Abstract
Group II intron ribozymes fold into their native structure by a unique stepwise process that involves an initial slow compaction followed by fast formation of the native state in a Mg2+-dependent manner. Single-molecule fluorescence reveals three distinct on-pathway conformations in dynamic equilibrium connected by relatively small activation barriers. From a most stable near-native state, the unobserved catalytically active conformer is reached. This most compact conformer occurs only transiently above 20 mM Mg2+ and is stabilized by substrate binding, which together explain the slow cleavage of the ribozyme. Structural dynamics increase with increasing Mg2+ concentrations, enabling the enzyme to reach its active state.
Keywords: multidomain RNA folding, single-molecule Förster resonance energy transfer, splicing, structural dynamics, metal ions
RNA molecules exhibit very diverse and fascinating folding pathways from random coil to native state. Smaller RNAs fold rapidly and smoothly after an apparent two-state process (1), whereas large RNAs sample intermediate conformations before reaching the active structure (2). Generally, a fast (1–50 ms) overall compaction into a stable but misfolded kinetic trap is found in an early stage of the folding pathway (1, 3–7) followed by slower rate-limiting folding to the native state (2).
Group II introns rank among the largest ribozymes in nature and display remarkable features distinct from other large RNAs (8, 9). These ribozymes are self-splicing multidomain RNA molecules exhibiting a rich evolutionary heritage, the parallels in structure and catalytic mechanism with spliceosomal RNA being only one example (9–11). Their overall secondary structure organization into six modular domains is generally preserved (Fig. 1A) (8). Only nucleotides involved in tertiary contacts are conserved, but these are dispersed throughout the secondary structure, unlike in other large RNAs (12, 13). Another specific property of group II intron ribozymes is the overall high Mg2+ requirement for maximal activity in vitro (14, 15).
Fig. 1.
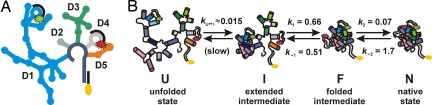
D135–L14 ribozyme and its folding pathway. (A) Scheme of the secondary structure of D135–L14. The individual domains (D1, blue; D2, gray; D3, green; D4, light gray; D5, orange) and the fluorophore labeled DNA-oligonucleotides (black; Cy3, green; Cy5, red) are shown. (B) Folding pathway of D135–L14. Compaction of D1 from the unfolded state U to the extended intermediate I is slow. The remaining domains pack onto D1 to give the folded species F, which itself rearranges to the native state N occurring only in small amounts. k1, k−1, k2, and k−2 refer to smFRET results at 100 mM Mg2+, and kU ↔ I was obtained from bulk FRET experiments and corresponds to earlier values of ≈1 min−1 (14, 18, 19). B is partially adapted from ref. 19. Folding rates are given in seconds−1.
The D135 ribozyme is derived from the Saccharomyces cerevisiae group II intron Sc.ai5γ and contains the necessary components for catalysis (9), allowing efficient and selective cleavage of RNA substrates with multiple turnover (14). This ribozyme has been used as a model system to study folding and metal ion binding of large RNAs (14, 16, 17). Until now, the folding pathway of D135 has been described as an apparent two-step process involving one obligatory intermediate (U ↔ I ↔ N) under optimal conditions for splicing (42°C, 500 mM KCl, 100 mM MgCl2) (17–19). The unfolded state U is an extended but structurally defined conformer of D135 in the presence of monovalent ions and possesses extensive secondary structure but no tertiary interactions (17, 19). The first folding step (kobs ≈1 min−1) consists of a critical rate-limiting compaction within domain 1 (D1, Fig. 1) to a folding intermediate I (17, 19, 20). The second step of domain assembly is not well characterized, but it is generally believed that the remaining domains fold rapidly into their native position onto the D1 scaffold (18).
Unlike other large RNAs, the group II intron folding is unique because the initial tertiary collapse of D135 is slow and I is not a kinetic trap but an obligatory folding intermediate (19). Structural and dynamic information about this intermediate is lacking. To elucidate the structural dynamics of the second folding step by single-molecule Förster resonance energy transfer (FRET), we have designed a fluorophore-labeled D135 construct (D135–L14) by introducing two modular loops (21) into D1 and D4 that specifically bind two DNA-oligonucleotides carrying the FRET pair Cy3 (Cy3-DNA) and Cy5 (Cy5-DNA), respectively [Fig. 1A and supporting information (SI) Figs. S1 and S2 ]. Our single-molecule experiments reveal three distinct conformations of D135–L14 in dynamic equilibrium under a wide range of Mg2+ concentrations and a previously undetected on-pathway folding intermediate. The native conformation is scarce and becomes apparent only at 20 mM Mg2+ or higher. However, this high-FRET state is stabilized by substrate binding, indicating that this is actually the catalytically active conformation in vitro. The folding rates between these conformations are fast and exhibit only a subtle Mg2+ dependence, in agreement with a recent ensemble averaged study (22), indicating that these conformations are connected by small activation barriers with similar, but not identical, Mg2+ affinities. This is the largest protein-free ribozyme studied by single-molecule fluorescence so far and exemplifies how single-molecule approaches can be applied to characterize the folding pathway of very large ribozymes in detail. Interestingly, the ribozyme dynamics increase with Mg2+ concentration, making the active state accessible.
Results
Fluorophore-Labeled D135–L14 Ribozyme Is Catalytically Competent and Requires Mg2+ for Folding.
