Many bacterial pathogens have evolved hair-like extensions that project from their surfaces and enable adhesion to host tissues, formation of biofilms, motility, and even transfer of genetic material (1). Gram-positive bacteria use sortases, membrane-anchored transpeptidases, to cleave precursor proteins at cognate recognition motifs within C-terminal sorting signals and assemble pili in their cell wall envelope (2, 3). The transient product of this reaction is an acyl enzyme that, depending on the sortase involved, is relieved by a specific nucleophile (4). Currently studied sortases accept only one nucleophile to complete their transpeptidation reaction (5–8). For example, sortase A of Staphylococcus aureus forms an acyl enzyme with surface proteins bearing LPXTG motif sorting signals, which are subsequently relieved by the nucleophilic attack of the amino group of lipid II, the precursor of peptidoglycan synthesis in bacteria (9, 10). In this issue of PNAS, Mandlik et al. (11) provide new insight into sortases and their complicated recipes for pilus assembly.
Starting with simple recipes, some sortases make pili from two ingredients (12, 13). Bacillus cereus sortase D cleaves the sorting signal of pilin precursors and accepts the side chain ε-amino group of lysine (K) within the YPKN motif of the major pilin protein as its nucleophile (14). Products of this reaction are covalently polymerized pilin subunits, glued together by an amide bond between the C-terminal carboxyl group of one subunit and the side-chain amino group of the YPKN motif of another (14). Conservation of sortase D, as well as the YPKN motif and cell wall sorting signal of major pilin proteins in many different species, suggests that this pilus assembly reaction is universal in Gram-positive bacteria (14, 15). A simple recipe for pili therefore requires only one chef, sortase D, and two ingredients, BcpA and BcpB (12). Sortase D acyl intermediates are relieved by the nucleophilic attack of the BcpA YPKN motif, thereby generating a pilus with BcpB at the tip joined to a BcpA homopolymer (14). Pilus assembly is terminated when sortase A cuts BcpA and accepts its nucleophile, lipid II. Because BcpA can still be joined to the pilus via its YPKN motif, incorporation of lipid II at the C-terminal end causes pili to be immobilized in the cell wall envelope. In chemical terms, anchoring of pili to the cell wall envelope occurs by a mechanism where another chef, sortase A, competes with sortase D for the same ingredient (BcpA substrate).
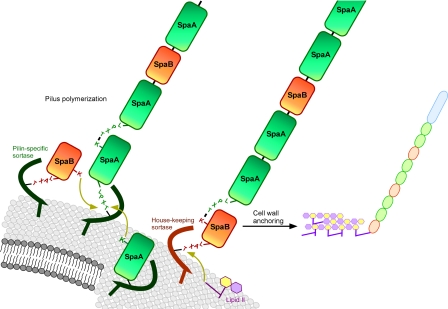
Pili made from three subunits thus far have presented an enigma to the pilus sortase hypothesis (3). The pilin-specific sortase of Corynebacterium diphtheriae (sortase A) is responsible for assembly of pili that harbor SpaC at the tip joined to a heteropolymer made of SpaA, the major pilin subunit with YPKN motif, and SpaB (3) (Fig. 1). Immune electron microscopy studies suggest an occasional incorporation of SpaB, a minor subunit that harbors a C-terminal sorting signal but no YPKN motif (3). Mandlik et al. (11) report that deletion of spaB does not abrogate pilus assembly; instead, the SpaC-SpaAn polymer is released into the extracellular medium, a phenotype similar to that of srtF mutants (the C. diphtheriae homolog of sortase A) (16). What is the nucleophilic attribute that makes SpaB an ingredient of pili? Mandlik et al. use a molecular genetic approach and identify the smallest domain that allows for incorporation of SpaB into SpaA pili. Substitution experiments identify lysine 139 (K139) as absolutely required and, similar to the YPKN motif (3), suggest that the side-chain amino group of this residue functions as the SpaB nucleophile (11). This discovery provides fresh and important views on pilus assembly in Gram-positive bacteria, because most sortase chefs use three ingredients to make pili and these pili are used for vaccine development (1, 17). If one assumed that C. diphtheriae sortase A cuts three subunits (SpaA, SpaB, and SpaC), the resulting acyl intermediates could be relieved by nucleophilic attack of two different substrates, SpaA or SpaB, which would be a true novelty of sortase chemistry. This recipe could get even more complicated because some pili involve two pilin-specific sortases for assembly, leaving open the possibility of multiple substrate specificities (18, 19). Whatever models turn out to be true, currently available data suggest that some pilin-specific sortases must accept more than one nucleophile to complete their transpeptidation reaction (11). Another attribute of recipes with three ingredients is that two chefs (sortases A and F) may compete for the minor pilin, whereas the major subunit is apparently only cut by sortase A (16). Similar mechanisms may apply for pili of Streptococcus agalactiae (group B streptococci), where deletion of the minor subunit also results in the release of pili (20). Unlike bacilli, corynebacteria, or group B streptococci, pili of Streptococcus pneumoniae were found localized to the cell wall in a mutant that lacked the housekeeping sortase A, the enzyme that accepts lipid II nucleophiles (21). If so, should we expect that pilin-specific sortases of S. pneumoniae accept two nucleophiles from different chemical groups, pilin precursor (a protein) and lipid II (a glycolipid)?
