Abstract
The induced fit and conformational selection/population shift models are two extreme cases of a continuum aimed at understanding the mechanism by which the final key-lock or active enzyme conformation is achieved upon formation of the correctly ligated enzyme. Structures of complexes representing the Michaelis and enolate intermediate complexes of the reaction catalyzed by phosphoenolpyruvate carboxykinase provide direct structural evidence for the encounter complex that is intrinsic to the induced fit model and not required by the conformational selection model. In addition, the structural data demonstrate that the conformational selection model is not sufficient to explain the correlation between dynamics and catalysis in phosphoenolpyruvate carboxykinase and other enzymes in which the transition between the uninduced and the induced conformations occludes the active site from the solvent. The structural data are consistent with a model in that the energy input from substrate association results in changes in the free energy landscape for the protein, allowing for structural transitions along an induced fit pathway.
Keywords: enzyme dynamics, population shift, phosphoenolpyruvate carboxykinase
It has been 50 years since the original presentation of the induced fit hypothesis by Daniel Koshland (1) that built upon Emil Fisher's key-lock principle (2) and introduced the idea that the inherent dynamic properties of enzymes play an essential role in the processes of molecular recognition and catalysis. Over the last decade, with the advent of new instrumentation and novel experimental approaches there has been a resurgence in investigations focused upon understanding the specific ways in that these dynamic properties influence the ability of enzymes to achieve enormous levels of selectivity and catalytic power (refs. 3–6 and references therein).
Presently there are two primary models used to explain the mechanism by which an enzyme can move toward adopting the final key-lock state (Fig. 1): (i) the induced fit model and (ii) the conformational selection/population shift model (7–14). As defined originally by Kosh land (1), the induced fit model states that the achievement of the final key-lock state, that is, the precise orientation of catalytic groups and residues necessary for catalysis, occurs only after changes in protein structure that are induced by the binding of a substrate. This pathway is represented by the equilibrium constants K1 and K2 in Fig. 1. Intrinsic to this model is the ability of the enzyme to form an “encounter complex”† (Fig. 1B). In this complex the enzyme is liganded identically to the eventual catalytically competent key-lock state; however, the conformational change leading to the active key-lock state has not yet occurred. In contrast, the conformational selection model states that the enzyme exists in multiple conformational states, of which the substrate has a high affinity for the active or key-lock state. In this model the ligand is suggested to bind to this lowly populated high-energy state (the active or key-lock state) (Fig. 1C). In a mechanism similar to that of the Monod–Wyman–Changeux model for allostery (15), to maintain equilibrium (Fig. 1, K3) binding to the active state conformation shifts the population of free molecules toward the active state, facilitating further substrate binding. NMR studies of enzymes such as RNase A (16, 17), cyclophilin A (18), and dihydrofolate reductase (19) provide evidence for conformational selection because the experimental data supports that even in the absence of ligand the enzyme samples multiple conformational states including the ligand-bound (active) state.
Fig. 1.
Thermodynamic cycle for the formation of the active key-lock state. The induced fit pathway is represented by the equilibrium constants K1 and K2 whereas the conformational selection pathway is represented by the equilibrium constants K3 and K4.
One issue with the conformational selection model that is often overlooked is that in enzymes such as triosephosphate isomerase (TIM) and tyrosyl-tRNA synthetase the formation of the active or key-lock complex results in the closure of an active site lid domain. This results in the occlusion of the active site from the bulk solvent (20–24). These conformational changes, often required to preclude solvent from competing with the catalzyed reaction (25, 26), also preclude substrate binding directly to this enzyme form. Therefore, even if we assume that the lowly populated “active” form of the conformational selection model is the same as the “key-lock” state and that the enzyme samples this state in solution, then substrate cannot bind directly to this state due to steric occlusion of the active site [supporting information (SI) Fig. S1]. This would exclude the conformational selection model in the mechanism of catalysis for enzymes in which such conformational changes occur. If the lowly populated active state of the conformational selection model is not the same as the final key-lock state, then “induction” from substrate binding is required for conformational selection to achieve the key-lock state, consistent with the key tenet of Koshland's induced fit model in these systems.
Our investigations of structures of the gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PEPCK) (Scheme S1) that mimic the Michaelis complexes for the forward and reverse reactions and the structure of an approximation of the enolate intermediate state (Fig. S2) provide continuing support for the induced fit hypothesis. This support is provided by direct evidence for the existence of the encounter complex as predicted by Koshland's induced fit model. Our studies suggest that the enzyme facilitates the formation of the key-lock conformation by making its formation more energetically favorable. This favorability increases as the reaction progresses from the holoenzyme state through the Michaelis complex and to the intermediate complex that is committed to catalysis. This suggests that the enzyme changes the thermodynamic landscape for the structural transition between the uninduced and key-lock states during progression through the catalytic cycle to facilitate catalysis. In the case of PEPCK, because closure of the active site lid domain precludes binding directly to the key-lock state, a change in the thermodynamic landscape after formation of the encounter complex results in the mechanism by which the induced fit is achieved.