We tested the activity of the in vitro transcribed (23) D135–L14 construct in a single-turnover cleavage assay with the substrate 17/7 under optimal conditions for self-splicing (17) (Fig. S3A and SI Results). The labeling scheme only minimally affects the cleavage activity of the ribozyme. D135–L14 yields a cleavage rate constant kobs = 0.37 ± 0.02 min−1, which is reduced in the presence of the fluorophore- and biotin-labeled DNA-oligonucleotides by only 27%. The final amount of product formation was 85% for all constructs. kobs depends on [Mg2+] giving a dissociation constant KMg = 52.7 ± 3.4 mM, comparable with reported values (9, 14).
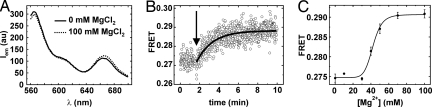
Folding in bulk was also measured by FRET (Fig. 2). Upon addition of Mg2+ ions to fluorophore labeled D135–L14, the donor intensity decreases while the acceptor increases (Fig. 2A). The overall FRET ratio increases exponentially with kobs = 0.90 ± 0.07 min−1 (100 mM MgCl2, Fig. 2B), in agreement with reported values (14, 18, 19). The FRET increase suggests that a conformational change takes place in the presence of Mg2+, during which the relative distance between the fluorophores on the RNA decreases. The observed FRET increase is Mg2+-dependent, giving again a KMg = 42.3 ± 1.3 mM (Fig. 2C).
Fig. 2.
Bulk fluorescence measurements of the fully labeled D135–L14 in dependence of Mg2+ concentration. (A) Emission spectrum of the ribozyme in the absence (0 mM, solid line) and presence (100 mM, dotted line) of MgCl2 showing correlating shifts in maxima at 565 nm (Cy3) and 665 nm (Cy5) on FRET. (B) FRET change upon addition of 100 mM Mg2+. The arrow indicates the addition of Mg2+. The single exponential fit of the experimental data is shown as a black line. (C) Maximal change in FRET upon increasing amounts of Mg2+ and fit to the Hill equation (S2) giving KMg = 42.3 ± 1.3 mM.
Finally, we confirmed efficient and specific binding between the DNA-oligonucleotides and D135–L14 by nondenaturing gel electrophoresis (Fig. S2).
Taken together, the above results demonstrate that the fluorophore-labeled D135–L14 behaves like the D135 ribozyme regarding catalysis, folding, and Mg2+ requirement.
Single-Molecule FRET Reveals (smFRET) Three Conformations in Equilibrium.
Single-molecule FRET offers the potential to monitor and identify transient folding intermediates without the need for synchronization and can help elucidate the folding pathways of ribozymes. Here, we have used an approach similar to our earlier studies with the hairpin ribozyme (24, 25) to study the folding dynamics of the much larger D135–L14 ribozyme.
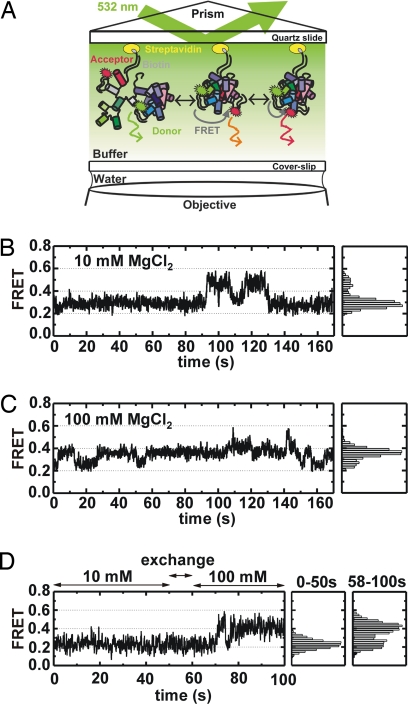
Fluorophore-labeled D135–L14 ribozymes were incubated in the presence of specific amounts of Mg2+ at 42°C to induce folding before immobilization on the quartz surface via a biotinylated DNA-oligonucleotide hybridized to the 3′ end tail (Figs. 1A and 3A and Fig. S1). Characteristic time trajectories of such surface immobilized D135–L14 ribozymes in 10 mM and 100 mM Mg2+ at room temperature are shown in Fig. 3 B and C. The observed FRET ratios jump stochastically between three values ≈0.25, ≈0.4, and ≈0.6. Hence, the D135–L14 ribozyme adopts at least three distinguishable structural conformations [extended intermediate (I), folded intermediate (F), and native (N)] in dynamic equilibrium. We assigned the low-FRET state to I and not U (Fig. 1B) for the following reasons: (i) a minimal FRET for U and low FRET for I are expected based on their hydrodynamic radii (17); (ii) very few molecules, if any, should be in the U state at [Mg2+] >20 mM because the RNA is incubated with Mg2+ before our measurements; (iii) no unfolding from I is observed in accordance with a much slower I to U transition than the duration of our time traces (<200 s, Fig. 3 B and C). In an attempt to assign a FRET value for U and to support our assignments, we measured single-molecule time trajectories of D135–L14 in the absence of Mg2+ and in the presence of 10 μM EDTA (Fig. S4). Under these conditions, only the U state is present (20). Indeed, in the absence of Mg2+, nearly all of the observed molecules remain static in a 0.1 FRET state, in agreement with our assignments. However, we cannot distinguish these molecules from a small fraction that lacks the acceptor fluorophore (photobleaching; Fig. 4 Top), making it impossible to quantify the fraction of molecules in the U state. Therefore, we did not include molecules in the U state in subsequent analysis.
