Abstract
We introduce a systematic computational methodology based on bioinformatics that has enabled us to identify and classify >120 endogenous peptide inhibitors of endothelial cell proliferation and migration. These peptides are derived from members of the type IV collagen, thrombospondin, and CXC chemokine protein families, as well as somatotropin hormones, serpins, and various kringle-containing proteins. Their activity in suppressing the proliferation and migration of endothelial cells in vitro provides proof of principle for the validity of this computational method. Interestingly, some of the peptides are derived from proteins known to be proangiogenic. By performing receptor neutralization studies, we have identified receptors to which these peptides bind. On the basis of this receptor-binding information, we evaluated several examples of peptide-based combinatorial screening strategies. In some cases, this combinatorial screening identified strong synergism between peptides. The current work provides a guideline for a computational-based peptidomics approach for the discovery of endogenous bioactive peptides.
Keywords: angiogenesis, bioinformatics, extracellular matrix
Excessive angiogenesis is a pathologic feature of >30 diseases, including cancer, eye diseases (e.g., age-related macular degeneration, diabetic retinopathy, retinopathy of prematurity), rheumatoid arthritis, atherosclerosis, diabetic nephropathy, pathologic obesity, asthma, cystic fibrosis, inflammatory bowel disease, psoriasis, endometriosis, vasculitis, and vascular malformations. The discovery of angiogenesis inhibitors would contribute to the development of therapeutic treatments for these diseases and might also point to endogenous inhibitors whose existence would further our understanding of the angiogenic balance between proangiogenic and antiangiogenic factors in vivo (1).
Two critical components of the angiogenic process are the proliferation and migration of activated endothelial cells, and agents capable of suppressing either of these processes can be considered as having antiangiogenic potential. To date, the experimental identification of endogenous peptide inhibitors of endothelial cell proliferation and migration has relied on an empirical process (2), in which selected proteins are proteolytically processed by proteases and the cleaved fragments are tested by using various in vitro and in vivo assays. Although this process has yielded important information regarding the localization of antiangiogenic active sites within proteins, it is both labor intensive and time consuming.
In the present study, we have introduced a systematic computational and experimental methodology that has enabled us to efficiently identify numerous endogenous peptide inhibitors of endothelial cell proliferation and migration. The identification of such peptides should lead to a better fundamental understanding of angiogenic homeostasis and the angiogenic switch, because these peptides are fragments of endogenous proteins that can potentially be released by proteases under various physiologic and pathologic conditions.
Results
Bioinformatic Analysis.
The amino acid sequences of 40 known potent antiangiogenic fragments were compiled from an extensive search of the relevant literature [Table 1 and supporting information (SI) Table S1]. The ability of these previously identified peptides to suppress both the proliferation and migration of endothelial cells in vitro and the process of angiogenesis in vivo has been well documented. In some cases, active epitopes of up to ≈25 aa within these fragments have been shown to exhibit potent antiangiogenic activity, inhibiting the proliferation and migration of endothelial cells; we refer to these fragments as “active domains.” Other sequences consist of protein fragments of 30–150 aa with identified activity; still others are larger protein domains or full-length proteins longer than 150 aa. We hypothesized that in all cases, the antiangiogenic potential of the proteins would reside within a short epitope, the active domain. We therefore used a combination of computational and experimental approaches to identify active domains within the whole human proteome, not only in the previously reported antiangiogenic proteins and fragments, but also in other proteins that had not previously been recognized as having antiangiogenic potential.
Table 1.
Query sequences and the associated conserved domains they contain
| Conserved domains | Antiangiogenic peptides |
|---|---|
| TSP1 Containing | METH-1(10), METH-2(10), TSP1/Mal2(3), TSP1/Mal3(3), Vasculostatin(11) |
| CXCs | BCA-1(33), Gro-beta(34), IP-10(35), ITAC(35), MIG(35), PF4(12, 35) |
| CIV CTR* | Arresten(18), Canstatin(19) |
| Tumstatin/T3(13), Tumstatin/T4(13) | |
| Serpins | Angiotensinogen(36), Antithrombin(37) |
| Kallistatin(38), Maspin(39), PEDF-34(5) | |
| PEDF-ERT(5), PEDF-TGA(5) | |
| Kringles | K1(40), K2(40), K3(40), K4(40), K5(41) |
| Somatotropins | Growth Hormone(42) Growth Hormone-tilted(4), Placental Lactogen(42), Prolactin(43), Prolactin-tilted(4) |
| LamininG | Endorepelin/LG3(44) |
| TIMP NTR† | TIMP2(45) |
| Complement C1q | Vastatin(46) |
| Vasostatin(7), Endostatin(8), Kininogen D5 (6) | |
| No Domains | Kininostatin(6), Restin(9) |
*Type IV collagen C-terminal.
†Tissue inhibitor of metalloproteinase N-terminal.
In addition to classifying these proteins according to their length, we also classified them into families according to the conserved domains contained within their sequences. This classification was based on the observation that most of the known antiangiogenic active fragments are localized within conserved protein domains or contain part of a conserved domain. A growing body of evidence supports this observation. For example, the thrombospondin 1 protein contains multiple thrombospondin type 1 (TSP1) domains, and short peptides contained within the second (Mal-2) and third (Mal-3) domains of the protein have been identified as antiangiogenic (3). The growth hormone (GH) and prolactin proteins contain the somatotropin-conserved domain, and short peptides derived from these domains have recently been identified as antiangiogenic (4). Pigment epithelium derived factor (PEDF) contains a serpin conserved domain, and short sequences within this domain have been recognized as the active antiangiogenic epitopes (5).
