Abstract
The earliest steps in the folding of proteins are complete on an extremely rapid time scale that is difficult to access experimentally. We have used rapid-mixing quench-flow methods to extend the time resolution of folding studies on apomyoglobin and elucidate the structural and dynamic features of members of the ensemble of intermediate states that are populated on a submillisecond time scale during this process. The picture that emerges is of a continuum of rapidly interconverting states. Even after only 0.4 ms of refolding time a compact state is formed that contains major parts of the A, G, and H helices, which are sufficiently well folded to protect amides from exchange. The B, C, and E helix regions fold more slowly and fluctuate rapidly between open and closed states as they search docking sites on this core; the secondary structure in these regions becomes stabilized as the refolding time is increased from 0.4 to 6 ms. No further stabilization occurs in the A, G, H core at 6 ms of folding time. These studies begin to time-resolve a progression of compact states between the fully unfolded and native folded states and confirm the presence an ensemble of intermediates that interconvert in a hierarchical sequence as the protein searches conformational space on its folding trajectory.
Keywords: protein folding, pulse labeling, rapid mixing
Most proteins fold rapidly from the highly heterogeneous conformational ensemble of the unfolded state into their well defined native conformations. For proteins with >100 residues, collapsed, partially folded intermediates are formed within hundreds of microseconds after the initiation of folding (1–4). Quench-flow pulse-labeling experiments have yielded considerable information about the development of secondary structure in such intermediates, on a time scale of milliseconds (5, 6). However, little is known about the processes that occur within the dead time (≈6 ms) of the conventional quench flow apparatus, nor about the dynamic behavior of the kinetic folding intermediates. To gain insights into the early folding processes for sperm whale apomyoglobin, we have performed pulsed hydrogen-deuterium (H/D) exchange experiments with submillisecond time resolution.
Apomyoglobin has been studied extensively by kinetic and equilibrium methods as a paradigm for understanding protein folding pathways and the structure of folding intermediates (7). The structure of apomyoglobin is similar to that of the holoprotein except that residues in the F helix and the C terminus of the H helix are disordered (8–10). During refolding, apomyoglobin forms an on pathway kinetic intermediate, in which major portions of the A, G, and H helices and part of the B helix are folded, within the 6-ms burst phase of conventional quench-flow H/D exchange experiments (11–14). These same regions adopt stable secondary structure in the equilibrium molten globule intermediate formed at pH 4.2 (10, 15–17). Recent H/D exchange experiments under a variety of conditions detected heterogeneity in the apomyoglobin kinetic intermediate, with the E helix partly folded in a subset of molecules in the conformational ensemble (12).
To obtain further information on the fluctuations and heterogeneity of the apomyoglobin intermediate, we adopted a rapid-mixing exchange protocol for determination of the elementary rate constants of H/D exchange (Scheme 1) (18–20):
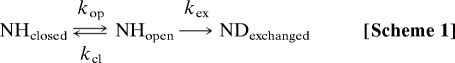 |
where NHclosed is a folded state in which amide protons are protected from exchange, and NHopen is an open state from which exchange occurs. Data were obtained by using a three-solution mixer that can be applied for pulsed H/D exchange experiments in the submillisecond time domain (21, 22). In contrast to previous pulse-labeling experiments (11–13), in which amides were exchanged by a single high pH pulse in the EX1 regime (kex ≫ kcl), our protocol uses a wide range of labeling pulse strengths in both the EX1 and EX2 regimes to measure the pH dependence of the proton occupancy (20). Because the pH dependence of kex is known (23), the proton occupancy curves can be analyzed to give the values of kop and kcl without assuming EX1 or EX2 exchange mechanisms. The fluctuations of the residues in the kinetic intermediate can be assessed from the rate constants, whose ratio (kcl/kop) corresponds to the stability of the closed conformation. Furthermore, accurate values of [NHopen] at the moment of applying the pulse can be estimated. By estimating [NHopen] as a function of refolding time, the evolution of secondary structure along the apomyoglobin folding pathway can be deduced. Fractional values of [NHopen] indicate heterogeneity in the intermediate. The application of this method to refolding of sperm whale apomyoglobin shows that the initial collapse event leads to formation of a folding core comprised of parts of the A, G, and H helices, with fluctuating secondary structure in the B and E helix regions, which becomes stabilized on a millisecond time scale as these helices dock to the core domain.
Results
Time-Dependent Changes in the Proton Occupancy Profiles.
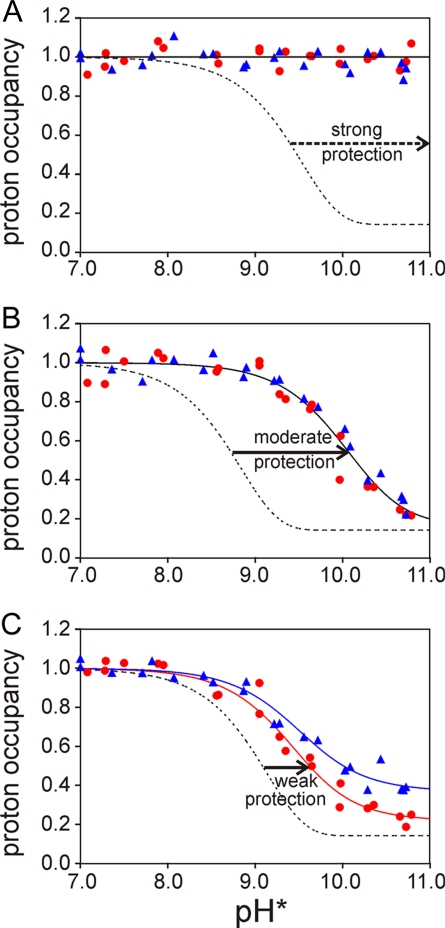
To obtain information on fluctuations and heterogeneity in the folding intermediate of apomyoglobin, we measured proton occupancies (Hobs; Fig. 1) at refolding times of 6 and 0.4 ms and pulse strengths ranging from pH 7 to 10.7, all with duration of 3.6 ms. The resulting exchange profiles can be classified in three groups, exemplified by the data shown in Fig. 1 (a complete set of plots for all residues is included in supporting information (SI) Fig. S1). The amide protons of residues in the first group (termed group S) are strongly protected and cannot be exchanged within the 3.6-ms exchange period even by the strongest labeling pulse (Fig. 1A). Many residues in helices A and G belong to this group. Amide protons in group M are moderately protected from exchange; however, a significant decrease in Hobs is observed at high pH (Fig. 1B). Assuming the EX2 scheme, in which exchange occurs at the rate of kex·kcl/kop, the magnitude of the shift of the sigmoid Hobs curve to higher pH compared to the curve for the free amide (kex; Fig. 1B, dashed line) gives a measure of the stability (kcl/kop) of the amide. The amide protons of many residues located in helices B and H belong to this group. The third group (W) shows only a small shift of the sigmoid Hobs curve to alkaline pH (Fig. 1C), indicating weak protection. