Abstract
Prp8 stands out among hundreds of splicing factors as a key regulator of spliceosome activation and a potential cofactor of the splicing reaction. We present here the crystal structure of a 274-residue domain (residues 1,822–2,095) near the C terminus of Saccharomyces cerevisiae Prp8. The most striking feature of this domain is a β-hairpin finger protruding out of the protein (hence, this domain will be referred to as the β-finger domain), resembling many globular ribosomal proteins with protruding extensions. Mutations throughout the β-finger change the conformational equilibrium between the first and the second catalytic step. Mutations at the base of the β-finger affect U4/U6 unwinding-mediated spliceosome activation. Prp8 may insert its β-finger into the first-step complex (U2/U5/U6/pre-mRNA) or U4/U6.U5 tri-snRNP and stabilize these complexes. Mutations on the β-finger likely alter these interactions, leading to the observed mutant phenotypes. Our results suggest a possible mechanism of how Prp8 regulates spliceosome activation. These results also demonstrate an analogy between a spliceosomal protein and ribosomal proteins that insert extensions into folded rRNAs and stabilize the ribosome.
Pre-mRNA splicing is a critical step for gene expression in all eukaryotes. In eukaryotes, DNA is first transcribed to pre-mRNAs whose introns have to be accurately removed before mRNA export and translation. Introns are removed through two transesterification steps. In the first step, the 2′-OH group of a critical adenosine residue in the branch point sequence (BPS) attacks the 5′ end of the intron and forms a lariat intermediate. In the second step, the newly freed 3′-OH group of the 5′-end exon attacks the 3′-end of the intron, releasing the lariat and ligating the two exons.
Pre-mRNA splicing is catalyzed by the spliceosome, a large RNA/protein complex that contains five snRNAs (U1, U2, U4, U5, and U6) and over 100 different protein factors. The spliceosome appears to assemble on pre-mRNA in a stepwise manner (1), although evidence also exists that the spliceosome may preassemble before encountering a pre-mRNA substrate (2). During spliceosome assembly, the 5′ splice site (ss), BPS, and 3′ ss of pre-mRNA are first recognized by the U1 snRNP, SF1/BBP, and U2AF65/35, respectively. Next, U2 snRNP replaces SF1 and base-pairs with the BPS. Subsequently, the U4/U6.U5 tri-snRNP joins the spliceosome. Next, extensive structural rearrangements occur to form the catalytically active spliceosome complex (first-step complex), which contains U2, U5, U6, and the pre-mRNA (3). During this activation process, the base-pairing between the 5′ ss and U1 snRNA is disrupted, and the 5′ ss interacts with the ACAGA box of U6 instead, using largely the same nucleotides that base-paired with U1 snRNA. The base-pairing between U4 and U6 is also disrupted, and new interactions between U2 and U6, which are mutually exclusive with those in the original U4/U6 complex, are formed. In addition, the BPS interacts with U2 snRNA, and both exons interact with U5 snRNP in this complex. After the first catalytic step, the spliceosome further changes conformations to perform the second catalytic reaction (4, 5). The exact nature of the second-step complex is unclear but may involve disruption of the U6–5′ ss interaction and repositioning of the pre-mRNA substrate (5). These conformational changes can be considered an activation process for the second catalytic step.
Prp8 occupies a central place in the spliceosome and is a key regulator of spliceosome activation. It is the only spliceosomal protein that extensively cross-links with the 5′ ss, BPS, 3′ ss, U5, and U6, all key components of the splicing reaction (6). A large number of mutations in PRP8 were identified that suppress the U4-cs1 cold-sensitive mutant, illustrating the role of Prp8 in the spliceosome activation mediated by U4/U6 unwinding (7–9). U4-cs1 is a mutation in U4 snRNA that resides opposite the U6 ACAGA box in the U4/U6 complex and prevents the ACAGA box from interacting correctly with the 5′ ss when yeast is grown at low temperatures (10). Consequently, U4-cs1 spliceosomes can assemble at low temperatures, but U4/U6 unwinding and the release of U1 snRNA from the 5′ ss are blocked. Over 40 Prp8 substitutions have been isolated that suppress the U4-cs1 cold sensitive mutant (8), which fall into five regions (a–e) in Prp8's primary structure. Regions a, d, and e demonstrate allele-specific genetic interactions with mutations in Prp28, Brr2, and U6 snRNA, respectively, suggesting at least three different mechanisms of U4-cs1 suppression (9).
Prp8 also regulates the equilibrium between the first catalytic step and the second catalytic step spliceosomal conformation. The two-state model proposed by Query and Konarska (11, 12) suggests that the conformation of the spliceosomal complex that favors the first step and the conformation that favors the second step are in competition. Modulation of the relative stabilities of the two conformations improves one of the catalytic steps and decreases the efficiency of the other. Two classes of Prp8 mutants (first- and second-step alleles) were identified that have opposite effects on the first and second catalytic steps of splicing, reflecting Prp8's ability to modulate the transition between the two catalytic steps (12).