Results and Discussion
Michaelis Complex States.
PEPCK catalyzes the conversion of oxaloacetic acid (OAA) to phosphoenolpyruvate (PEP) as the first committed step of gluconeogenesis (Scheme S1). Previous work has demonstrated that the monocarboxylic acids 2-phosphoglycolate (PGA) and β-sulfopyruvate (β-SP) are competitive inhibitors of PEPCK with respect to PEP and they are excellent stereoelectronic analogues of PEP and OAA respectively (Fig. S2) (27, 28). It also has been demonstrated previously that in the ternary PEPCK–Mn2+–PGA complex PGA binds in a similar fashion to PEP (28). On the basis of these principles, we sought to crystallize PEPCK in complexes with β-SP and PGA and the correct nucleotide counterpart that mimic the Michaelis complexes for both the forward and the reverse reaction catalyzed by the enzyme.
The Michaelis Complexes: Lid Closure Is Necessary for Catalysis.
Previous structural analysis of PEPCK has demonstrated that PEP and PGA bind to the enzyme in the ternary complex in a conformation that would not allow for inline phosphoryl transfer about the bimetal center, as the stereochemistry for phosphoryl transfer and the individual PEPCK–Mn2+–OAA and PEPCK–Mn2+–Mn2+GTP complexes suggest (28–32). This observation resulted in the hypothesis that a dynamic active site “lid” domain closes upon formation of the Michaelis complex, stimulating PEP/PGA to coordinate directly to the active site manganese ion and placing the phosphate in a position similar to that of the γ-phosphate of GTP in the PEPCK–Mn2+–Mn2+GTP complex (29). This lid domain has an omega-loop-type structure similar to the lid domain found in TIM, tyrosyl-tRNA synthetase, and other enzymes (20–24, 33, 34).
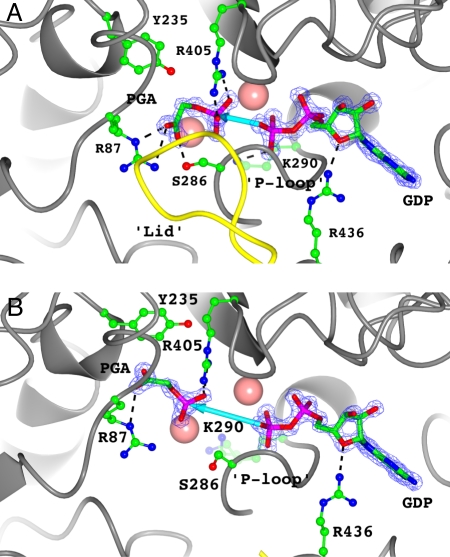
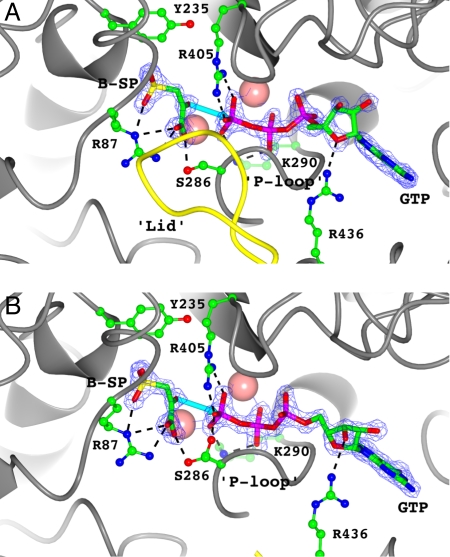
Analysis of molecule A of the crystallographic dimer of the PEPCK–Mn2+–PGA–Mn2+GDP complex mimicking the Michaelis complex for the reverse reaction clearly supports the previous hypothesis. In this molecule, the lid is observed in a fully closed conformation, capping the active site binding pocket and resulting in the direct coordination of PGA to the active site metal (Fig. 2A). This orientation of PGA in the active site places the phosphate as a bridging ligand between the two metals in a nearly identical position to that of the γ-phosphate of GTP in the GTP (32), β-γ-methylene–GTP (35), β-SP–GTP (this work), and oxalate–GTP (this work) structures. An identical closed-lid conformation is seen in molecule A of the PEPCK–Mn2+–β-SP–Mn2+GTP structure. Again, this arranges the phosphoryl donor and acceptor about the bimetal active site in an identical fashion to the PGA–GDP complex, consistent with the mechanism of phosphoryl transfer (Figs. 2 and 3). Further, the closing of the lid results in a slight closure of the active site cleft between the N- and the C-terminal lobes (Fig. S3). This motion, coupled with the slight closure of the P-loop (phosphate-binding loop/kinase-1a motif) mediated through the establishment of a hydrogen bond between S286 and the C1 carboxylate oxygen of PGA, results in a shift in the position of the bound nucleotide toward PGA through interactions between the backbone amides of the P-loop and the β-phosphate of GDP (Fig. 2 and Fig. S3B). The movement in the position of GDP from the location it occupies in the open-lid state (Fig. 2B) shortens the distance for phosphoryl transfer by ≈1 Å. The movement of the P-loop toward the metal center to a fully closed conformation described above appears necessary for the lid to occupy the closed conformation. This is due to steric conflict between A287 and T465 on the P-loop and lid domains, respectively, when the P-loop occupies any of the other more open conformations (Fig. S4) (32). This suggests that the closure of the lid and the P-loop are coupled processes.
