576stop were stained with the N-terminal Sla2p antibody. The remaining cells were stained with the C-terminal Sla2p antibody.
Figure 5.
Sla2p domain analysis summary. The phenotypes of the Sla2p C-terminal truncation and N-terminal deletion mutants are summarized. The schematics indicate the regions of Sla2p expressed in each case and reflect the domain arrangement shown in Figure 1A. The phenotypic analysis was performed in sla2Δ cells expressing the different Sla2p mutants, from either a genomic copy (the three C-terminal mutants were integrated into the genome) or from a high- or low-copy-number plasmid (the five N-terminal deletion mutants).
sla2Δ mutant cells expressing Sla2pΔ501–968 (Figure 4C) or Sla2pΔ33–359 (Figure 4E) were also round, but they exhibited largely polarized cortical actin patches. These patches did look “chunkier” (i.e., larger and less discrete) than those in wild-type cells. When these mutant cells were grown at 37°C, they still retained a polarized actin cytoskeleton, but the actin patches became even chunkier (our unpublished results).
sla2Δ cells expressing Sla2pΔ768–968, lacking the talin-like domain, do not exhibit any obvious defects in actin organization at 25°C (Figure 4B). These cells retain a polarized actin cytoskeleton at 37°C but exhibit fainter cables and chunkier cortical patches (our unpublished results). On the other hand, expression of the talin domain alone (Sla2pΔ33–750) not only failed to restore wild-type morphology to sla2Δ cells but resulted in a more abnormal actin cytoskeleton (Figure 4G). These cells seem to contain more cortical actin, and some cells also contain actin “cables,” which appear much thicker than those seen in wild-type cells. The exaggerated actin cables could be stained by rhodamine–phalloidin (our unpublished results), indicating that they contain filamentous actin. Thus, they are different from the actin bars that are found in some actin cytoskeleton mutants and that are thought to consist of aggregates of monomeric or denatured actin. Effects of expression of the talin-like domain alone on actin organization were seen in wild-type background and therefore are dominant.
sla2Δ cells exhibit an abnormal budding pattern (Yang et al., 1997). Of the eight mutants, only Sla2pΔ768–968 restored the wild-type bipolar budding pattern in MATa/MATα cells. Again, no dominant effects were observed.
Endocytosis in sla2 Mutants
We tested the ability of the sla2 truncation and deletion mutants to undergo fluid phase endocytosis as assayed by the ability to take up LY an concentrate it the vacuole. The ability of the sla2 mutants to endocytose closely followed their ability to grow at high temperatures. Cells expressing Sla2pΔ768–968, Sla2pΔ501–968, or Sla2pΔ33–359 were able to endocytose LY at both 25 and 37°C, whereas cells expressing the five remaining mutants were unable to do so even at 25°C. Additionally, overexpression of the five plasmid-based sla2 mutants in wild-type cells did not affect their ability to endocytose LY; therefore, the mutants did not confer any dominant effects.
Cellular Localization of the Sla2p Deletion Proteins
The generation of polyclonal antibodies capable of detecting Sla2p localization by indirect immunofluorescence allowed us to determine the effect of the eight sla2 mutations on the cellular localization of Sla2p. Double labeling with Sla2p and actin antibodies was performed for each mutant and is shown in Figure 4.
Sla2pΔ768–968 (Figure 4B) exhibited a localization indistinguishable from that of full-length Sla2p. sla2Δ mutants expressing either Sla2pΔ501–968 (Figure 4C) or Sla2pΔ33–359 (Figure 4E) contain chunkier actin cortical patches near or in the bud, and the mutant Sla2p colocalizes with these patches. Although there seems to be more Sla2pΔ360–968 in the cytoplasm, this protein is still present in cortical patches but at apparently reduced levels (Figure 4D). Sla2pΔ360–575 (Figure 4H) still localizes to cortical patches, but these patches were evenly spread over both mother and bud, and, intriguingly, many seem not to contain actin. Sla2pΔ33–750 (Figure 4G) colocalizes with actin cortical patches and the aberrant thick actin cables. The remaining two deletion proteins, Sla2pΔ33–501 (Figure 4F) and Sla2pΔ33–359,576stop (Figure 4I) both exhibited cytoplasmic localization and not cortical patch localization.
Full-length Sla2p displays partially polarized cortical localization in cells treated with the actin depolymerizing drug latruculin A (latA) (Ayscough et al., 1997). We assayed the localization pattern of the three Sla2p C-terminal truncation mutants (Sla2pΔ768–968, Sla2pΔ501–968, and Sla2pΔ360–968) when cells were grown in the presence of latA. All three mutants displayed some partially polarized cortical localization (our unpublished results).