After compiling all of the conserved domains within protein sequences that are known to have antiangiogenic effects, we identified nine protein families: (i) the type 1 thrombospondin repeat-containing proteins, (ii) CXC chemokines (containing the SCY domain), (iii) type IV collagens (containing the collagen IV C-terminal domain), (iv) serpins (containing the serpin domain), (v) somatotropin hormones (containing the somatotropin or somatomammotropin hormone domain), (vi) laminin globular domain-containing proteins, (vii) kringle-containing proteins, (viii) tissue inhibitors of metalloproteinases (TIMPs, containing the TIMP N-terminal domain), and (ix) complement component C1q-containing proteins. When a protein contained multiple different conserved domains, we based our classification on the domain that colocalized with the previously identified antiangiogenic site. There is a very small population of proteins for which the ability to suppress the proliferation and migration of endothelial cells has been precisely mapped to specific active domains within the sequence of the protein, but these active domains do not coincide with any conserved domains. Examples include kininogen (6), calreticulin (7), and the collagen XVIII (8) and XV (9) endostatins. These examples did not fall into our classification scheme.
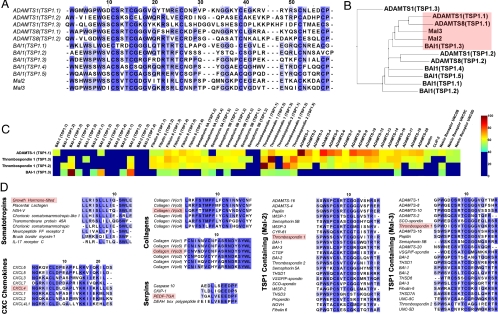
Our initial approach to identifying bioactive peptides was based on asking whether the conserved domains used for the classification of the antiangiogenic proteins are actually similar and whether there are other such domains that exist in proteins that have not yet been shown to exhibit antiangiogenic potency. To answer these questions, we performed multiple sequence alignments to calculate the similarities between the amino-acid sequences of the conserved domains of the known antiangiogenic proteins and the corresponding conserved domains of all proteins that contain these particular domains. Here, we show an example of such a calculation using various TSP1 domains (Fig. 1A).
Fig. 1.
Outline of the computational bioinformatic algorithm. (A) The similarities between conserved domains from previously identified antiangiogenic proteins were calculated after performing multiple sequence alignments of their amino-acid sequences. The blue color designates the amino acid conservation (the darker the color, the more conserved the amino acid). (B) In cases in which a protein contained more than one copy of a conserved domain, those that were primarily similar to the known antiangiogenic domains (pink highlight) were used. (C) Using the sequences from the known conserved domains, we determined their similarity to other such domains from the whole human proteome. (D) In addition to identifying large domains showing sequence similarity, we used the information from shorter known antiangiogenic fragments (pink highlight) and searched for sequences similar to those of the fragments. The blue highlight designates conservation as in A. These short fragments were derived from a variety of protein families, including the somatotropins, CXC chemokines, collagen IV fibrils, serpins, and TSP1 repeat-containing proteins.
Because such domains are abundant in the human proteome, we were able to take advantage of the information already available regarding the amino-acid sequences of TSP1 domains that have been shown to have antiangiogenic activity. We used the sequences from four previously identified antiangiogenic proteins: Meth-1 (10), which contains three TSP1 domains; Meth-2 (10), with two TSP1 domains; the brain angiogenesis inhibitor 1 (BAI-1) (11), with five TSP1 domains; and the type 1 thrombospondin protein, with three TSP1 domains. In the case of type 1 thrombospondin, we used the information from only two of the three domains that are already known to be antiangiogenic (Mal-2 and Mal-3) (3). In total, we collected sequences from 12 TSP1 domains.
Next, we performed a multiple-sequence alignment using these 12 sequences (Fig. 1A) and calculated which of these domains in the already-known antiangiogenic proteins are similar to Mal-2 and Mal-3 (Fig. 1B). In this case, we demonstrated that the first TSP1 domains of both Meth-1 and Meth-2 (TSP1.1) and the third domain of BAI-1 (TSP1.3) are more similar to Mal-2 and Mal-3 than are the rest of the tested TSP1 domains. These results indicated that these domains have antiangiogenic potential and that it was within these sequences that we should search for active domains and identify shorter active sequences.
Assuming that the antiangiogenic activity resides within these domains, we then asked whether it was possible to use these sequences, together with the sequences from the known Mal-2 and Mal-3 domains of our initial query, to identify other similar TSP1 domains within the human proteome. To answer this question, we collected the sequences of all of the TSP1 domains within the human proteome and calculated their similarities to those that we predicted to be primarily antiangiogenic (Fig. 1C). This calculation identified specific TSP1 domains with antiangiogenic potential in proteins not known to be antiangiogenic, including fibulin 6; semaphorins 5A and 5B; various ADAMTS proteins; papilin; cartilage intermediate layer protein; and the netrin receptors UNC5B, 5C, and 5D. It was then possible to repeat this calculation for all of the classified conserved domains and to predict antiangiogenic fragments in proteins not yet shown to exhibit antiangiogenic activity (see SI Materials and Methods and Fig. S1).