In contrast to the clear functional importance of Prp8 in splicing, the molecular mechanism of how Prp8 fulfills these functions remains elusive, partly because of its large size, low sequence similarity with other proteins, and lack of structural information. Human Prp8 is 2,335 residues and the yeast Prp8 is 2,413 residues in length. The two proteins share 61% sequence identity, but both have remarkably low sequence similarity with other known proteins. To understand the molecular mechanism of Prp8's function, we used a multipronged strategy combining structural, genetic, and biochemical methods. We present here the 2.05-Å resolution crystal structure of a 274-residue domain (residues 1,822–2,095) near the C terminus of S. cerevisiae Prp8 (yPrp8). The most striking feature of this domain is a β-finger that protrudes from the protein. This domain will henceforth be referred to as the β-finger domain. We found that mutations at the β-finger affect the conformational equilibrium between the first and second catalytic step, demonstrating first-step and second-step allele phenotypes. Mutations at the base of the β-finger suppress U4-cs1 cold sensitivity. These observations lead us to propose that Prp8 inserts this β-finger into the first-step complex (U2/U5/U6/pre-mRNA) or U4/U6.U5 tri-snRNP and stabilizes these complexes. Mutations on the β-finger alter these interactions, resulting in the observed phenotypes. Our results provide a possible molecular mechanism for Prp8's function in spliceosome activation. These results also present an intriguing analogy between a spliceosome protein and many globular ribosomal proteins with protruding extensions that insert into folded rRNAs and stabilize the ribosome.
Results
The Prp8 β-Finger Domain Contains an RNase H Fold Without Its Active Sites.
We determined the crystal structure of the β-finger domain (residues 1,822–2,095) of yPrp8 to 2.05-Å resolution. The β-finger domain is immediately upstream of the C-terminal domain (CTD, residues 2,143–2,413) structure we determined earlier (13, 14) (Fig. 1A). The β-finger domain (274 residues) forms a compact single-domain structure that can be further divided into the N-terminal and C-terminal subdomains (Fig. 2A). The N-terminal subdomain (170 residues from amino acids 1,833–1,992) forms a α/β structure, whereas the C-terminal subdomain (95 residues from amino acids 1,993–2,087) is exclusively α-helical (Fig. 1B and Fig. 2A). The extreme N terminus (11 residues) and C terminus (8 residues) of the β-finger domain are disordered in the crystal structure. This domain contains a unique β-hairpin finger formed by β2 and β3 (residues 1,859–1,875). There are two molecules in the crystallographic asymmetric unit, which have essentially identical conformations except for the β-finger (Fig. 2B). The significance of the β-finger will be discussed later in this paper.
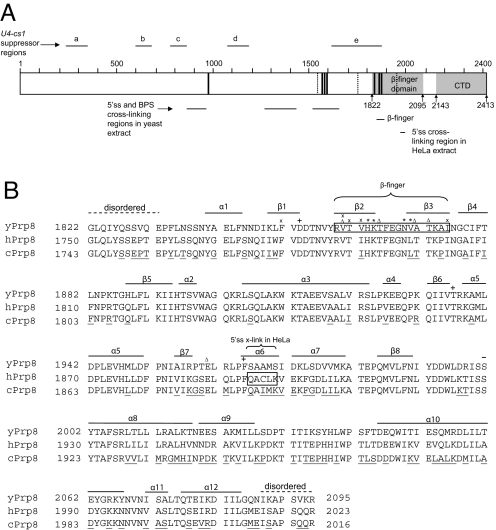
Fig. 1.
Position of the β-finger domain in the full-length yPrp8 primary structure and sequence alignment of the β-finger domain from different species. (A) A schematic diagram showing positions of the β-finger domain, the β-finger, and other functional regions, including the CTD, 5′ ss and BPS cross-linking regions in yeast extract, 5′ ss cross-linking region in HeLa extract, U4-cs1 suppressor regions, first-step alleles (dashed vertical line), and second-step alleles (solid vertical line). Only the first and second-step alleles that carry single site mutations are shown. Horizontal bars indicate the cross-linking and suppressor regions. (B) Sequence alignment of yeast Prp8 (yPrp8), human Prp8 (hPrp8), and Caenorhabditis elegans Prp8 (cPrp8) in the β-finger domain region. Alignment was performed by using MultAlin (40). Secondary structures as seen in the crystal structure of the β-finger domain of yPrp8 are labeled on top of the sequence. Dashed lines indicate residues that are disordered in the crystal structure. Boxed residues correspond to residues that compose the β-finger or are observed to cross-link with the 5′ ss in HeLa extract. Black underlines indicate residues that are not completely conserved among all three species. Δ, *, and x indicate first-step, second-step allele, and U4-cs1 suppressors, respectively. + indicates residues that correspond to the active sites in RNase H.