Thus, both the N-terminal 360 residues and the C-terminal talin-like domain of Sla2p are capable of localizing to cortical actin patches independent of the rest of the protein. In addition, the N-terminal third of Sla2p seems to achieve its localization independently of filamentous actin, whereas evidence discussed below points to actin as the localization signal for the C-terminal talin-like domain.
Synthetic Lethality with abp1Δ and sac6Δ
sla2Δ is synthetic lethal with abp1Δ and sac6Δ mutants (Holtzman et al., 1993). We tested the sla2 deletion and truncation mutants for synthetic lethality with either abp1Δ or sac6Δ mutations. Seven of the eight deletion mutants were synthetic lethal with both abp1Δ and sac6Δ, the exception being sla2Δ768–968, which was not synthetic lethal with either abp1Δ or sac6Δ. The sla2Δ768–968 abp1Δ double mutant exhibited no growth defects. However, the sla2Δ768–968 sac6Δ double mutant exhibited a narrower temperature range permissive for growth than that of the sac6Δ single mutant. Although the sac6Δ mutant can grow at 34°C, the sla2Δ768–968 sac6Δ double mutant was unable to grow at 34°C and grew only poorly at 30°C. This synergistic genetic interaction is striking, because the sla2Δ768–968 single mutant is very healthy and exhibits few discernible growth defects. Thus, the functionality provided by both the C-terminal and the N-terminal regions of Sla2p becomes important for viability in the absence of either Abp1p or Sac6p, with the talin-like domain being only partially dispensable.
Interaction of Sla2p Domains
The Sla2p talin-like domain (expressed and purified from E. coli) binds to actin in vitro (McCann and Craig, 1997) and is similar to a domain of talin that binds to actin (Hemmings et al., 1996). Therefore, it is not surprising that Sla2pΔ33–750, which encompasses the talin-like domain, localizes to actin structures in vivo. What is harder to explain is the cytoplasmic localization of Sla2pΔ33–501, which contains the entire talin-like domain plus the 250 amino acids N-terminal to this domain. The presence of the additional 250 amino acids somehow precludes binding of the talin-like domain to the actin cytoskeleton. This observation could be explained if the 250–amino acid region directly binds to the talin homology region, thereby occluding its actin localization signal. An alternative possibility is that the mutant protein aberrantly binds to another protein, which then obstructs the actin-binding signal in the talin homology region. Because we had observed that Sla2p residues 503–968 expressed and purified from E. coli do not bind to actin in vitro (our unpublished results), the self-interaction hypothesis was tested.
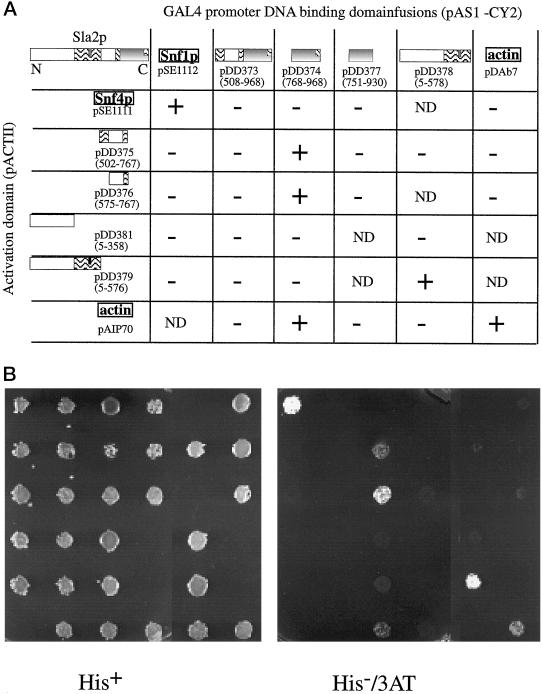
The two-hybrid system was used to test whether different domains of Sla2p could interact with each other. The following Gal4p DNA-binding domain fusions were made: Sla2p residues 503–968 (pDD373), 768–968 (pDD374), 751–930 (pDD377), and 5–576 (pDD378). The following Gal4p activation domain fusions were also created: Sla2p residues 502–767 (pDD375), 575–767 (pDD376), 5–358 (pDD381), and 5–576 (pDD379). Interactions with actin were also tested using actin fusion proteins. A pair of protein chimeras known to interact with each other, SNF1 (pSE1112) and SNF4 (pSE1111), were used as a positive control (Celenza et al., 1989). All the pair-wise plasmid combinations were tested to determine whether they could activate HIS3 expression (Figure 6) and also activate lacZ expression (our unpublished results).