Although this calculation is useful for identifying bioactive protein fragments, its verification is difficult because it requires the recombinant expression of each of these large protein domains and screening of these domains for activity. A more direct way to test our two hypotheses (i.e., use information from the amino-acid sequences to predict similar peptides, and that the activity resides within specific conserved domains) would be to perform similar calculations for shorter peptides already known to be active. We could then synthesize these putative peptides by using standard solid-phase synthesis techniques and screen them in established endothelial cell proliferation and migration assays.
There are a number of short peptides (<25 aa) from different protein families that could be used in such calculations. These peptides include the short fragments of GH and prolactin (4), a short peptide derived from platelet factor 4 (PF-4) (12), the short tumstatins (T3 and T4) (13) derived from the C-terminal of type IV collagen, the short fragments of the serpin PEDF (5), and short peptides from Mal-2 and Mal-3 of the thrombospondin 1 protein (3).
After searching within the human proteome for sequences similar to these short peptides and filtering the results by using a Monte Carlo algorithm (Materials and Methods and SI Materials and Methods) to identify the statistically significant similarities, we identified approximately 90 similar peptides (Table S2). If we also included the longer fragments in our query, the total number of identified peptides exceeded 120 from 82 different proteins (Table 2). Fig. 1D shows several similar short peptides from a range of protein families, including somatotropins, CXC chemokines, type IV collagen, serpins, and TSP1 repeat-containing proteins. We named the peptides according to the proteins from which they were derived (14–17). The names and associated amino-acid sequences of the peptides are included in Table S3.
Table 2.
The 82 different proteins from which our newly identified peptides are derived
| Protein family | Proteins with identified fragments |
|---|---|
| TSP-1-containing proteins | ADAM-9, ADAM-12, ADAMTS-1, ADAMTS-2, ADAMTS-3, ADAMTS-4, ADAMTS-5, ADAMTS-6, ADAMTS-7, ADAMTS-8, ADAMTS-9, ADAMTS-10, ADAMTS-12, ADAMTS-13, ADAMTS-14, ADAMTS-15, ADAMTS-16, ADAMTS-18, ADAMTS-19, ADAMTS-20, BAI-1, BAI-2, BAI-3, C6, CILP, CILP-2, CTGF, CYR61, fibulin-6, NOVH, papilin, properdin, ROR-1, ROR-2, semaphorin 5A, semaphorin 5B, SCO-spondin, THSD1, THSD3, THSD6, TSP-2, TSRC1, UNC5C, UNC5D, VSGP/F-spondin, WISP-1, WISP-2, WISP-3 |
| Collagens | α1CIV, α2CIV, α4CIV, α5CIV, α6CIV |
| CXC chemokines | Gro-α/CXCL1, Gro-β/CXCL2, Gro-γ/MIP-2β/CXCL3, ENA-78/CXCL5, GCP-2/CXCL6, THBG-β/CXCL7, IL-8/CXCL8, IP-10/CXCL10 |
| Kringle-containing proteins | AK-38 protein, hageman fct/cf XII, HGF, hyaluronan binding, KREMEN-1, KREMEN-2, Lp(a), macrophage stim. 1, thrombin/cf II, tPA |
| Somatotropins | Placental lactogen, hGH-V, chorionic somatommamotropin, chorionic somatommamotropin-like 1, transmembrane protein 45A, neuropeptide FF receptor 2, brush border myosin-1, IL-17 receptor C |
| Serpins | Caspase 10, CKIP-1, DEAH-box polypeptide 8 |
There are two important observations regarding this dataset. First, we have identified short fragments within various known antiangiogenic proteins or within very large protein fragments in which short active epitopes have not yet been identified. These proteins include arresten (18) and canstatin (19), derived from the α1 and α2 fibrils of type IV collagen, respectively; they also include active epitopes within Meth-1, Meth-2 (10), BAI-1 (11), and thrombospondin 2 (20). Second, we have identified a number of short peptides within sequences of proteins that are known to promote, rather than inhibit, angiogenesis. Several of these short peptides are derived from the Cyr61-Ctgf-Nov (CCN) protein family, which contains TSP1 domains. These proteins are traditionally considered proangiogenic, and they promote endothelial cell proliferation and migration (21). Examples of peptides derived from this protein family include cyrostatin, derived from CYR-61 (CCN1); connectostatin, derived from CTGF (CCN2); nephroblastostatin, derived from NOVH (CCN3); and the wispostatins -1, -2, and -3, derived from WISP-1, -2, and -3 (CCN-5), respectively. In addition to the peptides derived from the CCN protein family, we have also identified peptides derived from the Glu-Leu-Arg (ELR)-motif-containing CXC chemokines, which are known to be potent promoters of angiogenesis (22). Examples of such peptides include chemokinostatin-1, -3, -5, -6, -7, and -8, which are derived from CXCL1, 3, 5, 6, 7, and 8, respectively.
Experimental in Vitro Validation.
Our computational bioinformatic analysis was based on the hypothesis that there is an underlying sequence-based correlation between the activity of the known endogenous protein fragments and the predicted peptides. To verify the predicted effects of the peptides and provide proof of principle for this hypothesis, we evaluated the antiproliferative and antimigratory potency of our candidate peptides by using in vitro proliferation and migration assays of human umbilical vein endothelial cells (HUVECs), comparing the potency of our peptides with those of positive and negative controls (14–16). To date, we have screened 72 of the 120 predicted peptides for in vitro activity. A statistically significant effect on either proliferation or migration (or both) was exhibited by 58 of the 72 peptides, indicating a success rate for the algorithm of ≈80%. Examples of such screenings of 20 peptides derived from three protein families, the type IV collagen fibrils, CXC chemokines, and TSP1 repeat-containing proteins, are included in Fig. S2.