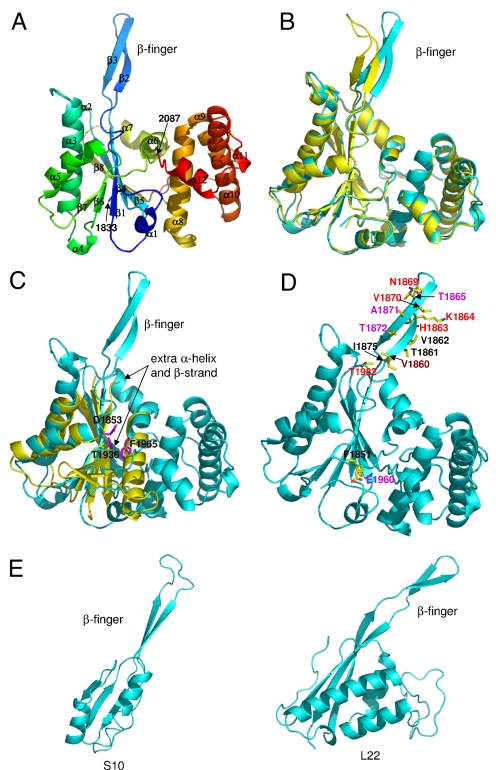
Fig. 2.
Structure of the β-finger domain of yPrp8. (A) The β-finger domain structure is colored in a rainbow spectrum from the N terminus to the C terminus, with secondary structures labeled. (B) The most striking feature of the β-finger domain is a protruding β-finger which adopts different conformations in the two molecules (yellow and cyan) in the asymmetric unit of the crystal. (C) The N-terminal α/β subdomain of the β-finger domain (cyan) is topologically similar to the RNase H fold as exemplified by the RNase H domain in the C-terminal of UvrC (yellow) (PDB ID code 2NRR). The extra α-helix and β-strand that are present in the β-finger domain but not in UvrC are labeled with arrows. Residues in Prp8 that correspond to the DDE active sites in RNase H are shown in purple. (D) The β-finger domain contains first-step alleles (purple), second-step alleles (red), and U4-cs1 suppressors (black). Brown designates the residue that confers both the first-step allele and U4-cs-1 suppressor phenotypes. (E) Examples of two ribosomal proteins (S10 and L22) with β-finger extensions.
A structural homology search using the DALI server (15) found that the N-terminal subdomain of the β-finger domain shares significant topological similarity with the RNase H fold in UvrC, a DNA repair enzyme (PDB ID code 2NRR) (Fig. 2C). The RNase H fold exists in a number of enzymes with nuclease or polynucleotide transferase activities, such as RNase HI (16), retrovirus integrase (17), Argonaute (18), RuvC (19), and DNA transposase (20). A typical RNase H fold has approximately 100 residues containing a five-stranded mixed β-sheet and three α-helices. The N-terminal subdomain of the β-finger domain has the same topology as a typical RNase H fold with two modifications (Fig. 2C). There is a β-finger insertion (β2 and β3 in the β-finger domain) between the first and the second β-strands of the RNase H fold and an extra short α-helix and β-strand after the last helix of the RNase H fold.
RNase-H-related enzymes typically contain three highly conserved active site residues, DDE, forming a carboxylate triad which coordinates divalent cations (i.e., Mg2+) critical for catalysis (21). In the β-finger domain of Prp8, residues D1853, T1936, and F1965 correspond to the DDE motif of RNase H fold in three-dimensional space (Fig. 2C). To examine the importance of D1853, which is highly conserved in Prp8 from different species, we generated a D1853A mutant of full-length yPrp8. Plasmid carrying prp8-D1853A was transformed into yJU75, which contains WT PRP8 on an URA3-marked plasmid, with chromosomal PRP8 deleted (22). The transformed strain failed to grow on medium containing 5-FOA, which selects against the URA3 plasmid carrying WT PRP8, indicating that D1853A is lethal (Fig. 3A).
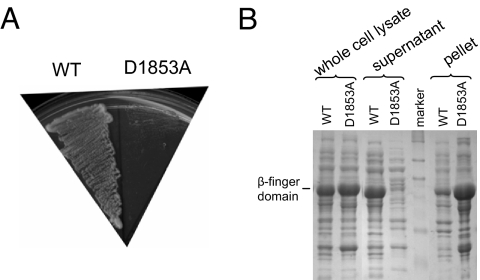
Fig. 3.
The lethality of D1853A is likely caused by protein misfolding/instability. (A) D1853A and WT Prp8 plasmids were transformed into yJU75 and plated on 5-FOA plates. D1853A is lethal and does not grow on 5-FOA plates at 30°C. (B) SDS-PAGE of WT and D1853A β-finger domain shows that D1853A is insoluble when expressed in E. coli.
To understand the mechanism of the D1853A mutant, we first examined whether the β-finger domain binds any metal ion with only one of the three canonical carboxylates present. We soaked the β-finger-domain crystals with 1 mM TbCl3 and collected anomalous data at the peak wavelength of Tb3+. Tb3+ has similar binding properties as Mg2+ (23) and a strong anomalous signal, which can be used to identify Mg2+ binding sites. An anomalous Fourier map revealed that D1853, T1936, and F1965 do not coordinate Tb3+ ion (data not shown). Instead, we found that the D1853A substitution leads to misfolding/instability of the β-finger domain. D1853A is almost exclusively in the insoluble fraction when expressed in Escherichia coli (Fig. 3B). The lethality of D1853A is likely caused by misfolded/unstable Prp8, illustrating again the importance of having functional Prp8 in the cell. This hypothesis is also consistent with the recessive lethality of D1853A (i.e., yeast carrying both WT PRP8 and D1853A plasmids grow similarly to those carrying WT PRP8 alone) (data not shown). A mutant that still allows the assembly of Prp8 into the spliceosome but blocks the function of Prp8 will likely have a dominant phenotype.