Fig. 2.
The active site of the PEPCK–Mn2+–PGA–Mn2+GDP complex in the (A) closed-lid and (B) open-lid/disordered states. Catalytic residues discussed in the text are rendered as ball-and-stick models whereas the PGA and GDP ligands are rendered as thick sticks. Potential hydrogen bonds and ionic interactions between the ligands and the active site residues are indicated with dashed lines. The protein backbone is rendered as a gray ribbon except for the region corresponding to the active site lid domain (463–474) that is colored yellow. The active site and nucleotide-associated metals are rendered as pink spheres at 0.8 of their van der Waals radius. 2Fo − Fc density rendered at 2σ is shown for PGA and GTP as a blue mesh. The P-loop motif containing S286 and K290 is also labeled. A thick blue arrow indicates the potential phosphoryl transfer distance between the two ligands. This distance is 3.2 and 5.9 Å in A and B, respectively.
Fig. 3.
The active site of the PEPCK–Mn2+–β-SP–Mn2+GTP complex in the (A) closed-lid and (B) open-lid/disordered states. The coloring, rendering, and labeling of the figure is identical to that described for Fig. 2. The potential phosphoryl transfer distances between the two ligands are 3.2 and 3.7 Å in A and B, respectively.
Structural Evidence for the Encounter Complex.
The second molecule in the asymmetric unit (ASU) (molecule B) of both the β-SP–GTP and the PGA–GDP complexes has an identical ligation state to its closed-lid counterpart (molecule A) described above. Despite this fact, in both structures the lid of this second molecule remains in its open/disordered conformation (Figs. 2B and 3B). As a result, the substrates are found in the identical orientation as they are in their respective ternary complexes (Figs. 2B and 3B) (28, 29). Therefore, the complexes present in molecule B of both Michaelis complex states clearly represent the encounter complex as predicted by the induced fit model (Fig. 1B). The dramatic difference in the positioning of PGA and the nucleotide substrates in the closed-lid conformation compared with that of the open-lid conformation of the enzyme decreases the phosphoryl transfer distance from one that is unreasonable for direct inline phosphoryl transfer (molecule B, PGA–GDP = 5.9 Å; Fig. 2; molecule B, β-SP–GTP = 3.9 Å, Fig. 3) to one that is almost within van der Waals contact distance (molecule A, 3.2 Å, Figs. 2 and 3). By extrapolation of the results from the PEPCK–Mn2+–PGA–Mn2+GDP complex to the complex that would be formed with the substrate PEP, the structural data clearly demonstrate that lid closure triggers the transition of PEP from its outer-sphere coordinated position to a catalytically competent state necessary for phosphoryl transfer, as was previously hypothesized (29). In addition, the closed conformation results in repositioning of the nucleotide substrate that is also necessary to close the distance for phosphoryl transfer to one that is reasonable.
These structures, representative of the open- and closed-lid conformations of the Michaelis complexes for the forward and reverse reactions, demonstrate the intimate relationship between the closed-lid state and catalysis. Further support for this conclusion comes from the observation of electron density consistent with a minor population of the open-lid conformation of the GTP nucleotide present in the closed-lid molecule (molecule A) of the PEPCK–Mn2+–β-SP–Mn2+GTP complex (Fig. S5). The population distribution is coincident with 80% of the protein molecules composing molecule A having the lid closed. Therefore, 20% of the lids in all A molecules composing the crystal are in the open/disordered state, resulting in 20% of the GTP nucleotide in molecule A being in the conformation equivalent to that found in molecule B (lid open). Thus, even within molecule A of the β-SP–GTP complex, the structural data confirm the direct relationship between lid closure and the observed conformational changes.
On the basis of these and other structural studies of PEPCK, it is apparent that the closed-lid state represents the final key-lock complex necessary for chemical transformation. What is clear from the nature of these structural complexes is that the direct binding of substrate to the closed-lid state, which is required by the conformational selection model, is impossible (Fig. S1). Thus, the conformational selection model cannot explain the coupling of the conformational change and catalysis in this instance. Furthermore, the observation of the open-lid complex under identical ligation conditions to that of the closed-lid state provides direct structural evidence for the existence of the encounter complex. This observation is a key tenet in Koshland's induced fit model and provides evidence that is contrary to the requirements of a pure conformational selection model.
Formation of the Enolate Intermediate Results in Further Stabilization of the Closed-Lid State.