Fig. 3.
smFRET analysis of D135–L14 ribozymes. (A) Principle of total internal reflection fluorescence spectroscopy (TIRF). Molecules are immobilized on a quartz slide and placed in the microscope. The fluorophores are excited by the laser's evanescent wave above the critical angle, giving emission signals collected through the objective with a high-quantum yield camera. (B) smFRET time trace and histogram at 10 mM Mg2+ showing two conformational states at 0.25 and 0.4. (C) smFRET time trace and histogram at 100 mM Mg2+ sampling the three FRET states 0.25, 0.4, and 0.6. (D) (Left) smFRET time trace shows a dynamic behavior only upon increase in MgCl2 concentration from 10 to 100 mM at 50–58 s. (Right) Distributions of the states before and after the addition of Mg2+.
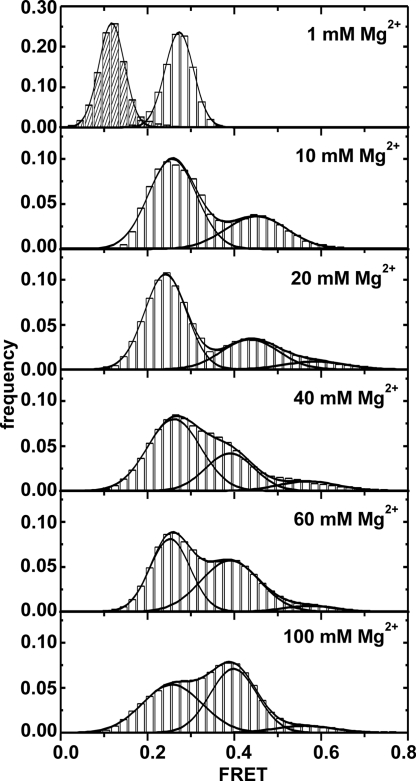
Fig. 4.
Histograms of FRET efficiency values from single-molecules traces in dependence of Mg2+ concentration. All traces (static and dynamic) were used to build the histogram at 1 mM, whereas all other histograms were created from traces showing dynamics only. Three distributions centered at FRET ≈0.25, ≈0.4, and ≈0.6. The peaks of the 0.25 and 0.4 states show a slight shift to lower FRET with higher Mg2+ concentration. The 0.6 distribution arises above 15–20 mM and is only minimally but steadily populated. Gaussian fits for the individual and the sum of all distributions are shown as solid lines. The dashed bars at 0.12 FRET correspond to the distribution in absence of acceptor (Top).
These conformational states could not be resolved in the bulk FRET experiment because it yields an ensemble-averaged result. Furthermore, the single-molecule data show that a large fraction of molecules remain static in the low FRET state (see below), explaining the observed low FRET in bulk.
At low [Mg2+], only the two lower FRET states are observed. The highest FRET state becomes apparent at [Mg2+] >15–20 mM (Fig. 4) but is populated transiently, and the few molecules that reach it return promptly to the intermediate state. Among the >10,000 total transitions observed, only 1% go directly from the I state to the N state, consistent with the contribution from very short dwell times in the F state, which we miss because of our 33-ms time resolution. This observation is thus direct evidence that the F state is an obligatory intermediate along the folding pathway and is devoid of any kinetic traps.
Magnesium Ions Affect Folding Equilibrium of the Group II Intron Ribozyme.
Mg2+ ions affect the relative stability of each conformation, as evidenced by the FRET time trajectories and the dwell time spent in each state. At 10 mM Mg2+ and below, dynamic molecules stay predominantly in the I state, whereas at 100 mM Mg2+ the F state prevails (Fig. 3B and C). To quantify the effect of Mg2+ on the stability of each state, we calculated FRET distributions from ≈150 single-molecule trajectories at Mg2+ concentrations ranging from 1 to 100 mM (Fig. 4). At 1 mM Mg2+ the D135–L14 ribozyme resides exclusively in the I state, and no structural dynamics are observed. We can easily distinguish this state from complexes lacking the acceptor fluorophore (caused by photobleaching or blinking) because the observed FRET ratio for the latter molecules is ≈0.1 (Fig. 4, Top). The distributions for the F and N states become apparent only above 10 and 20 mM Mg2+, respectively (Figs. 3B and C and 4). The N state is never populated for extended times, which is reflected as a low-amplitude distribution in the histograms. The amplitude of the F state distribution increases from 0.04 to 0.08 with increasing [Mg2+], whereas the amplitude of the low FRET decreases from 0.23 to 0.06. This indicates that the relative stability of the F state increases concomitantly with the decrease of the I state. This antiparallel behavior, however, is not very dramatic because both states are still present between 10 and 100 mM Mg2+. The N state distribution amplitude remains constant above ≈50 mM Mg2+ (see below).
To learn more about the Mg2+ requirement for these structural dynamics, we monitored the folding of a single D135–L14 ribozyme while changing the buffer conditions from 10 to 100 mM Mg2+ (Fig. 3D). During the first 50 s, the ribozyme kept in 10 mM Mg2+ displays a FRET ratio ≈0.25 (I state) matching the FRET histogram obtained under steady-state conditions (Fig. 4, Top). Between 50 and 58 s we exchanged the buffer in the microscope slide by using a syringe pump to increase the Mg2+ concentration to 100 mM. After a dead time of ≈10 s, the FRET ratio jumps to 0.4, indicating that a Mg2+-induced conformational change to the F state takes place. The structural dynamics of the ribozyme increase as evidenced by FRET jumps between the 0.25 and 0.4 FRET states. The FRET histogram corresponding to the high-[Mg2+] period also matches the FRET histogram obtained in steady-state conditions (Fig. 4Bottom). This experiment clearly shows that the individual FRET states are directly dependent on the Mg2+ concentration and that the D135–L14 ribozyme readily reacts to an increased Mg2+ background by reaching a higher FRET state.