Figure 6.
Sla2p two-hybrid interactions. Yeast strain Y190 containing the indicated plasmids was replica plated either onto medium containing histidine (His+ plates), or onto medium lacking histidine and containing 3-aminotriazole (His−/3AT plates). If the DNA-binding domain fusion protein and DNA activation domain fusion protein interact with each other, the cells are able to grow on media lacking histidine. The Snf1p-Snf4p pair serves as a positive control. Plates were scored for growth after 2–3 d of growth at 25°C. Positive interactions were also verified by detection of lacZ expression (our unpublished results).
Of the internal Sla2 interactions tested, only Sla2768–968 in combination with Sla2502–767 or Sla2575–767, as well as Sla25–578 in combination with Sla25–578, were positive (Figure 6, A and B). Note that amino acids 768–968, but not amino acids 503–968, interacted with amino acids 502–767. This suggests that the binding site for the Sla2502–767 fusion present in Sla2768–968 is occluded in Sla2503–968. Sla2503–968 is similar to the deletion mutant Sla2pΔ33–501, which exhibits cytoplasmic localization in vivo despite containing an actin-binding site. These observations are consistent with the hypothesis that the site in Sla2p that mediates localization to actin is masked in Sla2pΔ33–501 by an intra- or intermolecular interaction.
Furthermore, although Sla2751–930 in combination with Sla2575–767 did not activate HIS3 expression, Sla2768–968 in combination with Sla2575–767 was able to do so (Figure 6). Therefore, the predicted coiled-coil forming residues 930–968 are essential for the association of the talin-like domain with residues 575–767, which themselves contain a second region of predicted coiled-coil that is evolutionarily conserved (Figure 1).
We also tested the ability of the various Sla2p domains to interact with actin in the two-hybrid assay and found that only the talin-like domain (Sla2768–968) was capable of doing so. In addition, residues 930–968 (a predicted coiled-coil region) were essential for this interaction. This result agrees with the in vitro results of McCann and Craig (1997), who found that residue 957 is essential for actin binding. Thus, the same predicted Sla2p coiled-coil region that is necessary for actin binding is also necessary for self-association. No other region of Sla2p, including the construct containing residues 503–968, was capable of interacting with actin in our assay.
Finally, we observed that Sla2p fragment 1–575 could interact with itself, and that residues 360–575, the central predicted coiled-coil region, are essential for this interaction. A clone containing this coiled-coil domain alone was also retrieved from a two-hybrid library when residues 5–576 of Sla2p were used as bait (our unpublished results).
Evidence That Sla2p Is a Dimer In Vivo
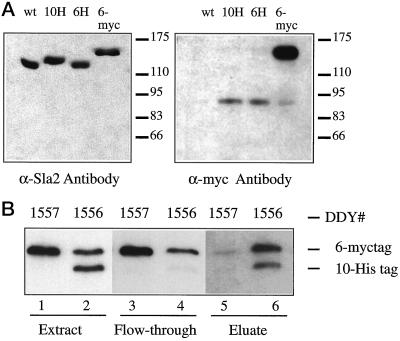
Wesp et al. (1997) demonstrated that the central coiled-coiled domain of Sla2p mediated the formation of a complex that sediments at ∼220 kDa. This complex might have resulted from dimerization of Sla2p or from the binding of another protein(s) to Sla2p. Our two-hybrid analysis showed that this coiled-coil domain is capable of interacting with itself, suggesting that the 220-kDa complex might be an Sla2p dimer. To test this result biochemically, Sla2p was tagged with either multiple histidines or multiple myc epitopes (see Figure 7A). Extracts were prepared from diploid yeast expressing either one copy of 10-His-Sla2p and one copy of 6-myc-Sla2 (DDY1156) or two copies of 6-myc-Sla2 (DDY1157). From these strains, the His-tagged Sla2p was isolated on a Ni-agarose column (see Figure 7B). Membrane-extracted 10-His-Sla2p was found to be associated with 6-myc-Sla2p, indicating that the two tagged versions of Sla2p form a complex with each other in vivo. A similar result was obtained with Sla2p extracted from the cytoplasm. This result, together with the evidence described above, strongly suggests that Sla2p dimerizes in a manner mediated by its central coiled-coil domain.