Oxalate has long been used as a mimic of the putative enolate intermediate in the reaction catalyzed by PEPCK (Fig. S2). This enolate mimic has been demonstrated previously both to be an effective competitive inhibitor and to bind directly to the active site Mn2+ ion in a fashion identical to that of the central skeleton of OAA (27, 28, 36, 37).
Examination of the crystal structure of the intermediate complex mimic shows little difference from that of the β-SP–GTP complex, with the phosphate of GTP positioned perfectly for inline phosphoryl transfer to the C1 carboxylate oxygen of oxalate (Fig. S6). The only significant difference is the rotation of Y235 forward, toward the metal in an identical fashion to that observed in the closed conformation of the PGA–GDP complex. In this case Y235 occupies the void left by what would be the decarboxylation of OAA. The most striking observation in the oxalate–GTP complex is that in both the cocrystal complex (P212121, 1 molecule/ASU) and the complex obtained by soaking of oxalate into the PEPCK–Mn2+-Mn2+GTP crystal (P21, 2 molecules/ASU) all of the molecules composing the crystal are in the closed state. As discussed below, this stabilization of the closed-lid state upon formation of the reactive intermediate species is consistent with the mechanism of catalysis for the enzyme and the coupling of binding energy to the stabilization of reactive intermediates put forth previously by Fersht (38).
Induced Fit and Conformational Selection.
Recently there have been issues raised with the notion of induced fit as originally described, particularly as it applies to antibody–antigen interactions (7–9, 11, 13). One concern is the idea that the equilibrium represented by K1 (Fig. 1) is unfavorable, resulting in only a small chance for the induced fit to occur. Thus, with reasonable values for the transitions along the induced fit pathway, this process is postulated to occur too slowly to account for most biological processes (8). The open-lid conformations in the Michaelis-like complexes presented here provide direct structural evidence for the encounter complex that is a requirement of the induced fit hypothesis. These complexes demonstrate, at least in the case of PEPCK, that the formation of a thermodynamically favorable encounter complex with the substrates of the reaction is possible (the equilibrium represented by K1 in Fig. 1 and Fig. S1) in the absence of the induced fit conformational change. As is evident from our studies and discussed below, there is no reason to assume that the uninduced complex is weak. Those interactions facilitating the formation of the encounter complex can be significant, although not of a magnitude necessary to cause a shift in the thermodynamic favorability of the enzyme adopting the final key-lock state. Therefore, we conclude that the formation of the encounter complex is possible, allowing for transitions along the induced fit pathway.
Energetic Implications for the Open/Closed Transition in Catalysis.
It has been demonstrated previously in several enzyme systems that the motion of active site loop regions is either rate limiting or intimately coupled to the chemical step (39–42). Previous structural studies have demonstrated that the predominant state for the active site lid domain in most complexes of PEPCK studied to date is the open/disordered state (28, 29, 32, 35). On the basis of the structural studies of complexes mimicking the Michaelis and enolate intermediate states presented here, the thermodynamic favorability of the closed state appears to increase as the enzyme commits to catalysis by formation of the enolate intermediate. Taken together, the structural information suggests a mechanism in which the closed conformation is stabilized as the enzyme proceeds from a predominantly open state in the holoenzyme and individual substrate complexes, to equal probability of the open and closed states in the Michaelis complex (represented by the β-SP–GTP and PGA–GDP complexes), and finally to a predominantly closed state upon the formation of the enolate intermediate (represented by the oxalate–GTP complex) (Fig. S7). This change in the energetic favorability of the enzyme adopting the closed state can be rationalized mechanistically. The holoenzyme, individual substrate/product, and Michaelis complexes need to promote substrate association and product dissociation and thus must sample the open state with some frequency. That frequency is likely to be consistent with data demonstrating that lid motion in TIM occurs on a similar time scale to kcat (39, 43). In PEPCK, once decarboxylation has occurred, the enzyme is committed to catalysis and must remain closed until after phosphoryl transfer has occurred to form the product. If the lid were to open after decarboxylation and before phosphoryl transfer, then the enolate intermediate would be protonated, resulting in the formation of pyruvate. Metabolically, this would have dire consequences by decoupling OAA consumption from PEP synthesis. A similar function has been ascribed to the active site lid domain in TIM, which is thought to sequester the intermediate of that reaction, preventing its hydration, which would lead to the toxic side product methylglyoxal (25, 26). Thus, our model for PEPCK catalysis suggests a mechanism by which the closed conformation of the enzyme becomes increasingly thermodynamically favorable as the enzyme progresses along the reaction coordinate.