The most potent antiproliferative agents were the collagen IV-derived peptides: tetrastatins derived from the α4 fibrils, pentastatins from the α5 fibrils, and hexastatins from the α6 fibrils. Some of these peptides completely inhibited proliferation (100% inhibition) in the in vitro proliferation assay. The most surprising results were obtained for the CXC chemokine-derived peptides (chemokinostatins). Although these peptides are derived from ELR-containing chemokines that are known as potent stimulators of angiogenesis in their full-length form, these chemokinostatins were effective in reducing the proliferation of the endothelial cells. A similar inhibitory effect was seen for some of the peptides (the wispostatins, nephroblastostatin, and connectostatin) derived from type I thrombospondin domain-containing proteins belonging to the CCN protein family.
We also examined the ability of the same 20 peptides to suppress the migration of a population of activated endothelial cells in an in vitro system. For this purpose, we used a modified Boyden chamber assay (Fig. S2). In most cases, the peptides effectively inhibited the migration of the endothelial cells through the chamber by 40–100%. The most potent antimigratory peptides were those derived from the TSP1 repeat-containing proteins. The peptides derived from the proangiogenic ELR motif-containing CXC chemokines were also potent antimigratory agents in this in vitro assay.
To look for possible correlations between the amino-acid sequences of the peptides and their activity in the in vitro assays, we treated the sequence similarities of the peptides and their potencies in the proliferation assay as independent datasets. We then performed a clustering analysis to search for relationships within each of the two datasets. If there was an underlying relationship between the individual amino-acid sequences and the corresponding potency profiles, we would expect that the sequences that clustered only because of sequence similarities would behave similarly in the assays and that their potency profiles would also cluster. When we performed such analyses using peptides from different protein families (14–16), we found that the activity of the peptides did indeed correlate with the similarity in their sequences (14–16).
Our proliferation and migration experiments demonstrated the potency of our predicted peptides in the in vitro assays, but also raised a further question: What is the mechanism by which these newly identified peptides affect proliferation and migration? For example, do they bind to cell-surface receptors? There is growing evidence that specific receptors are involved in the binding of known antiangiogenic peptides to endothelial cells. Tumstatin has two binding sites for αvβ3 integrins (13), although its antiangiogenic activity has been connected to a site in the N-terminal portion of the fragment. Tumstatin has also been shown to interact with α6β1 integrins (23). The major receptor that has been identified for the antiangiogenic CXC chemokines is CXCR3 (24). The CXC chemokine ligands of CXCR3 inhibit the proliferation and migration of human microvascular endothelial cells in response to a variety of angiogenic factors. In the case of thrombospondin 1, the prototypical type 1 thrombospondin repeat-containing protein, extensive studies have implicated CD36, a 88-kDa transmembrane glyco protein, as the cell-surface receptor that mediates its effects on endothelial cells (25). CD47 and various integrins have also been mechanistically implicated in the effects of thrombospondin 1 on endothelial cells (26).
Therefore, we investigated the possibility that our newly identified peptides share common receptors with some of the previously identified antiangiogenic peptides by performing neutralization studies against these receptors. We preincubated the endothelial cells with a range of concentrations of neutralizing monoclonal antibodies known to target single receptors, then compared the activity of our peptides in the proliferation assays with that seen in the absence of neutralizing antibody. We again examined peptides from the three protein families: the CXC chemokines, the collagen IV, and the TSP1 repeat-containing peptides. The results of the proliferation assays are included in Fig. S3. In each case, we included a negative control in which the cells were incubated in the presence of the appropriate antibody alone (no peptide). As expected, none of the antibodies in the tested concentrations showed any effect on the endothelial cells in the absence of peptide (data not shown).
In these neutralization studies, we first asked whether the CXCR3 receptor might be responsible for the binding of the chemokinostatins we had identified. To answer this question, we performed the proliferation assays in the presence of a CXCR3-neutralizing antibody (Fig. S3A) at 1 and 10 μg/ml (below and above the ED50 suggested by the manufacturer). In most cases, the inhibitory activity of the individual chemostatins was totally abrogated in the presence of the higher concentration of the neutralizing antibody against the CXCR3 receptor. Interestingly, when the peptide exhibited a biphasic dose response (in the case of chemokinostatin-1), the monoclonal antibody did not entirely neutralize the activity of the peptide. We conclude that in this case, more than one receptor or more than one mechanism is responsible for the activity of the peptide.
In the case of collagen-derived peptides, we performed similar neutralization experiments by using two monoclonal antibodies, one directed against β1 integrins and the other against β3 integrins, because the effects of tumstatins are primarily attributed to peptide binding to β1 and β3 integrins (13, 27) (Fig. S3B). We again tested two antibody concentrations, 1 and 10 ng/ml. The activity of our highly potent pentastatin-1 was abrogated after preincubation with either anti-integrin antibody.