Figure 7.
Sla2p intramolecular interaction in vivo. (A) Haploid yeast expressing Sla2p, tagged with multiple histidines, or multiple myc epitopes, were extracted as described in MATERIALS AND METHODS, and the supernatants from high-speed centrifugations were subjected to SDS-PAGE. The proteins were transferred to nitrocellulose and probed with either anti-Sla2p (C-terminal antibody) or anti-myc antibodies, as shown. The anti-myc antibody detects only the myc-tagged Sla2p and a background band at ∼90 kDa. (B) Heterozygous diploid 6myc-SLA2/10His-SLA2 yeast (DDY1556) and homozygous diploid 6myc-SLA2/6myc-SLA2 yeast (DDY1557) were extracted as described in MATERIALS AND METHODS. Solubilized membrane extract was applied to a Ni-agarose column, which binds the histidine-tagged Sla2 (lane 6) but not the myc-tagged Sla2 (lane 5). The Western blot was probed with anti-Sla2 antibodies. When His-tagged Sla2 from the heterozygous yeast binds to the column (and is thus lost from the flow-through; lane 4), myc-tagged Sla2 is brought with it (lane 6), indicating that Sla2 is capable of complexing with itself in vivo.
Fractionation of Sla2p and Sla2p N-terminal Deletion Mutants
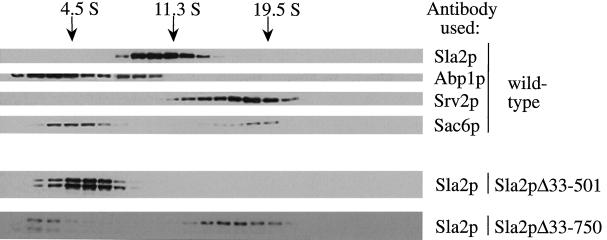
We determined the fractionation profile of some of the actin-binding proteins encoded by genes that when mutated exhibit synthetic lethality with sla2 mutants (see Figure 8). We performed this analysis to test the possibility that these proteins exist in complexes with Sla2p. Abp1p eluted in fractions ranging from ∼4.5 S (65 kDa) to 11.3 S, with more protein present in the earlier fractions. Srv2p eluted in fractions ranging from 11.3 to 19.5 S (670 kDa), with more protein present in the later fractions. Sac6p eluted as two distinct peaks, one at 4.5 S and the other at 19.5 S. Because Sla2pΔ33–750 (a protein that associates with actin patches and cables) and Sla2pΔ33–501 (a protein with cytoplasmic localization) showed vastly different cellular localizations when localized by immunofluorescence (Figure 4), the distribution of these mutant proteins was also analyzed by velocity sedimentation (see Figure 8) and gel filtration (our unpublished results).
Figure 8.
Sedimentation behavior of Sla2p and other cytoskeletal proteins. Velocity sedimentation was performed on a wild-type yeast extract using a 3–30% sucrose gradient as described in MATERIALS AND METHODS section. Fractions were immunoblotted with the indicated antibodies. (For Sla2p, the C-terminal antibody was used.) Where indicated, extracts were derived from sla2Δ cells expressing either Sla2pΔ33–750 or Sla2pΔ33–501. The size standards shown are BSA (4.5 S), catalase (11.3 S), and thyroglobulin (19.5 S).
The presence or absence of full-length Sla2p had no effect of the fractionation properties of either deletion mutant (our unpublished results). Sla2pΔ33–501 sedimented at 4.5 S (66 kDa), in accordance with the size of the monomeric protein (see Figures 3 and 8). Intriguingly, although a portion of Sla2pΔ33–750 sedimented below 4.5 S (as expected for its monomeric size), most of this protein was found at 19.5 S, suggesting that it is in a large, ∼670-kDa complex. (see Figure 8). Thus, Sla2pΔ33–750 is present in a significantly larger complex than the one containing full-length Sla2p (220 kDa). As described above, two other actin-binding proteins, Sac6p and Srv2p, also sediment at 19.5 S.
DISCUSSION
Yeast cells lacking Sla2p have previously been demonstrated to contain a defective actin cytoskeleton, wherein the cortical actin patches are no longer concentrated at the surface of the bud (Holtzman et al., 1993). These cells also exhibit defects in actin-mediated functions, such as endocytosis (Raths et al., 1993), the bipolar budding pattern (Yang et al., 1997), and abnormal post-Golgi secretory vesicle accumulation (Mulholland et al., 1997).