Our experiments suggest that the population distribution of the open- and closed-lid states observed in the crystals is not an artifact of lattice constraints. We reach this conclusion based on several lines of evidence: The structure of the PEPCK–Mn2+–GTP complex, obtained from the same crystals that are used in the formation of the β-SP–GTP and oxalate–GTP complexes, has one molecule in the ASU with an open lid (32). When these crystals are soaked in a solution containing β-SP, there is a change in the space group with two nonequivalent molecules, one containing an open lid and the other in a closed-lid conformation. When an identical experiment is carried out replacing β-SP with oxalate, both molecules in the ASU adopt an identical closed-lid state. These results demonstrate that lid motion in both molecules in the ASU is permitted in the crystal lattice. Finally, the oxalate–GTP complex obtained by cocrystallization in a different space group demonstrates a shift in the solution population favoring the closed conformation. Therefore, our crystallographic studies support the idea that the energy landscape for lid motion changes as the enzyme proceeds through catalysis from the holoenzyme to the Michaelis complex and the enolate intermediate, consistent with prior studies carried out on the lid domain of TIM (43). This notion of a change in the free energy landscape for motions in the enzyme between the differently liganded states has been suggested recently by computational studies on adenylate kinase, although the authors concluded that their results were consistent with a conformational selection process in that enzyme (44). Further, we should note that the idea of substrate binding energy being used for catalysis is not new and is a widely held view (45, 46). Additionally, the utilization of that binding energy in the sequestering of reactive intermediates has been suggested previously by Fersht as a mechanism by which enzymes can evolve to optimally stabilize reactive intermediates rather than preferentially bind the transition state to facilitate catalysis (38). The structural studies of PEPCK presented here provide an explanation of how that interaction energy is used by PEPCK along an induced fit pathway.
In 1975, Jencks proposed the “circe effect” to explain the utilization of the free energy of substrate binding in enzyme catalysis (46). As related to the induced fit model in hexokinase, Jencks demonstrated that most of the free energy from the interaction between hexokinase and glucose was used in the formation of the induced conformation from the uninduced state with the remainder manifested in the observed binding energy (46). In a mechanism consistent with Jencks' circe effect, our data are consistent with the idea that the enzyme stabilizes the closed-lid conformation during the catalytic cycle by offsetting the entropic penalty for lid closure through the favorable Gibbs free energy available from the formation of the Michaelis and enolate intermediate complexes, respectively. The simplest interpretation of the data is that the available enthalpy from substrate association increases with the addition of substrates as the enzyme forms the Michaelis complex. The enzyme can use this available enthalpy to offset the unfavorable entropy of lid closure that the structural studies demonstrate is necessary for catalysis. A favorable entropic contribution may also contribute, because inspection of the structure of holo-PEPCK shows the presence of well ordered water molecules in the positions corresponding to the oxygen atoms of bound PEP and OAA. At a minimum this could provide entropy–entropy compensation upon substrate binding and may even result in an increase in favorable entropy upon complex formation. On the basis of this model, the observation of only a closed-lid state in the enolate intermediate complex suggests that the interaction free energy is maximal in this enzyme form. This is consistent with Fersht's idea of maximum stabilization of the enolate intermediate and predicts that upon formation of the products the necessary reduction in complementarity would result in a decrease in the available interaction energy. This would have the effect of shifting the thermodynamic landscape for lid motion in a direction favoring the open state allowing for product release and a continuation of the catalytic cycle.
Conclusion
We do not refute the conformational selection model's tenet that enzymes are dynamically sampling multiple conformational states in the absence of ligand, and some of these conformations will have a greater affinity for ligand than others. Similar to the dynamic behavior of the lid domain in TIM (43), PEPCK most likely samples both the open- and the closed-lid states in all enzyme complexes although the crystallographic data demonstrate that the closed-lid conformation is negligibly populated in the absence of a fully ligated active site. In the case of PEPCK and TIM (43), the induction is simply the effect of the altered ligation state, resulting in changes in the free energy surface for lid motion. As a consequence, the populations of the two states move in a direction favoring the closed state over the open conformation. Clearly, in other systems where the “active” conformation does not result in steric occlusion of the active site, direct binding to an activated enzyme conformation is possible, consistent with the conformational selection model. However, if this state is not identical to the final catalytic key-lock complex, then an associated substrate molecule can influence the positions of active site residues through an induced fit mechanism. Therefore, we propose that the conformational selection and induced fit mechanisms are complementary rather than mutually exclusive models. In our view, induced fit is the general result of ligand association changing the thermodynamic landscape for the protein. The effect that ligand association has on the free energy surface for the protein will depend on both the magnitude of the energy input into the system through ligand association and the shape of the free energy landscape for the protein. In this way, ligand association can alter the landscape such that it alters the population distribution of already sampled states as demonstrated here for our simple two-state model of lid opening and closing in PEPCK catalysis. However, in a similar fashion one could envision ligand binding or the chemical transformation of substrates to products resulting in a significant restructuring of the energy landscape for the protein such that new conformations that were not previously accessible are now sampled.
Materials and Methods
Materials.
Glutathione Uniflow Resin was purchased from Clontech. HiQ and P6DG resins were from Bio-Rad. β-SP was synthesized and purified as previously described (47). All other materials were of the highest grade available.
Enzyme Expression and Purification.
Recombinant rat cPEPCK was expressed and purified as previously described (32).
Crystallization.