In the case of the TSP1 repeat-derived peptides, an antibody that neutralizes CD36, the main TSP1 repeat receptor, was able to abolish the peptides' activity. With increasing antibody concentration, we observed increased endothelial cell proliferation relative to the control (Fig. S3C). It is noteworthy that at these two antibody concentrations for which no direct effect on endothelial cells was observed, the antibodies were potent enough to neutralize the peptide activity. In contrast, blocking CD47, the integrin-associated receptor, only partially neutralized the activity of the TSP1 repeat-containing protein-derived peptide. As was observed for chemokinostatin-1 and the anti-CXCR3 receptor antibody, the anti-CD36 monoclonal antibody did not entirely neutralize the activity of netrinstatin-5C, a peptide that exhibited a biphasic dose response in the proliferation assay.
To determine whether the peptides share similar or neighboring binding sites with the antibodies used in the neutralization experiments, we also performed antibody-peptide competition assays. In these assays, we first preincubated the cells with varying concentrations of peptide, then removed the peptide solution and incubated the cells with fluorescently labeled antibody at the same concentrations used in the neutralization experiments. If the antibodies share binding epitopes on the associated receptors, or at least sterically interfere with peptide binding, we would expect that after washing away the unbound antibody and measuring the fluorescent signals from the seeded cells, we would see either no fluorescence or a decrease in the strength of the fluorescent signal with increasing peptide concentration. Given that the binding kinetics of the peptides and the antibodies were of the same order of magnitude, we reasoned that if the antibody and peptide shared neighboring binding sites, preincubation of the cells with the peptides would inhibit the antibody binding. The results of these experiments indicated that the peptides did indeed inhibit the antibody binding (Fig. S4). These results strongly suggest that the peptides and antibodies share neighboring, if not overlapping, binding sites.
Based on the information obtained from the neutralization experiments, we then developed a systematic way to create combinations of individual peptides and test their effectiveness as inhibitors in the functional assays. By using combinations of peptides that bound to different receptors, we attempted to target different pathways and assess the combined activity in our functional assays. To evaluate these combinations, we chose the more sensitive proliferation assay. Using multiple peptides targeting multiple targets with different mechanisms or modes of action created the opportunity for a variety of favorable outcomes, including an increased efficacy of the therapeutic effect, the ability to obtain an analogous or increased level of efficacy with a lower dosage (as a strategy to avoid toxicity), and a minimization of, or delay in, the development of resistance (28).
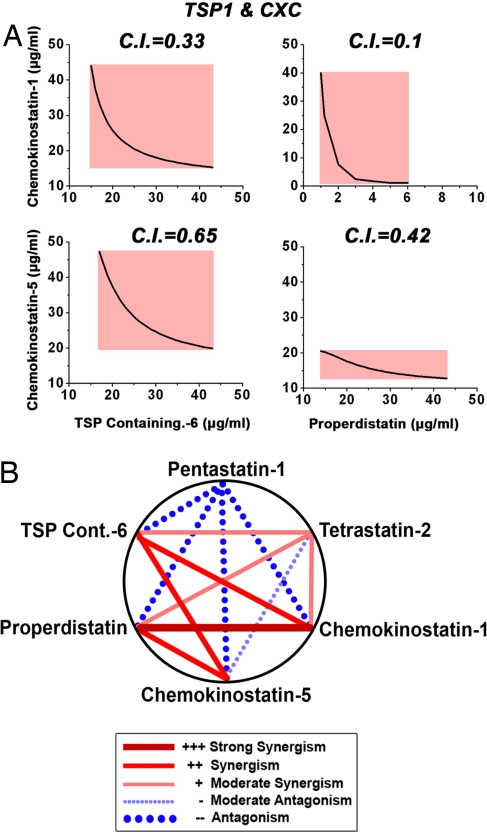
Using the proliferation assay, we tested combinations of two peptides from each of the three major protein families (the CXC chemokines, type IV collagen fibrils, and TSP1 repeat-containing proteins), combining one peptide from each family at four different concentrations (0.1, 1, 10, and 30 μg/ml). The two peptides were applied in series to avoid possible interactions between them, and the viability of the cells was then evaluated. Using the information from the dose response curves, we fit the data to sigmoidal Hill curves (29). Based on the estimated Hill curves, we calculated the resulting isobolograms, the state space of peptide concentrations with equipotent sums of doses, and created graphs of equally effective dose pairs (isoboles) with the same level of effectiveness as that observed for a single peptide. In addition to the isobolograms, we also calculated the combination indices (C.I.) for different peptide combinations (30) to compare the relative efficacy of the various combinations (Fig. 2 and Fig. S5).
Fig. 2.
Evaluation of peptide combinations from different protein families. Two peptides from three different protein families were combined serially in the proliferation assay, and the efficiency of the various peptide combinations was evaluated after calculating the isobolograms and C.I. for each of the combinations. (A) The isobolograms for the combination of a CXC chemokine-derived peptide and a TSP1 repeat-derived peptide. For each of the tested conditions (the individual peptides and the peptides tested in combination), proliferation dose response sigmoidal curves were estimated by fitting the data to sigmoidal Hill curves. Equally effective dose pairs in the combination experiments were then matched to the same level of inhibition obtained with a single peptide. The red highlighted areas designate peptide concentrations at which the activity is synergistic. The isobolograms for the combination of a collagen IV-derived peptide and a CXC chemokine and for a collagen IV-derived peptide and a TSP1 repeat-containing protein-derived peptide are included in Fig. S5. (B) Graphical interpretation of the most potent synergistic strategies, based on the calculations from the combinatorial experiments. The red lines indicate strong synergism and the blue lines antagonism.