In this study, we demonstrate that Sla2p is a component of the cortical actin cytoskeleton. Sla2p localizes to only a subset of the actin cortical patches, as well as to cortical patches apparently free of actin (Figure 2), despite containing an actin-binding site (McCann and Craig, 1997). This is in contrast to previously characterized actin-binding proteins, such as Abp1p or cofilin, which are present in all actin patches and absent from patches devoid of actin (Drubin et al., 1988; Moon et al., 1993). (These proteins also maintain their actin patch localization even in sla2Δ mutant cells [our unpublished results].) An additional difference is that these actin-binding proteins (Abp1p and cofilin) became localized to the cytoplasm when cells were treated with the actin-depolymerizing drug latA (Ayscough et al., 1997). In contrast, Sla2p still exhibited cortical localization, and this localization appeared appropriately polarized to one end of the cell in ∼20% of the latA-treated cells (Ayscough et al., 1997). This and additional observations discussed below suggest that Sla2p interacts with cortical proteins in addition to actin.
The colocalization of Sla2p with actin is most evident early in the cell cycle, in unbudded and small-budded cells, suggesting that its interaction with actin might be especially important during these stages and that its localization and functionality is regulated during the cell cycle. This suggestion is reinforced by the observation that Sla2p is localized almost exclusively to the cytoplasm in stationary phase yeast cells (our unpublished results). A number of hypotheses to explain the observation that Sla2p patches do not always contain actin can be posited. First, Sla2p may play a regulatory role in actin cortical patch formation. For example, Sla2p might help nucleate actin polymerization or recruit and stabilize the correct components of cortical patches as they initially form; thus the patches containing Sla2p but no detectable actin may mark the future sites of actin patch formation. This hypothesis is supported by the observation that sla2Δ mutants are defective in nucleation of actin assembly in permeabilized cells (Li et al., 1995). Alternatively, Sla2p may remain behind after an actin patch has been disassembled. A third possibility is that the cortical patches that contain Sla2p alone (compared with those that contain both Sla2p and actin) constitute separate functional entities. Further studies using differentially colored green fluorescent protein–tagged fusions of Sla2p and actin in living cells might shed light on this issue.
To elucidate first the function of Sla2p in actin assembly and distribution and second to investigate the regulation of Sla2p distribution, we generated eight Sla2p truncation or deletion mutants (Figures 3 and 5). Previously, Wesp et al. (1997) had also generated and characterized some Sla2p deletion mutants. They reported that only the N-terminal domain (specifically residues 114–284) is required for growth at 37°C, for endocytosis, and for polarized actin. In addition, a portion of the predicted central coiled-coil domain of Sla2p becomes necessary for the above activities when Srv2p is absent from the cell or when the Src homology 3 domain of Abp1p is deleted. In contrast, the C-terminal talin-like domain of Sla2p was dispensable under all conditions tested. We have created a different set of deletion proteins (the only overlapping one is Sla2pΔ768–968) and subjected them to a complementary set of analyses, including actin localization, Sla2p localization, and two-hybrid interactions. These experiments have produced novel insights regarding the regulation and important functional regions of the protein.
We found that Sla2p missing large portions of either its N- or C-terminal domain was still largely functional. Both Sla2pΔ501–968 and overexpressed Sla2pΔ33–359 could fully rescue the temperature sensitivity of sla2Δ cells and restore the polarized placement of the cortical actin patches and endocytosis (Figures 3 and 4). (Unlike full-length Sla2p, these mutants did not allow growth in the absence of Sac6p or Abp1p and did not restore the bipolar budding pattern.) These Sla2p mutants colocalized with the cortical actin patches (Figure 4). Thus, both the N- and C-terminal domains of Sla2p contain a cortical patch localization signal, and this localization is probably key to the functionality of these Sla2 deletion proteins. In addition, the N terminus of Sla2p seems to localize to the cell cortex independently of actin, because this domain still exhibits partially polarized cortical localization in cells lacking filamentous actin.
A closer examination of the indispensable functional regions of Sla2p demonstrates that there is a surprising amount of redundancy within Sla2p. As stated above, we observed that either the N or C terminus (but not both) of Sla2p can be removed without greatly perturbing its functional activity. These two deletion mutants have in common the central predicted coiled-coil region (as well as the first 32 residues). Although we found that a Sla2p deletion mutant entirely lacking this domain (Δ360–575) exhibited no functional activity, Wesp et al., (1997) characterized a slightly less complete deletion of the predicted coiled-coil domain (Δ376–573) and found that it was capable of complementing most of the sla2 null mutant’s deficiencies. The different boundaries likely contributed to the different functionalities of the deletion proteins and could also explain the discrepancy between our observations and those of Wesp et al. (1997) concerning the importance of the N terminus of Sla2p.