The presence of both the F and N states only above 20 mM Mg2+ coincides with the onset of catalytic activity (compare Fig. 4 and Fig. S3), suggesting that one of these two conformations corresponds to the catalytically active structure. It has been suggested that D135 collapses directly to the active state with a midpoint of ≈20–40 mM Mg2+ (14).
Peaks of the FRET Histogram Distributions Decrease Slightly with Mg2+.
Close inspection of the histograms (Fig. 4) reveals that the peak distributions of the I and F states shift to slightly lower FRET values with increasing Mg2+. The I state distribution shifts from ≈0.27 (1 mM Mg2+) to 0.24 (20 mM Mg2+), whereas the F state distribution shifts from 0.45 (10 Mg2+) to 0.40 (≥40 mM Mg2+). At lower Mg2+ concentrations, the F and N states are possibly in fast dynamic equilibrium (lifetime <30 ms), and as a result we observe only a weighted-average FRET ratio between F and N. Alternatively, the I and F states each become structurally less compact above 1 mM and 20 mM Mg2+, respectively, giving rise to a shorter distance between the two fluorophores. Neither of the two hypotheses can be ruled out at the moment, but this result supports the idea that the group II intron conformations are connected by relatively small activation barriers.
Substrate Binding Stabilizes the 0.6-FRET Conformation.
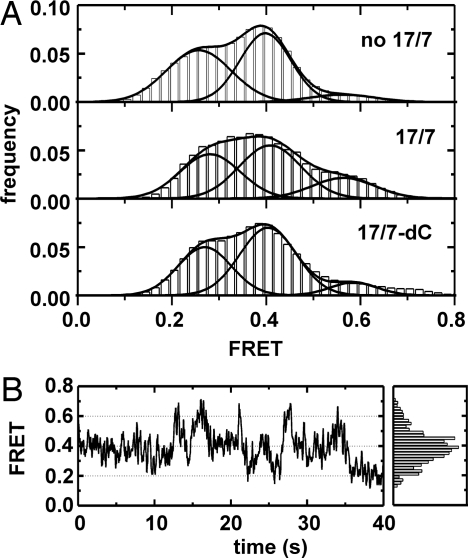
To determine which state (F or N) represents the active conformation, we performed smFRET studies in the presence of the substrates 17/7 and 17/7-dC (2′-deoxy at cleavage site) (26). The slow cleavage of 17/7-dC (kobs = 0.05 ± 0.01 min−1) enabled us to observe the substrate-bound D135–L14 molecules for longer periods of time. The presence of either 17/7 or 17/7-dC clearly stabilizes N (Fig. 5), suggesting that the high-FRET state corresponds to the active conformation. Consequently, F represents a hitherto undetected obligatory intermediate, or near-native state, from which only few molecules reach the active state at a time. The formation of the ε–ε′ and λ–λ′ tertiary contacts and the two intron and exon binding sequences (IBS-EBS) interactions between the substrate and D1 appear to be essential to stabilize the active conformation. The wild-type (WT) substrate 17/7 leads to a higher occurrence of the 0.6 state compared with the 17/7-dC oligonucleotide (Fig. 5A). A plausible explanation for this observation could be that the 2′-OH at the cleavage site makes contacts that contribute to assembly of the catalytic core and hence also to the overall architecture of the ribozyme.
Fig. 5.
Effect of substrate binding on the dynamics and native state stabilization. (A) Histograms of FRET efficiency of fully labeled D135–L14 alone (Top), in the presence of WT substrate 17/7 (Middle), and in the presence of the slower cleaving substrate 17/7-dC (Bottom). The 0.6 state is clearly stabilized by substrate binding. (B) (Left) Typical smFRET time trace in the presence of 17/7-dC. The 0.6 state occurs at high frequency (compare with Fig. 3). The drop to 0.25 at 35 s might indicate cleavage. (Right) Distribution.
The assignment of the 0.25 state to I (D1-folded), the 0.4 state to F (compact D135), and the 0.6 state to the native structure is further supported by a recent study (22). Under low-salt conditions, D1 folds to the near-native state to which D3 and D5 can dock. However, docking is not stable, and the folding pathway arrests primarily at the I state. This lack of structural stability can be overcome by increasing the Mg2+ concentration in vitro or the participation of proteins in vivo (9). The occurrence of the 0.4 state only at higher [Mg2+] clearly assigns the I and F state to folded D1 and compact D135, respectively.
Mg2+ Ions Increase Structural Dynamics to Form the Active State.