Crystals of PEPCK used for data collection were obtained as previously described (32). The complexes with PGA, oxalate, and β-SP were obtained by soaking the PEPCK–Mn2+–Mn2+GDP(GTP) crystals for 1 h in a cryoprotectant solution (25% PEG 3350, 10% PEG 400, 0.1 M Hepes pH 7.5, 2 mM MnCl2, and 10 mM GDP(GTP)) containing 10 mM of the analogue before crycooling in liquid nitrogen. Cocrystals of the PEPCK–Mn2+–oxalate–Mn2+GTP complex were obtained under similar conditions as described previously and were cryoprotected in an identical fashion to the complexes obtained by soaking.
Data Collection.
Data on the cryocooled crystals of the PEPCK–Mn2+–PGA–Mn2+GDP, and PEPCK–Mn2+–oxalate–Mn2+GTP complexes maintained at 100 K were collected at the Stanford Synchrotron Radiation Laboratory, Beamline 11–1, Menlo Park, CA. Data on the PEPCK–Mn2+–β-SP–Mn2+GTP complex and the cocrystal of the PEPCK–Mn2+–oxalate–Mn2+GTP complex maintained at 100 K were collected at the Advanced Photon Source, BioCars 14-BM-C, Argonne, IL. All data were integrated and scaled with HKL-2000 (48). Data statistics are presented in Table S1.
Structure Determination and Refinement.
The structures of the rat cytosolic enzyme were determined by molecular replacement using MOLREP (49) in the CCP4 (50) package and the previously determined structure of rat cPEPCK [PDB ID: 2QEW (32)]. This molecular replacement solution was refined using Refmac5 followed by manual model adjustment and rebuilding using COOT (51). Ligand, metal, and water addition and validation also were performed in COOT. A final round of translation/libration/screw (TLS) refinement was performed for all models in Refmac5. A total of five groups per chain were used for all models as refinements using greater than five groups per chain did not significantly improve R/Rfree. The optimum TLS groups were determined by submission of the PDB files to the TLSMD server [http://skuld.bmsc.washington.edu/∼tlsmd/ (52, 53)]. Final model statistics are presented in Table S1. In the PEPCK–Mn2+–β-SP–Mn2+GTP complex, inspection of the Fo − Fc map after modeling of the major conformation of GTP in molecule A revealed positive density in the location of the nucleotide observed in molecule B (Fig. S5). However, due to the low occupancy the alternate conformation for the nucleotide was not included in the final model. The occupancy of the major conformer was reduced to 0.8 and resulted in B-factors for GTP that were similar to the B-factors for the coordinating protein residues. The occupancy of the closed-lid domain (residues 464–474) was manually adjusted to 0.8 (identical to that of the major conformation of the nucleotide), resulting in B-factors consistent with the neighboring residues and eliminating the observed negative Fo − Fc difference density for the lid domain.
Supplementary Material
Acknowledgments.
We thank Drs. Gerald Carlson, Aron Fenton, Owen Nadeau, and Mark Wilson for their helpful discussions during the preparation of this paper. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory supported by the Department of Energy and the National Institutes of Health. Use of the Advanced Photon Source was supported by the U.S. Department of Energy. Use of BioCARS Sector 14 was supported by National Institutes of Health Grant RR07707.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3DT2, 3DT4, 3DT7, and 3DTB).
The encounter complex is the enzyme complex with all substrates and cofactors required for catalysis bound however, the conformational change necessary for catalysis has not occurred. The holoenzyme is the PEPCK–Mn2+ complex. The Michaelis complex again defines the fully ligated state of the enzyme, such that all substrates and cofactors that are required for catalysis are bound, however, it does not take into consideration the conformational state of the enzyme. The intermediate complex is the PEPCK–Mn2+–enolate–Mn2+GTP state that is formed after decarboxylation of OAA.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805364105/DCSupplemental.