Both of our analyses indicated a significant synergism between CXC chemokines and TSP1 repeat-containing protein-derived peptides (Fig. 2A). Thus, it appeared that by using certain specific peptide combinations, we could achieve activity levels similar to those obtained when each of the peptides were used alone, but at significantly lower dosages. When we combined chemokinostatin-1 with properdistatin, for example, we achieved an effective decrease in dosage of one order of magnitude. Furthermore, we found that when these two peptides were applied at higher concentrations in combination, the level of activity obtained was much higher than that observed when either one was applied alone. When we combined collagen IV-derived peptides with either CXC-derived (Fig. S5A) or TSP1-derived (Fig. S5B) peptides, we observed a synergistic effect only at the lower collagen peptide concentrations; at higher concentrations, the collagen-derived peptides were apparently antagonized by the CXC and TSP1 repeat-derived peptides. This finding raised the question of whether the order in which the peptides were applied might have affected their combined activity, which is a question that could be of particular relevance to an in vivo therapeutic setting in which the peptides might be applied in an alternating manner multiple times. We therefore repeated the experiment, varying the order in which the peptides were applied (Fig. S6). The peptide combinatorial activity differed slightly when the order was reversed, but the variation was not significant. Thus, the synergism was preserved and was independent of the order in which the peptides were applied.
Discussion
Angiogenesis is regulated by the orchestrated expression of endogenous regulatory elements (31, 32), which include various growth factors that stimulate and control the proliferation and migration of endothelial cells. There is also a growing population of endogenous regulatory elements that work competitively by inhibiting both of these processes, thus suppressing angiogenesis. The fine control of these two opposing elements, referred to as the angiogenic balance (1), is vital for the homeostasis of a physiologic tissue and is disrupted during pathologic conditions such as cancer. Using the systematic bioinformatic approach and in vitro assays described here, we have identified a variety of bioactive peptides and demonstrated that these peptides can act as negative regulators of endothelial cell proliferation and migration. By doing so, we have substantially increased the number of known endogenous peptide regulators of these processes. In addition to the potential usefulness of these peptides for therapeutic purposes, further analysis of the expression and fine regulation of the identified peptides in vivo should contribute to a better understanding of the essential angiogenic balance within tissues.
Materials and Methods
Peptide Synthesis and Handling.
The peptides used were produced by a commercial provider (Abgent) using a solid phase synthesis technique. HPLC and MS analyses of each peptide were performed. In each case, the synthetic procedure yielded >95% pure peptide. In the case of highly hydrophobic peptides, DMSO was used as a solvent at a maximum concentration of 0.1% (vol/vol); we verified experimentally that at this concentration, the solvent had no effect on the experimental results. In all of the other cases, the peptides were solubilized in PBS. The peptides were stored at −80°C and thawed at room temperature just before use.
Cell Culture.
Primary HUVECs from a single donor were purchased from Cambrex. The cells were propagated in EGM-2 medium, consisting of a basal cell medium with 2% FBS, growth factors human basic FGF and VEGF, and antibiotics (gentamicin/amphotericin B). All of the cells used were from passages 3–6.
Statistical Analysis.
Statistical significance was assessed by using the Student's t test, with P values <0.001 defined as significant. The question being tested was whether the scaled experimental result was different from the negative control.
Monoclonal Antibody Neutralization Assays.
For the monoclonal antibody neutralization experiments, 2 × 103 endothelial cells per well were grown overnight in 96-well microplates in 100 μl of complete basal cell medium with growth factors and serum (EGM-2). The medium was then removed and replaced with 100 μl of growth-factor- and serum-free medium containing various final dilutions of monoclonal antibody-recognizing β1 integrins (R&D Systems FAB17781), αvβ3 integrins (R&D Systems FAB3050), CXCR3 (R&D Systems FAB1685), CD36 (BD PharMingen CB38/NL07), or CD47 (BD PharMingen B6H12). The cells were incubated for 2 h with the antibody solution, after which each of the tested peptides in growth-factor- and serum-free medium were added to the wells to give the desired final concentration. As a control, a set of cells was incubated only in the presence of each monoclonal antibody, without peptide. The cells were incubated for 3 d, and cell viability was assessed with the colorimetric cell proliferation reagent WST-1 (Roche) in a manner analogous to that for the proliferation assays (SI Materials and Methods).
Peptide Combination Assays.
For the combination experiments, 2 × 103 cells per well were seeded into 96-well microplates and allowed to attach overnight in complete medium with growth factors and serum. The medium was then removed, and a solution of a single peptide in growth-factor- and serum-free medium was applied to give a final concentration of 0.1, 1, 10, or 30 μg/ml. After 2 h, the medium was removed without any washing and the second peptide was applied in growth-factor- and serum-free medium to give the same final concentration as was used for the first peptide; if the first peptide was applied at 10 μg/ml, the second was also applied at 10 μg/ml. Each of the peptides was also applied alone to serve as a reference. After 24 h, the WST-1 dye was applied, and the number of live cells was estimated. Dose responses were estimated by fitting the data to sigmoidal Hill curves. Details of the quantification of the synergism are provided in the SI Materials and Methods.
Supplementary Material
Acknowledgments.