Interestingly, the β-finger domain is the second domain in Prp8 that has a fold similar to an enzyme but does not have its catalytic site(s). The C-terminal domain structure we determined earlier (13, 14) has the MPN fold present in several Zn2+-dependent isopeptidases that remove ubiquitin or ubiquitin-like molecules from target proteins (24). However, the partial JAMM-motif in the C-terminal domain of Prp8 does not bind Zn2+ and is unlikely to serve as a metalloenzyme. The β-finger domain reported here contains the structural fold of RNase H but does not have its active site. The evolutionary implication of these results will be interesting for further investigation.
The Protruding β-Finger of the β-Finger Domain Resembles Many Ribosomal Proteins.
The most striking feature of the β-finger domain is the β-hairpin finger protruding away from the protein (Fig. 2A). The β-finger contains both hydrophobic (V, I, and F) and hydrophilic residues interspersed with each other (Figs. 1B and 2D). The β-finger of both molecules in the asymmetric unit makes extensive contacts with multiple neighboring molecules in the crystal but is not part of any β-sheet. Therefore, the β-finger conformation does not seem to be induced by crystal contacts. It is not uncommon for proteins to have extended termini (such as viral capsid proteins and histone proteins), but it is highly unusual for a β-finger to protrude from the middle of a protein in isolation. It inevitably will interact with other proteins or RNA. The interaction between the β-finger and multiple neighboring molecules in the crystal exemplifies the tendency of the β-finger to interact with other partners.
The protruding β-finger is strongly reminiscent of many ribosomal proteins (over one-third of proteins in the large subunit and over half of proteins in the small subunit) with extensions protruding from globular protein bodies (25, 26). These protrusions in ribosomal proteins insert into folded 16S and 23S rRNAs. The protrusion often interacts with multiple domains in the 16S and 23S rRNAs, stabilizing the RNA structure, particularly the relative orientations of adjacent RNA domains. The extensions in ribosomal proteins can be extended loops, α-helix, or β-hairpin fingers. Fig. 2E presents examples of a protein from the large subunit (L22) and a protein from the small subunit (S10) with β-fingers. L22 inserts its β-finger into the 23S rRNA and interacts with all six domains of 23S rRNA (25). S10 interacts with both the 16S RNA and a neighboring protein S14 (26). The striking structural similarity between the β-finger domain and ribosomal proteins prompted us to examine the functional significance of the β-finger.
The β-Finger Regulates the Equilibrium Between the First and Second Catalytic Step.
Our first clue of the function of the β-finger comes from genetic mutations that affect the first- and second-step equilibrium, particularly the second-step alleles of Prp8. These alleles favor the second-step spliceosomal conformation and suppress a number of mutations at the 5′ ss (i.e., U2A or A3C), BPS (i.e., BSC or BSG), and 3′ ss (i.e., UuG) (collectively referred to as SS/BS mutations) (11, 12, 27, 28). The tip of the β-finger houses a number of known second-step alleles, including prp8–151 (N1869D) and prp8–162 (V1870N) (Fig. 2D). Another mutant located at the tip of the β-finger, prp8-D143 (K1864E), suppresses multiple 5′ ss and 3′ ss mutations (27) and is likely also a second-step allele. These observations suggest that the β-finger may stabilize the first-step complex and the above mutations destabilize the first-step complex, demonstrating second-step allele phenotypes.
To further understand the role of the β-finger in the first- and second-step equilibrium, we set out to examine whether other mutations on the β-finger demonstrate second-step allele phenotypes, using copper-resistance and primer extension assays. These assays use the yJU75 yeast strain harboring a deletion of the endogenous CUP1 gene and carrying ACT1-CUP1 splicing reporter plasmids with different SS/BS mutations (29). These strains normally do not grow well in copper-containing plates because defects in ACT1 splicing caused by SS/BS mutations lead to defective CUP1 function, which is responsible for copper resistance. The SS/BS suppression effect of second-step alleles can be observed as increased copper resistance in these strains (12, 29). Primer extension analyses demonstrate that the second-step allele has reduced lariat intermediate and increased mRNA product in these strains. The first-step alleles have the opposite phenotypes, showing decreased copper resistance in ACT1-CUP1 reporter strains carrying SS/BS mutations, increased lariat intermediate, and decreased mRNA product in primer extension assays.
We examined other β-finger mutants by generating the following single site mutations on full-length yPrp8: H1863E, T1865K, F1866K, E1867K, A1871E, T1872E, and K1873E. We mutated these residues to dramatically different amino acids to maximize our chance of obtaining mutants that will have any phenotype. None of these mutants confers any obvious growth phenotype at any temperature in yeast carrying WT ACT1-CUP1 reporter (Fig. 4A). Mutant H1863E suppresses BSG (BS mutation) and UuG (3′ ss mutation) in a copper-resistance assay (Fig. 4B). Primer extension demonstrates that this mutant leads to decreased lariat intermediate and increased mature mRNA product, characteristic of second-step alleles (Fig. 4C). H1863E, together with previously identified second-step alleles, points to the tip of the β-finger as a particularly important region in stabilizing the first-step conformation (Fig. 2D).