References
- 1.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher E. The influence of configuration on enzyme activity. (Translated from German) Dtsch Chem Ges. 1894;27:2984–2993. [Google Scholar]
- 3.Antoniou D, Basner J, Nunez S, Schwartz SD. Computational and theoretical methods to explore the relation between enzyme dynamics and catalysis. Chem Rev. 2006;106:3170–3187. doi: 10.1021/cr0503052. [DOI] [PubMed] [Google Scholar]
- 4.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 5.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 6.Warshel A, et al. Electrostatic basis for enzyme catalysis. Chem Rev. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 7.Berger C, et al. Antigen recognition by conformational selection. FEBS Lett. 1999;450:149–153. doi: 10.1016/s0014-5793(99)00458-5. [DOI] [PubMed] [Google Scholar]
- 8.Bosshard HR. Molecular recognition by induced fit: How fit is the concept? News Physiol Sci. 2001;16:171–173. doi: 10.1152/physiologyonline.2001.16.4.171. [DOI] [PubMed] [Google Scholar]
- 9.Foote J, Milstein C. Conformational isomerism and the diversity of antibodies. Proc Natl Acad Sci USA. 1994;91:10370–10374. doi: 10.1073/pnas.91.22.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: Dynamic landscapes and population shifts. Protein Sci. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leder L, et al. Spectroscopic, calorimetric, and kinetic demonstration of conformational adaptation in peptide-antibody recognition. Biochemistry. 1995;34:16509–16518. doi: 10.1021/bi00050a035. [DOI] [PubMed] [Google Scholar]
- 12.Ma B, Shatsky M, Wolfson HJ, Nussinov R. Multiple diverse ligands binding at a single protein site: A matter of pre-existing populations. Protein Sci. 2002;11:184–197. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai CJ, Ma B, Nussinov R. Folding and binding cascades: Shifts in energy landscapes. Proc Natl Acad Sci USA. 1999;96:9970–9972. doi: 10.1073/pnas.96.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durr E, Bosshard HR. A monoclonal antibody induces opening of a coiled coil. Global protection of amide protons from H/D exchange decreases by up to 1000-fold in antibody-bound triple-stranded coiled coil. Eur J Biochem. 1997;249:325–329. doi: 10.1111/j.1432-1033.1997.t01-1-00325.x. [DOI] [PubMed] [Google Scholar]
- 15.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 16.Beach H, Cole R, Gill ML, Loria JP. Conservation of μs-ms enzyme motions in the apo- and substrate-mimicked state. J Am Chem Soc. 2005;127:9167–9176. doi: 10.1021/ja0514949. [DOI] [PubMed] [Google Scholar]
- 17.Kovrigin EL, Loria JP. Enzyme dynamics along the reaction coordinate: Critical role of a conserved residue. Biochemistry. 2006;45:2636–2647. doi: 10.1021/bi0525066. [DOI] [PubMed] [Google Scholar]
- 18.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 19.Osborne MJ, Schnell J, Benkovic SJ, Dyson HJ, Wright PE. Backbone dynamics in dihydrofolate reductase complexes: Role of loop flexibility in the catalytic mechanism. Biochemistry. 2001;40:9846–9859. doi: 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- 20.Alber T, et al. On the three-dimensional structure and catalytic mechanism of triose phosphate isomerase. Philos Trans R Soc Lond B Biol Sci. 1981;293:159–171. doi: 10.1098/rstb.1981.0069. [DOI] [PubMed] [Google Scholar]
- 21.Banner DW, et al. Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 angstrom resolution using amino acid sequence data. Nature. 1975;255:609–614. doi: 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- 22.Fersht AR, Knill-Jones JW, Bedouelle H, Winter G. Reconstruction by site-directed mutagenesis of the transition state for the activation of tyrosine by the tyrosyl-tRNA synthetase: A mobile loop envelopes the transition state in an induced-fit mechanism. Biochemistry. 1988;27:1581–1587. doi: 10.1021/bi00405a028. [DOI] [PubMed] [Google Scholar]
- 23.Lolis E, Petsko GA. Crystallographic analysis of the complex between triosephosphate isomerase and 2-phosphoglycolate at 2.5-Å resolution: Implications for catalysis. Biochemistry. 1990;29:6619–6625. doi: 10.1021/bi00480a010. [DOI] [PubMed] [Google Scholar]
- 24.Jogl G, Rozovsky S, McDermott AE, Tong L. Optimal alignment for enzymatic proton transfer: Structure of the Michaelis complex of triosephosphate isomerase at 1.2-Å resolution. Proc Natl Acad Sci USA. 2003;100:50–55. doi: 10.1073/pnas.0233793100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pompliano DL, Peyman A, Knowles JR. Stabilization of a reaction intermediate as a catalytic device: Definition of the functional role of the flexible loop in triosephosphate isomerase. Biochemistry. 1990;29:3186–3194. doi: 10.1021/bi00465a005. [DOI] [PubMed] [Google Scholar]
- 26.Sampson NS, Knowles JR. Segmental motion in catalysis: Investigation of a hydrogen bond critical for loop closure in the reaction of triosephosphate isomerase. Biochemistry. 1992;31:8488–8494. doi: 10.1021/bi00151a015. [DOI] [PubMed] [Google Scholar]