Because very few residues of Sla2p were present in all of our active Sla2p deletion proteins, the results of the Sla2p domain analysis suggest that the protein comprises several functional modules. Removal of any one portion causes relatively little perturbation, but more extensive deletion leads to loss of function. The basic requirements for Sla2p function would appear to be 1) a cortical localization signal and 2) an additional domain, perhaps for contacting another constituent of the cortical patches.
In addition, every deletion mutation we made resulted in some loss of function. For example, we observed that removal of the C-terminal talin homology region of Sla2p had little effect on the actin cytoskeleton or cell growth under optimal conditions (Figures 3 and 4), consistent with the results reported by Wesp et al., (1997). At 37°C, however, we observed that the actin cytoskeleton had fainter cables and chunkier cortical patches if the talin-like domain was deleted (our unpublished results). In addition, this truncation mutation showed a synthetic negative synergism with a sac6Δ mutant (causing inviability at 34°C). Intriguingly, expression of the C-terminal talin homology region (Sla2pΔ33–750) by itself resulted in a dominant phenotype. Cells appeared to exhibit more filamentous actin structures, larger and more abundant actin cortical patches, and thick actin cables (Figure 4). Thus, the Sla2p talin homology domain may promote actin polymerization or stabilize F-actin structures.
Consistent with the observations that the Sla2p talin-like domain pellets with F-actin (McCann and Craig, 1997), we found that this domain interacts with actin in the two-hybrid system (Figure 6). However, as assayed through the two-hybrid system, no other region of Sla2p exhibited an interaction with actin, supporting the view that Sla2p makes actin-dependent and independent interactions with the cell cortex. Furthermore, a construct containing an additional 265 amino acids N-terminal of the talin-like domain did not interact with actin, suggesting that the actin-binding site may be masked, somehow, in this construct.
High overexpression of the Sla2p talin-like domain results in death in sla2Δ cells. In contrast, overexpression of full-length Sla2p, or of a large C-terminal fragment of Sla2p (Sla2pΔ33–501), has no detectable dominant effect on cell growth or on the actin cytoskeleton. The excess Sla2p appears to be mainly cytoplasmic and not present in cortical patches, as judged by indirect immunofluorescence (our unpublished results). These results suggest that sequences N-terminal to the talin homology domain may be able to mask the talin-like region of Sla2p, through either a direct inter- or intramolecular interaction (see Figure 9). There are precedents in which intramolecular interactions affect the accessibility of functional domains in other cytoskeletal proteins, including vinculin, a component of focal adhesions, and ezrin, a membrane-cytoskeletal linking protein. The N-terminal domains of ezrin and vinculin bind to their C-terminal domains, thereby masking an F-actin–binding sites (Gary and Bretscher, 1995; Johnson and Craig, 1994, 1995). Phosphatidyl inositol bisphosphate, which is also thought to regulate cytoskeletal proteins, such as profilin (Lassing and Lindberg, 1985), can disrupt vinculin self-association, thus exposing its talin- and actin-binding sites (Gilmore and Burridge, 1996).
Figure 9.
Model of Sla2p regulation and interactions. Sla2p exists in the cytoplasm and associated with cortical actin patches. Localization to the cortex is probably necessary for activity. Based on the N-terminal deletion and C-terminal truncation analysis of Sla2p, both termini of the protein contain a cortical actin patch localization signal. The unknown proteins that mediate Sla2p’s cortical patch localization are denoted X and Y. The C terminus has been demonstrated to contain an actin-binding site, so Y might be actin. The large, central coiled-coil domain potentially interacts with other proteins (Z). Indeed, Rvs167p has been reported to bind this domain of Sla2p in two-hybrid studies (Wesp et al., 1997). Rvs167 binds to Abp1p, which itself binds to Srv2p and to actin. When expressed alone, however, the coiled-coil domain is cytoplasmic, and thus this link seems to be insufficient for Sla2p localization. Regulation of the distribution and function of Sla2p could potentially be accomplished through interaction with any of a number of proteins. We have shown, however, that the actin-binding site located in the C-terminal talin-like domain is likely to be inactivated through a self-association with the upstream small coiled-coil domain of the protein. This interaction may also be sufficient to block the N-terminal and coiled-coil domain binding sites for proteins X and Z, as shown ii, or additional switches and signals may be involved, as depicted in i.