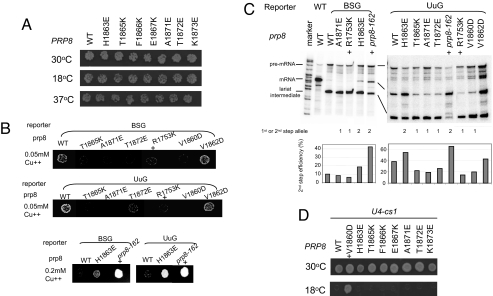
Fig. 4.
Copper-resistance and U4-cs1 suppression analyses of various β-finger mutations. (A) All β-finger mutations grow similarly to the WT at 30°C, 18°C, and 37°C. Only one concentration point in the serial dilution is shown. (B) Copper-resistance assay indicates that V1860D, T1865K, A1871E, and T1872E grow worse than the WT in both the BSG and UuG reporters at 0.05 mM Cu++ concentration, characteristic of first-step alleles. Mutant H1863E grows better than the WT in both the BSG and UuG reporters at 0.2 mM Cu++ concentration, characteristic of second-step alleles. V1862D behaves similarly to the WT and does not demonstrate a clear first- or second-step allele phenotype. Known first-step allele R1753K and second-step allele prp8–162 (V1870N) are used as positive controls and labeled with +. (C) Primer extension experiment indicates that V1860D, T1865K, A1871E, and T1872E are first-step alleles, which demonstrate increased lariat intermediate, reduced mRNA product, and reduced second-step efficiency compared with the WT. H1863E is a second-step allele, which demonstrates decreased lariat intermediate, increased mRNA product, and increased second-step efficiency compared to the WT. V1862D is neither a clear first- nor second-step allele. R1753K and prp8–162 (V1870N) are used as positive controls for first and second-step alleles, and the corresponding lanes are labeled with +. pBR322 DNA digested with MspI is used as a molecular weight marker. (D) V1860D (positive control, designated with +) but no other β-finger mutants tested suppress the U4-cs1 phenotype at 18°C. Only one concentration point in the serial dilution is shown.
Interestingly, three β-finger mutations, T1865K, A1871E, and T1872E, exhibit the first-step allele phenotype. They grow worse than WT in copper-resistance assays with BSG and UuG reporters (Fig. 4B). Primer extension experiments demonstrate that these mutants produce increased lariat intermediate and decreased mRNA product, typical of first-step alleles (Fig. 4C). In addition, we demonstrated that V1860D, a previously identified U4-cs1 suppressor (8), is also a first-step allele (Fig. 4B and C). These results indicate that substitution of some residues on the β-finger favor the first-step conformation, suggesting a more complicated role of the β-finger in the first- and second-step equilibrium instead of simply stabilizing the first-step complex to the fullest extent.
The β-Finger Regulates U4/U6 Unwinding-Mediated Spliceosome Activation.
Another important clue of the function of the β-finger comes from previously identified U4-cs1 suppressors (8). Five of the PRP8 U4-cs1 suppressor mutants in region e (F1851L, V1860D/N, T1861P, V1862A/D/Y, and I1875T) fall into the β-finger domain. F1851L is located inside the protein core and is not surface exposed (Fig. 2D). The effect of F1851L may be indirect through conformational changes of the protein. The other four U4-cs1 mutants are all located on the base of the β-finger on strand β2 or β3 (Fig. 2D). We found that none of the seven β-finger single-site mutants we generated suppresses the cold sensitivity of U4-cs1 (Fig. 4D). This finding is consistent with the notion that the large-scale U4-cs1 suppressor screen performed by Kuhn and Brow (8) has identified most or all of the regions of Prp8 involved in U4-cs1 suppression, because almost half of the suppressors were identified in two or more independent screens. These results suggest the β-finger is involved in the regulation of U4/U6 unwinding-mediated spliceosome activation.
We evaluated whether the U4-cs1 suppression phenotype overlaps with either the first-step or second-step allele phenotypes. We tested two U4-cs1 suppressors (V1860D and V1862D) in copper-resistance and primer extension assays (Fig. 4). V1860D demonstrates the first-step allele phenotype. V1862D behaves similarly to the WT and is neither a first-step nor second-step allele. Another U4-cs1 suppressor, T1861P (prp8–201), suppresses multiple SS/BS mutants (8, 27) and is likely a second-step allele. There appears to be no uniform overlap between the U4-cs1 suppression phenotype and either the first- or second-step allele phenotype.
The β-Finger Domain Binds ss and dsRNA Weakly.