The authors are grateful to Drs. Joel Bader and Akhilesh Pandey for constructive suggestions on bioinformatics methods; Drs. Zaver Bhujwalla and Roberto Pili for useful discussions; Drs. Venu Raman, Dmitri Artemov, David Qian, David Noren, Kristine Glunde, Noriko Mori, and Paul Winnard for their valuable advice on the experimental assays; and Dr. Deborah McClellan for editorial assistance. This work was supported in part by National Institutes of Health Grants R01 HL079653 (from the National Heart, Lung, and Blood Institute) and P50 CA103175 (from the National Cancer Institute).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803241105/DCSupplemental.
References
- 1.Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J. Angiogenesis: An organizing principle for drug discovery? Nat Rev Drug Discov. 2007;6:273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- 3.Tolsma SS, et al. Peptides derived from two separate domains of the matrix protein thrombospondin-1 have anti-angiogenic activity. J Cell Biol. 1993;122:497–511. doi: 10.1083/jcb.122.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen NQ, et al. Prolactin/growth hormone-derived antiangiogenic peptides highlight a potential role of tilted peptides in angiogenesis. Proc Natl Acad Sci USA. 2006;103:14319–14324. doi: 10.1073/pnas.0606638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filleur S, et al. Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer Res. 2005;65:5144–5152. doi: 10.1158/0008-5472.CAN-04-3744. [DOI] [PubMed] [Google Scholar]
- 6.Colman RW, Jameson BA, Lin Y, Johnson D, Mousa SA. Domain 5 of high molecular weight kininogen (kininostatin) down-regulates endothelial cell proliferation and migration and inhibits angiogenesis. Blood. 2000;95:543–550. [PubMed] [Google Scholar]
- 7.Pike SE, et al. Calreticulin and calreticulin fragments are endothelial cell inhibitors that suppress tumor growth. Blood. 1999;94:2461–2468. [PubMed] [Google Scholar]
- 8.O'Reilly MS, et al. Endostatin: An endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 9.John H, Radtke K, Standker L, Forssmann WG. Identification and characterization of novel endogenous proteolytic forms of the human angiogenesis inhibitors restin and endostatin. Biochim Biophys Acta. 2005;1747:161–170. doi: 10.1016/j.bbapap.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Vazquez F, et al. METH-1, a human ortholog of ADAMTS-1, and METH-2 are members of a new family of proteins with angio-inhibitory activity. J Biol Chem. 1999;274:23349–23357. doi: 10.1074/jbc.274.33.23349. [DOI] [PubMed] [Google Scholar]
- 11.Kaur B, Brat DJ, Devi NS, Van Meir EG. Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene. 2005;24:3632–3642. doi: 10.1038/sj.onc.1208317. [DOI] [PubMed] [Google Scholar]
- 12.Jouan V, et al. Inhibition of in vitro angiogenesis by platelet factor-4-derived peptides and mechanism of action. Blood. 1999;94:984–993. [PubMed] [Google Scholar]
- 13.Maeshima Y, et al. Extracellular matrix-derived peptide binds to alpha(v) beta(3) integrin and inhibits angiogenesis. J Biol Chem. 2001;276:31959–31968. doi: 10.1074/jbc.M103024200. [DOI] [PubMed] [Google Scholar]
- 14.Karagiannis ED, Popel AS. Identification of novel short peptides derived from the alpha 4, alpha 5, and alpha 6 fibrils of type IV collagen with anti-angiogenic properties. Biochem Biophys Res Commun. 2007;354:434–439. doi: 10.1016/j.bbrc.2006.12.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karagiannis ED, Popel AS. Anti-angiogenic peptides identified in thrombospondin type I domains. Biochem Biophys Res Commun. 2007;359:63–69. doi: 10.1016/j.bbrc.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karagiannis ED, Popel AS. Peptides derived from type I thrombospondin repeat-containing proteins of the CCN family inhibit proliferation and migration of endothelial cells. Int J Biochem Cell Biol. 2007;39:2314–2323. doi: 10.1016/j.biocel.2007.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karagiannis ED, Popel AS. Novel anti-angiogenic peptides derived from ELR-containing CXC chemokines. J Cell Biochem. 2008;104:1356–1363. doi: 10.1002/jcb.21712. [DOI] [PubMed] [Google Scholar]
- 18.Colorado PC, et al. Anti-angiogenic cues from vascular basement membrane collagen. Cancer Res. 2000;60:2520–2526. [PubMed] [Google Scholar]
- 19.Kamphaus GD, et al. Canstatin, a novel matrix-derived inhibitor of angiogenesis and tumor growth. J Biol Chem. 2000;275:1209–1215. doi: 10.1074/jbc.275.2.1209. [DOI] [PubMed] [Google Scholar]
- 20.Volpert OV, et al. Inhibition of angiogenesis by thrombospondin-2. Biochem Biophys Res Commun. 1995;217:326–332. doi: 10.1006/bbrc.1995.2780. [DOI] [PubMed] [Google Scholar]
- 21.Brigstock DR. The CCN family: A new stimulus package. J Endocrinol. 2003;178:169–175. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- 22.Belperio JA, et al. CXC chemokines in angiogenesis. J Leukoc Biol. 2000;68:1–8. [PubMed] [Google Scholar]