In support for a model invoking regulation of Sla2p association with the cell cortex via intramolecular interaction, we found using the two-hybrid system that the C-terminal talin homology domain of Sla2p binds specifically to the 250-residue segment immediately upstream of it (Figure 6). This interaction is dependent on Sla2p residues 930–968 and on residues 567–767, both of which contain evolutionarily conserved domains that are predicted to form alpha-helical coiled coils. As mentioned above, residues 930–968 are also necessary for actin binding. Thus, Sla2p can self-associate, and this self-association is predicted to mask the actin-binding site (see Figure 9). How the Sla2p intramolecular (or intermolecular) interactions may be regulated has not yet been investigated. Sla2p is phosphorylated (our unpublished results), and this provides one potential means of regulation.
The large central coiled-coil region mediates Sla2p dimerization (Figure 7). Dimerization may be necessary for full functionality. In addition, the central coiled-coil domain might interact with other cytoskeletal proteins; this region has been shown to interact with Rvs167p using the two-hybrid system (Wesp et al., 1997). Specific interaction between the putative alpha-helical coiled-coil regions of one protein with another protein has been reported previously (Spencer et al., 1997). In addition, the Sla2p human homologue Hip1p binds to huntingtin, the protein whose mutation results in HD, and this interaction is mediated by the conserved central putative coiled-coil region of Hip1p (Kalchman et al., 1997).
Crude fractionation of yeast extract suggested that the majority of Sla2p is present in a large, membrane-associated complex (Figure 8). Fractionation of wild-type yeast extract on a sucrose gradient showed that full-length Sla2p sediments at 11.3 S, which corresponds to ∼220 kDa (assuming a globularly shaped protein). The talin homology region, however, sedimented at 19.5 S (670 kDa). Two other cortical cytoskeletal proteins, Sac6p and Srv2p, also sediment at 19.5 S. Intriguingly, null alleles of all three genes, SLA2, SAC6, and SRV2, are synthetically lethal with each other. Thus, the 19.5 S complex might contain all three proteins, providing internal functional redundancy, and represent a subunit or a building block of a “cortical patch.”
Sla2p presumably contacts other proteins to promulgate its effects (shown as X, Y, and Z in Figure 9). Because the N- and C-terminal regions are independently capable of localization to cortical patches, they both must interact with cortically located components. Furthermore, because the majority of Sla2p is localized to the cytoplasm in stationary (Go) cultures, or when Sla2p is overexpressed, negative regulation and/or the requirement for a positive signal must occur at both termini. From our results, we believe that the switch at the C-terminus involves inhibition of self-association mediated by the two small predicted coiled-coil domains of Sla2p. Whether this interaction also regulates the N-terminal binding is unknown; however, this is unlikely because in the absence of the C terminus, the protein still seems to be correctly localized and functional. It is possible that a number of signals and switches are involved. Additionally, the binding partners for Sla2p might themselves be targets of regulation, providing an indirect method of regulating Sla2p distribution.
ACKNOWLEDGMENTS
This work was supported by a grant to D.G.D. from the National Institutes of Health (GM-50399) and by a Human Frontier Science Program fellowship to M.J.T.V.C.