To evaluate whether the β-finger domain binds ss or dsRNA in vitro, we performed electrophoresis mobility shift assay (EMSA) by using an arbitrary 13-nt RNA (GCUUUACGGUGCU, referred to as ssR1), its double-stranded (ds) form (dsR1), a 13-nt sequence from U6 that forms Helix II in the first-step complex (GAGAUUUAUUUCG, referred to as R2), and its ds form (dsR2). The β-finger domain binds to all four RNAs weakly, whereas the CTD of Prp8 does not [supporting information (SI) Fig. S1]. The low percentage of binding in Fig. S1 with 32–33 μM protein indicates a Kd >32 μM for all RNAs. Deletion of the β-finger (residues 1,860–1,874 deleted and replaced with Gly-Ser) does not affect the overall protein structure and stability (data not shown) but reduces the binding (Fig. S1). These results indicate that the β-finger domain and its β-finger may play a role in RNA binding.
Discussion
The combination of structural and genetic data points to an attractive model for the function of the β-finger. The β-finger seems to resemble the extensions on many ribosomal proteins structurally and functionally. Analogous to the extensions of ribosomal proteins that insert in folded rRNAs and stabilize the ribosome, the β-finger in the β-finger domain may insert into the first-step complex containing U2/U5/U6/pre-mRNA and stabilize this complex. Because the spliceosome is more protein rich than the ribosome, the β-finger can potentially contact RNA, protein, or both to stabilize the first-step complex. This possibility explains why mutations on the β-finger demonstrate second-step allele phenotypes, likely because of destabilization of the first-step complex. In general, the spliceosome (particularly its RNA components) likely needs stabilizers. It is not difficult to imagine that a significant number of spliceosomal proteins may resemble Prp8 and ribosomal proteins. These proteins may insert protruding extensions into spliceosomal complexes and serve as stabilizers for these complexes.
The interactions between the β-finger and the spliceosome are likely weak because of the dynamic nature of the spliceosome. For example, the first-step complex has to be disrupted before the second catalytic step. These weak interactions are consistent with the weak RNA binding affinity observed for the β-finger domain (Fig. S1) and the fact that the β-finger is not particularly Lys/Arg-rich as are extensions on ribosomal proteins (Figs. 1B and 2D). These potentially weak interactions also predict that some mutations on the β-finger can be first-step alleles. These residues may not originally contact U2/U5/U6/pre-mRNA, but mutations of these residues may establish additional contacts with U2/U5/U6/pre-mRNA, stabilizing the first-step conformation and demonstrating first-step allele phenotypes. Indeed, three of the seven β-finger mutations we generated (T1865K, A1871E, and T1872E) and a previously identified U4-cs1 mutation (V1860D) are first-step alleles (Fig. 4). It should be noted that we cannot rule out the possibility that the β-finger plays an additional role in stabilizing a second-step conformation. In this event, the first-step alleles may destabilize whereas the second-step alleles stabilize this conformation. The presence of first- and second-step alleles on the β-finger is in line with the previously observed mechanistic similarity between these alleles and the ram and restrictive mutants in ribosome, which affect tRNA recognition accuracy (12). The ram and restrictive mutants are located on the large- and small-subunit interface, influencing the open-close conformational equilibrium (30). In parallel, a significant number of first- and second-step alleles are located on the β-finger, influencing the first-second-step conformational equilibrium.
The β-finger is likely not the only region, but one of several regions, on Prp8 involved in stabilizing the first-step complex. This possibility is supported by the presence of a first-step allele (E1960K = prp8-101) in the globular region of the β-finger domain on the protein surface (on the loop connecting β7 and α6) (Fig. 2D). This mutation decreases the cross-linking between Prp8 and the 3′ ss in vitro (31). T1982A (prp8-153) suppresses multiple 5′ ss and 3′ ss mutations (28) and is potentially a second-step allele. T1982 is located on the protein surface of the globular region, close to the β-finger (Fig. 2D). Furthermore, in HeLa extract, 5′ ss has been observed to cross-link to a region close to residues 1,894–1,898 (corresponding to residues 1,966–1,970 in yPrp8) (32). Residues S1966, A1967, and S1970 (corresponding to Q1894, A1895, and K1898 in hPrp8) are indeed surface exposed in the β-finger domain structure, although two of the above residues are not conserved in yPrp8 (Fig. 1B) and it is unclear whether the equivalent residues in yeast contact RNA. Another region of Prp8 that very likely contacts RNA and contributes to the stabilization of the first-step complex is the central region of Prp8 where cross-linking between 5′ ss/BPS and Prp8 has been observed (33). Consistent with this notion, all of the other known first- and second-step alleles outside the β-finger domain (R1753K, L1557F, P986T, T1565A N1721Y V1752A, W1575R, E1576V, W1609R N1618D) (12) reside within or in close proximity to the three regions that cross-link with the 5′ ss and BPS (residues 871–970, 1,281–1,413, 1,503–1,673) (33) (Fig. 1A).