- 23.Maeshima Y, Colorado PC, Kalluri R. Two RGD-independent alpha v beta 3 integrin binding sites on tumstatin regulate distinct anti-tumor properties. J Biol Chem. 2000;275:23745–23750. doi: 10.1074/jbc.C000186200. [DOI] [PubMed] [Google Scholar]
- 24.Strieter RM, Burdick MD, Mestas J, Gomperts B, Keane MP, Belperio JA. Cancer CXC chemokine networks and tumour angiogenesis. Eur J Cancer. 2006;42:768–778. doi: 10.1016/j.ejca.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Dawson DW, et al. CD36 mediates the in vitro inhibitory effects of thrombospondin-1 on endothelial cells. J Cell Biol. 1997;138:707–717. doi: 10.1083/jcb.138.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao AG, et al. Integrin-associated protein is a receptor for the C-terminal domain of thrombospondin. J Biol Chem. 1996;271:21–24. doi: 10.1074/jbc.271.1.21. [DOI] [PubMed] [Google Scholar]
- 27.Maeshima Y, et al. Identification of the anti-angiogenic site within vascular basement membrane-derived tumstatin. J Biol Chem. 2001;276:15240–15248. doi: 10.1074/jbc.M007764200. [DOI] [PubMed] [Google Scholar]
- 28.Dorrell MI, Aguilar E, Scheppke L, Barnett FH, Friedlander M. Combination angiostatic therapy completely inhibits ocular and tumor angiogenesis. Proc Natl Acad Sci USA. 2007;104:967–972. doi: 10.1073/pnas.0607542104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 30.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 31.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 32.Nyberg P, Xie L, Kalluri R. Endogenous inhibitors of angiogenesis. Cancer Res. 2005;65:3967–3979. doi: 10.1158/0008-5472.CAN-04-2427. [DOI] [PubMed] [Google Scholar]
- 33.Spinetti G, et al. The chemokine CXCL13 (BCA-1) inhibits FGF-2 effects on endothelial cells. Biochem Biophys Res Commun. 2001;289:19–24. doi: 10.1006/bbrc.2001.5924. [DOI] [PubMed] [Google Scholar]
- 34.Cao Y, Chen C, Weatherbee JA, Tsang M, Folkman J. gro-beta, a -C-X-C- chemokine, is an angiogenesis inhibitor that suppresses the growth of Lewis lung carcinoma in mice. J Exp Med. 1995;182:2069–2077. doi: 10.1084/jem.182.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strieter RM, Belperio JA, Phillips RJ, Keane MP. CXC chemokines in angiogenesis of cancer. Semin Cancer Biol. 2004;14:195–200. doi: 10.1016/j.semcancer.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Celerier J, Cruz A, Lamande N, Gasc JM, Corvol P. Angiotensinogen and its cleaved derivatives inhibit angiogenesis. Hypertension. 2002;39:224–228. doi: 10.1161/hy0202.103441. [DOI] [PubMed] [Google Scholar]
- 37.O'Reilly MS, Pirie-Shepherd S, Lane WS, Folkman J. Antiangiogenic activity of the cleaved conformation of the serpin antithrombin. Science. 1999;285:1926–1928. doi: 10.1126/science.285.5435.1926. [DOI] [PubMed] [Google Scholar]
- 38.Miao RQ, Chen V, Chao L, Chao J. Structural elements of kallistatin required for inhibition of angiogenesis. Am J Physiol. 2003;284:C1604–C1613. doi: 10.1152/ajpcell.00524.2002. [DOI] [PubMed] [Google Scholar]
- 39.Zhang M, Volpert O, Shi YH, Bouck N. Maspin is an angiogenesis inhibitor. Nat Med. 2000;6:196–199. doi: 10.1038/72303. [DOI] [PubMed] [Google Scholar]
- 40.Cao Y, et al. Kringle domains of human angiostatin. Characterization of the anti-proliferative activity on endothelial cells. J Biol Chem. 1996;271:29461–29467. doi: 10.1074/jbc.271.46.29461. [DOI] [PubMed] [Google Scholar]
- 41.Cao Y, et al. Kringle 5 of plasminogen is a novel inhibitor of endothelial cell growth. J Biol Chem. 1997;272:22924–22928. doi: 10.1074/jbc.272.36.22924. [DOI] [PubMed] [Google Scholar]
- 42.Clapp C, Aranda J, Gonzalez C, Jeziorski MC, Martinez de la Escalera G. Vasoinhibins: Endogenous regulators of angiogenesis and vascular function. Trends Endocrinol Metab. 2006;17:301–307. doi: 10.1016/j.tem.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Clapp C, et al. Vasoinhibins: A family of N-terminal prolactin fragments that inhibit angiogenesis and vascular function. Front Horm Res. 2006;35:64–73. doi: 10.1159/000094309. [DOI] [PubMed] [Google Scholar]
- 44.Mongiat M, Sweeney SM, San Antonio JD, Fu J, Iozzo RV. Endorepellin, a novel inhibitor of angiogenesis derived from the C terminus of perlecan. J Biol Chem. 2003;278:4238–4249. doi: 10.1074/jbc.M210445200. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez CA, Butterfield C, Jackson G, Moses MA. Structural and functional uncoupling of the enzymatic and angiogenic inhibitory activities of tissue inhibitor of metalloproteinase-2 (TIMP-2): Loop 6 is a novel angiogenesis inhibitor. J Biol Chem. 2003;278:40989–40995. doi: 10.1074/jbc.M306176200. [DOI] [PubMed] [Google Scholar]
- 46.Xu R, et al. NC1 domain of human type VIII collagen (alpha 1) inhibits bovine aortic endothelial cell proliferation and causes cell apoptosis. Biochem Biophys Res Commun. 2001;289:264–268. doi: 10.1006/bbrc.2001.5970. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.