REFERENCES
- Amberg D, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Struct Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Ausubel F. Expression and purification of glutathione-S-transferase fusion proteins. In: Ausubel F, editor. Current Protocols in Molecular Biology. New York: Greene Publishing Associates; 1990. , 16.7. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. New York: Greene Publishing Associates–Wiley-Interscience; 1989. [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latruculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blader IJ, Cope MJTV, Jackson TR, Profit AA, Greenwood AF, Drubin DG, Prestwich GD, Theibert AB. GCS1, an Arf guanosine triphosphatase-activating protein in Saccharomyces cerevisiae, is required for normal actin cytoskeletal organization in vivo and stimulates actin polymerization in vitro. Mol Biol Cell. 1999;10:581–596. doi: 10.1091/mbc.10.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Eng FJ, Carlson M. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol Cell Biol. 1989;9:5045–5054. doi: 10.1128/mcb.9.11.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Miller KG, Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Mulholland J, Zhu ZM, Botstein D. Homology of a yeast actin-binding protein to signal transduction proteins and myosin-I. Nature. 1990;343:288–290. doi: 10.1038/343288a0. [DOI] [PubMed] [Google Scholar]
- Dulic V, Egerton M, Elguindi I, Raths S, Singer B, Riezman H. Yeast endocytosis assays. Methods Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associated with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Freeman NL, Chen Z, Horenstein J, Weber A, Field J. An actin monomer binding activity localizes to the carboxyl-terminal half of the Saccharomyces cerevisiae cyclase-associated protein. J Biol Chem. 1995;270:5680–5685. doi: 10.1074/jbc.270.10.5680. [DOI] [PubMed] [Google Scholar]
- Freeman NL, Lila T, Mintzer KA, Chen Z, Pahk AJ, Ren R, Drubin DG, Field J. A conserved proline-rich region of the Saccharomyces cerevisiae cyclase-associated protein binds SH3 domains and modulates cytoskeletal localization. Mol Cell Biol. 1996;16:548–556. doi: 10.1128/mcb.16.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary R, Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol Biol Cell. 1995;6:1061–1075. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore AP, Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4–5-bisphosphate. Nature. 1996;381:531–535. doi: 10.1038/381531a0. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies–A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. Immunizations; pp. 53–138. [Google Scholar]
- Hemmings L, Rees DJG, Ohanian V, Bolton SJ, Gilmore AP, Patel B, Priddle H, Trevithick JE, Hynes RO, Critchley DR. Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J Cell Sci. 1996;109:2715–2726. doi: 10.1242/jcs.109.11.2715. [DOI] [PubMed] [Google Scholar]
- Holtzman DA, Yang S, Drubin DG. Synthetic-lethal interactions identify two novel genes Sla1 and Sla2 that control membrane cytoskeleton assembly in Saccharomyces-cerevisiae. J Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. An intramolecular association between the head and tail domains of vinculin modulates talin binding. J Biol Chem. 1994;269:12611–12619. [PubMed] [Google Scholar]
- Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- Kalchman MA, et al. HIP1, a human homologue of S. cerevisiae Sla2p, interacts with membrane associated huntingtin in the brain. Nat Genet. 1997;16:44–53. doi: 10.1038/ng0597-44. [DOI] [PubMed] [Google Scholar]
- Lappalainen P, Kessels MK, Cope MJTV, Drubin DG. The ADF homology (ADF-H) domain: a highly exploited actin-binding module. Mol Biol Cell. 1998;9:1951–1959. doi: 10.1091/mbc.9.8.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature. 1985;314:472–474. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- Li R, Zheng Y, Drubin DG. Regulation of cortical actin cytoskeleton assembly during polarized cell growth in budding yeast. J Cell Biol. 1995;128:599–615. doi: 10.1083/jcb.128.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lila T, Drubin DG. Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein an the actin cytoskeleton. Mol Biol Cell. 1997;8:367–385. doi: 10.1091/mbc.8.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Muir RS, Lim E, McElver J, Weber SC, Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled-coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- McCann RO, Craig SW. The I-LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci USA. 1997;94:5679–5684. doi: 10.1073/pnas.94.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AL, Janmey PA, Louie KA, Drubin DG. Cofilin is an essential component of the yeast cortical cytoskeleton. J Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J, Wesp A, Riezman H, Botstein D. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretory vesicle. Mol Biol Cell. 1997;8:1481–1499. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na S, Hincapie M, McCusker JH, Haber JE. MOP2 (SLA2) affects the abundance of the plasma membrane H(+)-ATPase of Saccharomyces cerevisiae. J Biol Chem. 1995;270:6815–6823. doi: 10.1074/jbc.270.12.6815. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S, Dowbenko D, Cheng J, Li W, Brush J, Utzig S, Simanis V, Lasky LA. PSTPIP: a tyrosine phosphorylated cleavage furrow-associated protein that is a substrate for a PEST tyrosine phosphatase. J Cell Biol. 1997;138:845–860. doi: 10.1083/jcb.138.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanker EE, Rovira C, Scherzinger E, Hasenbank R, Walter S, Tait D, Colicelli J, Lehrach H. HIP-I: a huntingtin interacting protein isolated by the yeast two-hybrid system. Hum Mol Genet. 1997;6:487–495. doi: 10.1093/hmg/6.3.487. [DOI] [PubMed] [Google Scholar]
- Wesp A, Hicke L, Palecek J, Lombardi R, Aust T, Munn AL, Riezman H. End4p-S1a2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:2291–2306. doi: 10.1091/mbc.8.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Ayscough KR, Drubin DG. A role for the actin cytoskeleton of Saccharomyces cerevisiae in bipolar bud-site selection. J Cell Biol. 1997;136:111–123. doi: 10.1083/jcb.136.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]