The β-finger may also stabilize the tri-snRNP complex. This possibility will explain why mutations around the base of the β-finger suppress U4-cs1, likely because of destabilization of the tri-snRNP. The fact that U4-cs1 suppressors do not uniformly overlap with the first-step alleles (Fig. 4) argues against the possibility that these mutants indirectly suppress U4-cs1 through the stabilization of the first-step complex; instead, it argues for the possible role of the β-finger in a different event such as tri-snRNP stabilization. Consistent with this possible role, Prp8 is clearly present in both the U4/U6.U5 tri-snRNP (34) and the spliceosome (35). In yeast, nucleotides U97 of U5 snRNA and U54 of U6 in the tri-snRNP have been cross-linked to the central region of Prp8 (33), indicating the proximity of Prp8 to U5 and U6. Furthermore, three U4-cs1 suppressors at the base of the β-finger (V1860D, T1861P, and V1862D) are synthetically lethal with U6-UA (a mutant that hyperstabilizes the U6 intramolecular stem) (9), suggesting potential interactions between Prp8 and U6.
The β-finger can potentially play a role in regulating spliceosome activation. The interactions between the β-finger and the spliceosomal complexes are likely weak. In addition, the β-finger can readily change conformations, evidenced by the different conformations adopted by the two molecules in the asymmetric unit of the crystal (Fig. 2B). The conformational changes, potentially induced by other changes in the spliceosome, may serve as a tool for regulating spliceosome activation.
Materials and Methods
Yeast Strains and Plasmids.
Copper-reporter and primer extension experiments were performed in strain yJU75 [MATa, ade2 cup1Δ::ura3 his3 leu2 lys2 prp8Δ::LYS2 trp1; pJU169 (PRP8 URA3 CEN ARS)] carrying ACT1-CUP1 reporter plasmids (WT, BSG, and UuG) (12, 29) (gift of C. Query, Albert Einstein College of Medicine, Bronx, NY). U4-cs1-suppression experiments were carried out in strain ZRL102 (MATa snr14::TRP1 prp8Δ::ADE2 trp1 ura3 lys2 his3 ade2 [pRS314-U4-cs1] [YCp50-PRP8]) (8) (gift of D. Brow, University of Wisconsin, Madison, WI). Most Prp8 mutants were generated by using pRS313(HIS3)-PRP8(SacII) (8) (gift of D. Brow) as a template and the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Mutants were confirmed through DNA sequencing. Mutant plasmids prp8-V1860D and prp8-V1862D were gifts from D. Brow; prp8–162 was the gift of C. Query; and prp8-R1753K was the gift of B. Schwer (Weill Cornell Medical College, New York).
Protein Expression and Purification.
S. cerevisiae Prp8 residues 1,822–2,095 (the β-finger domain) were subcloned from pRS313(HIS3)-PRP8(SacII) (8) into a pGEX-6P-1 vector (GE Healthcare) and expressed in E. coli strain XA90 as a GST-fusion protein. The fusion protein was first purified by using glutathione Sepharose resin and cleaved by using PreScission protease. The resultant β-finger domain was further purified on a Superdex-200 gel-filtration column and concentrated to 16 mg/ml for crystallization trials.
Crystallization and Data Collection.
The β-finger domain was crystallized by the hanging drop vapor diffusion method by using a well solution containing 0.1 M Tris-HCl (pH 8.5), 10% PEG8000, and 0.2 M Li2SO4. All crystallographic data were collected by using the mail-in data collection program at the National Synchrotron Light Source. Data were processed by using the HKL2000 package (36), and data statistics are shown in the Table S1.
Structural Determination.
The structure of the β-finger domain was determined by using the Se-Met SAD method and the Solve and Resolve programs (37) with Se-Met positions identified by the HKL2MAP program (38). The initial electron density map was of excellent quality, and most of the main chain and side chains were traced without any ambiguity. Refinement was performed by using CNS (39), and refinement statistics are shown in the Table S1. The structure has been deposited to the Protein Data Bank (PDB ID code 3E66).
Copper-Resistance Assay.
ACT1-CUP1 reporter plasmids (WT, BSG, and UuG) were first transformed into S. cerevisiae strain yJU75. Then Prp8 mutants on pRS313 (HIS3) were transformed into yJU75 carrying different ACT1/CUP1 reporters. Transformed yeasts were plated on 5-fluoroorotic-acid plates (5-FOA) to shuffle out WT PRP8. The resulting strains were grown to mid-log phase in -Leu medium and diluted to OD600 = 0.1. Equal volumes of yeast culture were dropped onto -Leu plates containing 0–1.0 mM CuSO4. These plates were incubated at 30°C for 3 days and photographed. All mutants in yJU75 carrying WT reporter were used to evaluate whether these mutants have any general growth defects at 30°C, 18°C, and 37°C.
U4-cs1 Suppression Experiments.
Prp8 mutants were generated as detailed in the copper-resistance assay and transformed into ZRL102. Transformed yeasts were plated on 5-FOA plates to shuffle out WT PRP8. The resulting strains were plated on yeast extract/peptone/dextrose plates, grown at 18°C for 10 days, and photographed.
Primer Extension.
Yeast strains carrying PRP8 mutants and different ACT1-CUP1 reporters were grown in -Leu medium to OD600 = 1.0. Total RNA was extracted by using the MasterPure Yeast RNA purification Kit (Epicentre Biotechnologies), and primer extension was performed by using SuperScript III Reverse Transcriptase (Invitrogen) and primer YAC6 5′-GGCACTCATGACCTTC-3′ (complementary to nucleotides 31–46 of CUP1 fused downstream of ACT1 exon 2). Primer extension products were separated on a 7% polyacrylamide, 8 M urea gel, visualized, and quantified by autoradiography. Second-step efficiency was calculated as M/(M+LI), where M is mRNA and LI is lariat intermediate quantity.