- 27.Ash DE, Emig FA, Chowdhury SA, Satoh Y, Schramm VL. Mammalian and avian liver phosphoenolpyruvate carboxykinase—Alternate substrates and inhibition by analogs of oxaloacetate. J Biol Chem. 1990;265:7377–7384. [PubMed] [Google Scholar]
- 28.Stiffin R M, Sullivan SM, Carlson GM, Holyoak T. Differential inhibition of cytosolic PEPCK by substrate analogues. Kinetic and structural characterization of inhibitor recognition. Biochemistry. 2008;47:2099–2109. doi: 10.1021/bi7020662. [DOI] [PubMed] [Google Scholar]
- 29.Holyoak T, Sullivan SM, Nowak T. Structural insights into the mechanism of PEPCK catalysis. Biochemistry. 2006;45:8254–8263. doi: 10.1021/bi060269g. [DOI] [PubMed] [Google Scholar]
- 30.Konopka JM, Lardy HA, Frey PA. Stereochemical course of thiophosphoryl transfer catalyzed by cytosolic phosphoenolpyruvate carboxykinase. Biochemistry. 1986;25:5571–5575. doi: 10.1021/bi00367a034. [DOI] [PubMed] [Google Scholar]
- 31.Sheu KF, et al. Stereochemical course of thiophosphoryl group transfer catalyzed by mitochondrial phosphoenolpyruvate carboxykinase. Biochemistry. 1984;23:1779–1783. doi: 10.1021/bi00303a030. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan SM, Holyoak T. Structures of rat cytosolic PEPCK: Insight into the mechanism of phosphorylation and decarboxylation of oxaloacetic acid. Biochemistry. 2007;46:10078–10088. doi: 10.1021/bi701038x. [DOI] [PubMed] [Google Scholar]
- 33.First EA, Fersht AR. Mutational and kinetic analysis of a mobile loop in tyrosyl-tRNA synthetase. Biochemistry. 1993;32:13658–13663. doi: 10.1021/bi00212a034. [DOI] [PubMed] [Google Scholar]
- 34.Joseph D, Petsko GA, Karplus M. Anatomy of a conformational change: Hinged “lid” motion of the triosephosphate isomerase loop. Science. 1990;249:1425–1428. doi: 10.1126/science.2402636. [DOI] [PubMed] [Google Scholar]
- 35.Dunten P, et al. Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site. J Mol Biol. 2002;316:257–264. doi: 10.1006/jmbi.2001.5364. [DOI] [PubMed] [Google Scholar]
- 36.Cotelesage JJH, Prasad L, Zeikus JG, Laivenieks M, Delbaere LTJ. Crystal structure of Anaerobiospirillum succiniciproducens PEP carboxykinase reveals an important active site loop. Int J Biochem Cell Biol. 2005;37:1829. doi: 10.1016/j.biocel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Reed GH, Morgan SD. Kinetic and magnetic resonance studies of the interaction of oxalate with pyruvate kinase. Biochemistry. 1974;13:3537–3541. doi: 10.1021/bi00714a020. [DOI] [PubMed] [Google Scholar]
- 38.Fersht AR. Catalysis, binding and enzyme-substrate complementarity. Proc R Soc Lond B Biol Sci. 1974;187:397–407. doi: 10.1098/rspb.1974.0084. [DOI] [PubMed] [Google Scholar]
- 39.Desamero R, Rozovsky S, Zhadin N, McDermott A, Callender R. Active site loop motion in triosephosphate isomerase: T-jump relaxation spectroscopy of thermal activation. Biochemistry. 2003;42:2941–2951. doi: 10.1021/bi026994i. [DOI] [PubMed] [Google Scholar]
- 40.Falzone CJ, Wright PE, Benkovic SJ. Dynamics of a flexible loop in dihydrofolate reductase from Escherichia coli and its implication for catalysis. Biochemistry. 1994;33:439–442. doi: 10.1021/bi00168a007. [DOI] [PubMed] [Google Scholar]
- 41.Waldman AD, et al. The use of site-directed mutagenesis and time-resolved fluorescence spectroscopy to assign the fluorescence contributions of individual tryptophan residues in Bacillus stearothermophilus lactate dehydrogenase. Biochim Biophys Acta. 1987;913:66–71. doi: 10.1016/0167-4838(87)90233-0. [DOI] [PubMed] [Google Scholar]
- 42.Wang GP, Cahill SM, Liu X, Girvin ME, Grubmeyer C. Motional dynamics of the catalytic loop in OMP synthase. Biochemistry. 1999;38:284–295. doi: 10.1021/bi982057s. [DOI] [PubMed] [Google Scholar]
- 43.Williams JC, McDermott AE. Dynamics of the flexible loop of triosephosphate isomerase: The loop motion is not ligand gated. Biochemistry. 1995;34:8309–8319. doi: 10.1021/bi00026a012. [DOI] [PubMed] [Google Scholar]
- 44.Arora K, Brooks CL., III Large-scale allosteric conformational transitions of adenylate kinase appear to involve a population-shift mechanism. Proc Natl Acad Sci USA. 2007;104:18496–18501. doi: 10.1073/pnas.0706443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fisher HF. A unifying model of the thermodynamics of formation of dehydrogenase–ligand complexes. Adv Enzymol Relat Areas Mol Biol. 1988;61:1–46. doi: 10.1002/9780470123072.ch1. [DOI] [PubMed] [Google Scholar]
- 46.Jencks WP. Binding energy, specificity, and enzymic catalysis: The circe effect. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. doi: 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- 47.Griffith OW, Weinstein CL. β-Sulfopyruvate. Methods Enzymol. 1987;143:221–223. doi: 10.1016/0076-6879(87)43040-1. [DOI] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 50.Bailey S. The Ccp4 Suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. 2006;D62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 53.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39:109–111. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.