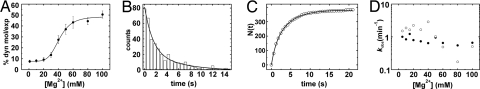
It is generally assumed that the addition of Mg2+ to RNA initiates folding and leads to a compact and more rigid structure. Here, we detect the opposite and so far unobserved behavior. Although Mg2+ clearly initiates folding of D135 leading to the more compact I, F, and N states, the structural dynamics increase considerably with higher [Mg2+]. At very low [Mg2+] the molecules remain static in the I state, but at higher concentrations an increasing number of transitions among the three states is observed. The static molecules depict constant FRET values of ≈0.25 (i.e., the I state) at all Mg2+ concentrations >1 mM tested (compare Fig. 4 and Fig. S5). It is possible that these static molecules correspond to a subpopulation of molecules that display a very slow k1 (≪0.005 s−1), slower than our ≈200-s observation window, but we cannot distinguish between these two possibilities. The fraction of dynamic molecules versus [Mg2+] (Fig. 6A) yields a dissociation constant KMg = 41.4 ± 1.4 mM, which coincides with the values for bulk folding and cleavage (Fig. 2C and SI Results). Even at high [Mg2+], the fraction of dynamic molecules does not exceed 50% in apparent discrepancy with the cleavage experiments of the same D135–L14 construct (see above). However, the cleavage assays are conducted at 42°C, whereas the smFRET experiments are done at 22°C for technical reasons. This finding suggests that the higher temperature is needed to enable the remaining molecules reach the active conformation. Taken together, these two results clearly link the structural dynamics of D135 to its function, raising the interesting possibility that domain motion contributes to successful reactivity.
Fig. 6.
Mg2+ dependence of single-molecule dynamics and the observed rates kobs. (A) The percentage of molecules on a slide showing dynamics are plotted versus the Mg2+ concentration. The ratio of dynamic molecules increases from 7 to 49% with KMg = 41.4 ± 1.4 mM. (B) Representative histogram of dwell times in the 0.25 FRET state at 50 mM Mg2+ showing the number of events at a particular dwell time range. The solid line corresponds to the double exponential decay fit characterizing the rate. (C) The number of dwell times in low FRET state N(t) shorter than time t, obtained by integration of the dwell time histogram in A. The double-exponential growth fit is shown as a solid line. (D) Effect of Mg2+ concentration on the observed single-molecule folding rates k1 (●) and k−1 (○).
Dwell-Time Analysis Determines the Rates of Folding.
To quantify the relative stability of each conformation, we determined the rates of folding k1, k−1, k2, and k−2 for the ribozyme. The dwell time in each state between two conformational transitions from ≈150 single-molecule time trajectories was measured, and dwell time distributions for each state were calculated. Static molecules in the I state could not be included in this analysis. Dwell times in the 0.4 FRET state were divided into two distributions depending on the final state (I or N) to determine k−1 and k2, respectively. Fig. 6B shows a characteristic dwell time (τ1) distribution for the I state in 50 mM Mg2+. The distribution decays exponentially as expected for a stochastic process. To obtain k1, we integrated the distribution into a cumulative time distribution (Fig. 6C), which was then fit to an exponential growth curve, as described (24, 25, 27). The integrated distribution reveals folding heterogeneity in the form of two distinguishable exponentials, as observed for other ribozymes (25, 28–30). The major component exhibits an exponential rate k1 = 0.66 s−1. The folding rate k−1 = 0.51 s−1 obtained similarly also exhibited heterogeneity. The measured rates in 100 mM Mg2+ are in agreement with the FRET histogram distributions (Fig. 4Bottom). The ratio k1/k−1 = 1.3 corresponds with the ratio of distribution amplitudes for the F and I states, A0.4/A0.2 = 0.78/0.56 = 1.4. Please note that in an ensemble experiment the observed folding rate would be slower than those reported here because of averaging between the static and dynamic subpopulations. This illustrates the strength of single-molecule experiments, which enable us to uncover heterogeneity in folding and thus elucidate details of the mechanism otherwise hidden by ensemble averaging.
Lacking a statistically significant number of I to N transitions because k2 ≪k−1 and the high FRET conformation is only rarely reached, k2 and k−2 could not be determined with dwell time distributions. To estimate their values, we measured the average dwell time in I before jumping to N, and in N before jumping back to I for 19 molecules (τav = 15.3 and 0.6 s, respectively). We calculated k2 = 0.07 s−1 and k−2 = 1.7 s−1 as the inverse of these values (Fig. 1B). The ratio k2/k−2 = 0.04 is in reasonable agreement with the ratio of relative amplitudes of the 0.6 and 0.4 FRET distributions (A0.6/A0.4 = 0.008/0.078 = 0.10, Fig. 4), confirming the low abundance of N in absence of the substrate. We also measured k2 and k−2 in presence of 17/7 and 17/7-dC from 6 and 17 single-molecule trajectories, respectively. Substrate binding increases k2 by nearly 10-fold (0.8 and 0.7 s−1, respectively) and decreases k−2 by ≈30% (1.1 and 1.2 s−1, respectively), further indicating that the substrate stabilizes folding into the N state.
Forward Rates Are Independent of Mg2+.
We determined the folding rates in the absence of substrate between 5 and 100 mM [Mg2+] (Fig. 6D). Below this concentration, the ribozyme is mostly static in the I state, and thus, not enough dynamic events were observed to quantify the rates with confidence. We found evidence of folding heterogeneity for k1 and k−1 at all concentrations (Fig. S6 and Tables S1 and S2). At 5 mM Mg2+, the major component of k−1 is 1.5-fold larger than the major component of k1, consistent with I being more stable than F at low Mg2+ (Fig. 6D). k1 remains approximately constant, whereas k−1 first increases and then decreases at [Mg2+] >40 mM, shifting the equilibrium from I toward F (Tables S1 and S2). This finding is consistent with the change in relative stability observed in the FRET histogram distributions (Fig. 4). The rate constants k2 and k−2 were also found to be approximately constant over the Mg range tested.