Fig. 1.
Pilus assembly in Corynebacterium diphtheriae. Pilin-specific sortase cleaves the sorting signal of SpaA and SpaB at the threonine residue (T) of the LPXTG motif, generating an acyl-enzyme intermediate. Nucleophilic attack by the side chain of lysine (K) in the YPKN motif of SpaA forms an amide bond between SpaA or SpaB and provides for pilus polymerization. SpaB is cleaved by the housekeeping sortase. Nucleophilic attack by lysine 139 (K139) of SpaB connects the minor subunit to the pilus. Nucleophilic attack of lipid II at the acyl intermediate of the housekeeping sortase with SpaB transfers the pilus to the cell wall envelope.
In sum, recipes for pili with three ingredients seem complicated and require sortases that cleave multiple substrates and accept multiple nucleophiles. What's needed now are biochemical approaches that recapitulate these assembly reactions in vitro and reveal the molecular details of substrate recognition and sortase catalysis. Looking deep into chefs' recipes will not only provide insight into Gram-positive pili, but should also generate menus of astonishing variety that can be used in imaginative ways for biological investigation (22).
Footnotes
The authors declare no conflict of interest.
See companion article on page 14147.
References
- 1.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 2.Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 4.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ton-That H, Faull KF, Schneewind O. Anchor structure of staphylococcal surface proteins. I. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J Biol Chem. 1997;272:22285–22292. doi: 10.1074/jbc.272.35.22285. [DOI] [PubMed] [Google Scholar]
- 6.Dhar G, Faull KF, Schneewind O. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry. 2000;39:3725–3733. doi: 10.1021/bi992347o. [DOI] [PubMed] [Google Scholar]
- 7.Marraffini LA, Schneewind O. Anchor structure of staphylococcal surface proteins. V. Anchor structure of the sortase B substrate IsdC. J Biol Chem. 2005;280:16263–16271. doi: 10.1074/jbc.M500071200. [DOI] [PubMed] [Google Scholar]
- 8.Marraffini LA, Schneewind O. Sortase C-mediated anchoring of BasI to the cell wall envelope of Bacillus anthracis. J Bacteriol. 2007;189:6425–6436. doi: 10.1128/JB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ton-That H, Mazmanian SK, Faull KF, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH2-Gly3 substrates. J Biol Chem. 2000;275:9876–9881. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- 10.Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem. 2002;277:16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- 11.Mandlik A, Das A, Ton-That H. The molecular switch that activates the cell wall anchoring step of pilus assembly in Gram-positive bacteria. Proc Natl Acad Sci USA. 2008;105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budzik JM, Marraffini LA, Schneewind O. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol Microbiol. 2007;66:495–510. doi: 10.1111/j.1365-2958.2007.05939.x. [DOI] [PubMed] [Google Scholar]
- 13.Mishra A, Das A, Cisar JO, Ton-That H. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J Bacteriol. 2007;189:3156–3165. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Budzik JM, et al. Amide bonds assemble pili on the surface of bacilli. Proc Natl Acad Sci USA. 2008;105:10215–10220. doi: 10.1073/pnas.0803565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 16.Swaminathan A, et al. Housekeeping sortase facilitates the cell wall anchoring step of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maione D, et al. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaspar AH, Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol. 2006;188:1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosini R, et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 20.Nobbs AH, et al. Sortase A utilizes ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect Immun. 2008;76:3550–3560. doi: 10.1128/IAI.01613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemieux J, Woody S, Camilli A. The roles of the sortases of Streptococcus pneumoniae in assembly of the RlrA pilus. J Bacteriol. 2008;190:6002–6013. doi: 10.1128/JB.00379-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: A versatile method for protein labeling. Nat Chem Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]