Protein and RNA Interaction Using EMSA.
RNA oligo was ordered from Integrated DNA Technologies and 5′-end-labeled with 32P-γ-ATP by using polynucleotide kinase (Fermentas). Labeled RNA was incubated on ice with 20 μg of protein in 20 μl of 10 mM Hepes (pH 7.6), 1 mM MgCl2, 100 mM NaCl, and 5% glycerol for 60 min. The reaction mixture was separated on a 4% native polyacrylamide gel in TBE buffer and visualized by using autoradiography.
Supplementary Material
Acknowledgments.
We thank D. Brow, C. Query, and B. Schwer for providing yeast strain and plasmids; D. Brow, J. Kieft, and R. Davis for helpful discussion and/or critical reading of the manuscript; the staff of the x-ray facility (supported in part by the University of Colorado Cancer Center) and Biophysics core facility at the University of Colorado Denver Anschutz Medical Campus. Financial support for National Synchrotron Light Source comes principally from the Offices of Biological and Environmental Research and from the Basic Energy Sciences of the U.S. Department of Energy and the National Center for Research Resources of the National Institutes of Health. This work was supported by a National Science Foundation Grant MCB-0718802 (to R.Z.). R.Z. is a Kimmel Scholar. K.Y. is supported in part by a Thorkildsen postdoctoral fellowship. L.Z. is supported by an American Heart Association postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805960105/DCSupplemental.
References
- 1.Burge CB, Tuschl TH, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, Cech T, Atkins JF, editors. The RNA World. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 2.Stevens SW, et al. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 3.Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 4.Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 5.Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Grainger RJ, Beggs JD. Prp8 protein: At the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn AN, Li Z, Brow DA. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol Cell. 1999;3:65–75. doi: 10.1016/s1097-2765(00)80175-6. [DOI] [PubMed] [Google Scholar]
- 8.Kuhn AN, Brow DA. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics. 2000;155:1667–1682. doi: 10.1093/genetics/155.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn AN, Reichl EM, Brow DA. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation; Proc Natl Acad Sci USA; 2002. pp. 9145–9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Brow DA. A spontaneous duplication in U6 spliceosomal RNA uncouples the early and late functions of the ACAGA element in vivo. RNA. 1996;2:879–894. [PMC free article] [PubMed] [Google Scholar]
- 11.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, et al. Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants. Protein Sci. 2007;16:1024–1031. doi: 10.1110/ps.072872007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pena V, Liu S, Bujnicki JM, Luhrmann R, Wahl MC. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell. 2007;25:615–624. doi: 10.1016/j.molcel.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 15.Holm L, Sander C. Dali: A network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 16.Yang W, Hendrickson WA, Crouch RJ, Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 17.Rice PA, Baker TA. Comparative architecture of transposase and integrase complexes. Nat Struct Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- 18.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 19.Ariyoshi M, et al. Atomic structure of the RuvC resolvase: A holliday junction-specific endonuclease from E. coli. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 20.Lovell S, Goryshin IY, Reznikoff WR, Rayment I. Two-metal active site binding of a Tn5 transposase synaptic complex. Nat Struct Biol. 2002;9:278–281. doi: 10.1038/nsb778. [DOI] [PubMed] [Google Scholar]
- 21.Haren L, Ton-Hoang B, Chandler M. Integrating DNA: Transposases and retroviral integrases. Annu Rev Microbiol. 1999;53:245–281. doi: 10.1146/annurev.micro.53.1.245. [DOI] [PubMed] [Google Scholar]
- 22.Umen JG, Guthrie C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brewer JM, Carreira LA, Irwin RM, Elliott JI. Binding of terbium (III) to yeast enolase. J Inorg Biochem. 1981;14:33–44. doi: 10.1016/s0162-0134(00)80012-4. [DOI] [PubMed] [Google Scholar]
- 24.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 26.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 27.Collins CA, Guthrie C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siatecka M, Reyes JL, Konarska MM. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 31.Umen JG, Guthrie C. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA. 1995;1:584–597. [PMC free article] [PubMed] [Google Scholar]
- 32.Reyes JL, Gustafson EH, Luo HR, Moore MJ, Konarska MM. The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5′ splice site. RNA. 1999;5:167–179. doi: 10.1017/s1355838299981785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner IA, Norman CM, Churcher MJ, Newman AJ. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA. 2006;12:375–386. doi: 10.1261/rna.2229706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens SW, Abelson J. Purification of the yeast U4/U6. U5 small nuclear ribonucleoprotein particle and identification of its proteins; Proc Natl Acad Sci USA; 1999. pp. 7226–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology. Vol 276. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 37.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Page T, Schneider TR. HKL2MAP: A graphical user interface for phasing with SHELX programs. J Appl Cryst. 2004;37:843–844. [Google Scholar]
- 39.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.