Discussion
Folding of Large Multidomain RNAs.
Here, we have used the group II intron ribozyme as a model system to study folding of large multidomain ribozymes. Our single-molecule experiments reveal important concepts for RNA folding in general resolve long-standing questions for group II intron catalysis.
The group II intron ribozyme folds sequentially in three steps from the unfolded to the native state (Fig. 1B), with a previously uncharacterized on-pathway folding intermediate, extending earlier studies that suggested a folding pathway devoid of kinetic traps. High [Mg2+] (>10 mM) is required in vitro to populate the near-native state F significantly. We propose that the newly discovered N state corresponds to the active state, but it can only be distinguished from F above 20 mM Mg2+. Because N is a transient conformer in the absence of substrate, it was not observed in bulk experiments.
We observe no Mg2+ dependence for the I to F transition (Figs. 4 and 6D), similarly to the Tetrahymena ribozyme (29) and the RNase P C domain (30). A possible explanation is that this step is dominated by a conformational capture mechanism, in which the F conformation is unstable and must be trapped by Mg2+ coordination. A recent study showing that the rate of D1 compaction does not depend on Mg2+, whereas the compaction amplitude does (20), supports this idea. Hydroxyl radical footprinting experiments also show that D5 becomes fully protected only above 100 mM Mg2+, in agreement with our observations (14). Mg2+ ions are, therefore, essential to capture D3 and D5 in their active conformation and to stabilize the D135 complex in vitro. Interestingly, we observed increased RNA structural dynamics upon Mg2+ binding (Fig. 6A), linking domain motion to catalysis.
The relative stability of the F and N states remains constant above 40 mM Mg2+, ruling out a capture mechanism involving Mg2+ for this step. Instead, substrate binding stabilizes the N state, making it the catalytically active species (Fig. 5). The FRET increase from F to N indicates that a major conformational change accompanies D135 compaction. Because the IBS-EBS and ε–ε′ interactions are located within D1 and thus, cannot be observed with our labeling scheme, it must be the λ–λ′–λ′ interaction, which links the intronic 5′ end with central nucleotides in D1 and the catalytically crucial D5 (31), that causes this major rearrangement of domains. A recently published crystal structure of a group IIB intron further supports our findings (32). This structure shows that the product form of the RNA folds into a highly compacted state where domains 1–4 interact with the catalytic domain 5. We propose that formation of this highly compacted state might correspond to our F to N transition. However, a quantitative structural comparison with our data is not possible because the loops where the fluorophores are attached are not present in the crystallized group II intron from Oceanbacillus iheyensis. In addition, there is no structural information available on the loop where the donor is located, which could easily introduce an error of 10–20 Å on any distance estimate from the crystal structure.
The transient nature of the native state raises the question of whether the rarely occurring transition from F to N, regulated by k2, is limiting for cleavage. To address this question we measured the cleavage kinetics of the ribozyme at room temperature and 100 mM Mg2+. We found a cleavage rate kobs = 0.024 ± 0.001 min−1 (see Fig. S7), 180-fold slower than k2. This suggests that the transition from F to N alone is not rate-limiting. Alternatively, a combination of folding, catalysis, and activation of the static population may be rate-limiting for group II intron catalysis. Testing this hypothesis will be the scope of future work.
Implications for Group II Intron Folding and Function in Vivo.
Our results show that large multidomain RNAs, such as D135–L14, can fold in a highly ordered stepwise pathway consisting only of on-pathway intermediates without kinetic traps. This strategy is very advantageous for a retroelement such as a group II intron that must fold cotranscriptionally in vivo. It allows folding to start in D1, which is transcribed first. Then, the slow transition from U to I ensures proper folding of this domain and subsequent docking of the downstream domains as soon as they are transcribed (e.g., D3 and D5).
Although Sc.ai5γ requires high salt and temperature to function in vitro, Mss116, a DEAD-box ATPase with helicase activity, promotes splicing in vivo (33, 34). It has been a matter of debate as to whether Mss116-unwinding activity is needed to resolve kinetic traps (35, 36). The absence of folding kinetic traps in vitro supports a model where Mss116 is needed only to stabilize the native fold at low [Mg2+] and not to help the RNA escape from misfolded intermediates.
Materials and Methods
Preparation of RNA.
The D135 ribozyme sequence from S. cerevisiae intron Sc.ai5γ on plasmid pT7D135 was modified by insertion of two loops with the length of 18 nt each. Sequence L1 (3′-CCCAAAUUAAUAACGCUCUUGGUAGGG-5′) was placed between residues 276 and 305 on the d2b stem of D1 (Fig. S1), and L2 (3′-CCCAAAUAUAUUUCGGACUACGUAGGG-5′) replaced the residues 681–806 in D4, giving the plasmid pT7D135–L14. Digestion with HindIII and subsequent in vitro transcription with home-made T7 RNA polymerase under standard conditions (23) gave D135–L14 RNA, which was stored in water at −20°C.
Biotinylated DNA strands, Cy3-DNA, Cy5-DNA, and the RNA substrates 17/7 (3′-UGGCGAGCUUUUACAGGGUGGUGC-5′, 2′-protected) and 17/7-dC (3′-UGGCGAG(dC)UUUUACAGGGUGGUGC-5′, 2′-protected) were purchased from Howard Hughes Medical Institution Biopolymer/Keck Foundation Biotechnology Resource Laboratory (Yale University, New Haven, CT). DNA strands were purified by denaturing 18% PAGE and subsequent C8 reversed-phase HPLC as described in ref. 37. The RNAs were deprotected as suggested by the manufacturer and subsequently gel- and HPLC-purified (37).
Design of a Fluorophore-Labeled Group II Intron Ribozyme for Single-Molecule Studies.
The D135 ribozyme is a 618-nt-long single-stranded RNA (Fig. S1) (16, 17, 19), which makes it highly impractical to label using a synthetic approach. Hence, we introduced two modular loops within the D135 sequence at locations within D1 and D4 dispensable for catalysis (9) as described (Fig. 1A and Fig. S1) (21). The resulting construct (D135–L14) contained all domains necessary for catalytic function of the intron and an expendable 37-nt extended 3′ tail (Fig. 1A). Two DNA-oligonucleotides specifically complementary to each extended loop in D1 and D4 carried the FRET pair Cy3 (Cy3-DNA) and Cy5 (Cy5-DNA), respectively. In addition, a biotinylated DNA-oligonucleotide that hybridizes specifically to the 3′ end tail of D135–L14 was used for surface immobilization in the single-molecule experiments (Fig. 1A). D135–L14 binds all three DNA-oligonucleotides as expected (Fig. S2).
Steady-State FRET Kinetic Assay.
Ratios of Cy3-DNA to ribozyme to Cy5-DNA were chosen to be 1:2.5:6 to minimize unbound amount of Cy3-DNA and maximize Cy5 binding to ribozyme for optimized fluorescence transfer. Cy3-DNA (0.1 μM), D135–L14 (0.25 μM), and Cy5-DNA (0.6 μM) were denatured at 90°C for 1 min in reaction buffer and subsequently incubated at 42°C for 15–20 min. Reaction mixture (100 μl final) was excited at 555 nm, and emission was recorded at 565 nm and 665 nm on a Cary Eclipse fluorescence spectrometer. Slits were set at 5 nm, and sample was thermostatted at 42°C. Emission time courses were measured for 15 min after the addition of varying amounts of MgCl2 in the range of 0–100 mM.
Normalized absolute FRET changes (FRETt(15) − FRETt(o)) were plotted against Mg2+ concentrations for analysis of overall Mg2+ dependence of folding by the Hill equation (S2) (SI Methods). Time courses of FRET ratio [defined as EFRET = IA/(ID + IA)] were fitted with a single exponential expression.
Single-Molecule FRET Experiments.
Cy3-DNA, Cy5-DNA, and T-biotin-DNA (10 μM each) were heat-annealed to D135–L14 (1 μM) in reaction buffer containing 0.5% mercaptoethanol. After the addition of MgCl2 (0–100 mM final), the reaction mixture was incubated at 42°C for 15–20 min. The FRET-labeled, biotinylated, and annealed ribozyme complex was diluted to ≈25–50 pM and bound to a streptavidin-coated quartz slide surface. Excess Cy3-DNA, Cy5-DNA, and T-biotin oligonucleotides were removed from the slide by a washing step with reaction buffer. An oxygen-scavenging system consisting of 10% (wt/v) glucose, 2% (vol/vol) 2-mercaptoethanol, 50 μg/ml glucose oxidase, and 10 μg/ml catalase was used in all experiments to reduce photobleaching. All smFRET experiments were carried out at room temperature according to described procedures (25, 27, 38). The donor (ID) and acceptor (IA) fluorescence signals of optically resolved single molecules were recorded on a total internal reflection fluorescence microscope as described (27, 38). Donor and acceptor signals were found to bleach in a single step, confirming the presence of single molecules. Single-molecule traces showing dynamics and before photobleaching of typically >150 molecules at each concentration were manually selected, and EFRET values for individual Mg2+ concentrations were accumulated in histograms. Histogram distributions were analyzed with a double or triple Gaussian equation to reveal reoccurring mean FRET values. Dwell times from single-molecule trajectories were calculated and plotted in dwell time histograms for calculation of rate constants as described in ref. 25. Control experiments in the absence of D135–L14 or biotin-DNA did not exhibit any conformational dynamics, ruling out nonspecific interactions between the label DNA or the RNA and the quartz surface. All RNA systems studied to date exhibit only negligible perturbations from surface immobilization (39).
Supplementary Material
Acknowledgments.
We thank Anna Marie Pyle for reviewing this manuscript, and Olga Fedorova and Nils G. Walter for many helpful and stimulating discussions and for being allowed to conduct preliminary experiments in the laboratory of N.G.W. This work was supported by the University of Zürich and the Swiss National Science Foundation SNF-Förderungsprofessur (to R.K.O.S.), Wayne State University, National Institutes of Health Grant GM GM085116 (to D.R.), and National Science Foundation CAREER Grant 0747285 (to D.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804034105/DCSupplemental.
References
- 1.Woodson SA. Recent insights on RNA folding mechanisms from catalytic RNA. Cell Mol Life Sci. 2000;57:796–808. doi: 10.1007/s000180050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woodson SA. Metal ions and RNA folding: A highly charged topic with a dynamic future. Curr Opin Chem Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Sosnick TR, Pan T. RNA folding: Models and perspectives. Curr Opin Struct Biol. 2003;13:309–316. doi: 10.1016/s0959-440x(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 4.Russell R, et al. Rapid compaction during RNA folding. Proc Natl Acad Sci USA. 2002;99:4266–4271. doi: 10.1073/pnas.072589599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang XW, Thiyagarajan P, Sosnick TR, Pan T. The rate-limiting step in the folding of a large ribozyme without kinetic traps. Proc Natl Acad Sci USA. 2002;99:8518–8523. doi: 10.1073/pnas.142288399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodson SA. Folding mechanisms of group I ribozymes: Role of stability and contact order. Biochem Soc Trans. 2002;30:1166–1169. doi: 10.1042/bst0301166. [DOI] [PubMed] [Google Scholar]
- 7.Das R, et al. The fastest global events in RNA folding: Electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J Mol Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann K, Schmidt U. Group II introns: Structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- 9.Fedorova O, Zingler N. Group II introns: Structure, folding and splicing mechanism. Biol Chem. 2007;388:665–678. doi: 10.1515/BC.2007.090. [DOI] [PubMed] [Google Scholar]
- 10.Gordon PM, Sontheimer EJ, Piccirilli JA. Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: Extending the parallels between the spliceosome and group II introns. RNA. 2000;6:199–205. doi: 10.1017/s1355838200992069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pyle AM, Lambowitz AM. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2006. pp. 469–505. [Google Scholar]
- 12.Michel F, Ferat JL. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 13.Cech TR. Conserved sequences and structures of group I introns: Building an active site for RNA catalysis. A review. Gene. 1988;73:259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- 14.Swisher JF, Su LJ, Brenowitz M, Anderson VE, Pyle AM. Productive folding to the native state by a group II intron ribozyme. J Mol Biol. 2002;315:297–310. doi: 10.1006/jmbi.2001.5233. [DOI] [PubMed] [Google Scholar]
- 15.Sigel RKO. Group II intron ribozymes and metal ions: A delicate relationship. Eur Inorg Chem. 2005;12:2281–2292. [Google Scholar]
- 16.Sigel RK, Vaidya A, Pyle AM. Metal ion-binding sites in a group II intron core. Nat Struct Biol. 2000;7:1111–1116. doi: 10.1038/81958. [DOI] [PubMed] [Google Scholar]
- 17.Su LJ, Brenowitz M, Pyle AM. An alternative route for the folding of large RNAs: Apparent two-state folding by a group II intron ribozyme. J Mol Biol. 2003;334:639–652. doi: 10.1016/j.jmb.2003.09.071. [DOI] [PubMed] [Google Scholar]
- 18.Su LJ, Waldsich C, Pyle AM. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res. 2005;33:6674–6687. doi: 10.1093/nar/gki973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pyle AM, Fedorova O, Waldsich C. Folding of group II introns: A model system for large, multidomain RNAs? Trends Biochem Sci. 2007;32:138–145. doi: 10.1016/j.tibs.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Waldsich C, Pyle AM. A kinetic intermediate that regulates proper folding of a group II intron RNA. J Mol Biol. 2008;375:572–580. doi: 10.1016/j.jmb.2007.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith GJ, Sosnick TR, Scherer NF, Pan T. Efficient fluorescence labeling of a large RNA through oligonucleotide hybridization. RNA. 2005;11:234–239. doi: 10.1261/rna.7180305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fedorova O, Waldsich C, Pyle AM. Group II intron folding under near-physiological conditions: Collapsing to the near-native state. J Mol Biol. 2007;366:1099–1114. doi: 10.1016/j.jmb.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallo S, Furler M, Sigel RKO. In vitro transcription and purification of RNAs of different size. Chimia. 2005;59:812–816. [Google Scholar]
- 24.Bokinsky G, et al. Single-molecule transition-state analysis of RNA folding. Proc Natl Acad Sci USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rueda D, et al. Single-molecule enzymology of RNA: Essential functional groups impact catalysis from a distance. Proc Natl Acad Sci USA. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin EA, Jr, Qin Z, Michels WJ, Jr, Pyle AM. Group II intron ribozymes that cleave DNA and RNA linkages with similar efficiency and lack contacts with substrate 2′-hydroxyl groups. Chem Biol. 1995;2:761–770. doi: 10.1016/1074-5521(95)90104-3. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang X, et al. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 28.Tan E, et al. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc Natl Acad Sci USA. 2003;100:9308–9313. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodak JH, Downey CD, Fiore JL, Pardi A, Nesbitt DJ. Docking kinetics and equilibrium of a GAAA tetraloop-receptor motif probed by single-molecule FRET. Proc Natl Acad Sci USA. 2005;102:10505–10510. doi: 10.1073/pnas.0408645102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Z, Srividya N, Sosnick TR, Pan T, Scherer NF. Single-molecule studies highlight conformational heterogeneity in the early folding steps of a large ribozyme. Proc Natl Acad Sci USA. 2004;101:534–539. doi: 10.1073/pnas.2636333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boudvillain M, de Lencastre A, Pyle AM. A tertiary interaction that links active-site domains to the 5′ splice site of a group II intron. Nature. 2000;406:315–318. doi: 10.1038/35018589. [DOI] [PubMed] [Google Scholar]
- 32.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang HR, et al. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seraphin B, Simon M, Boulet A, Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989;337:84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- 35.Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 36.Del Campo M, et al. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter NG. Structural dynamics of catalytic RNA highlighted by fluorescence resonance energy transfer. Methods. 2001;25:19–30. doi: 10.1006/meth.2001.1212. [DOI] [PubMed] [Google Scholar]
- 38.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 39.Zhuang X. Single-molecule RNA science. Annu Rev Biophys Biomol Struct. 2005;34:399–414. doi: 10.1146/annurev.biophys.34.040204.144641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.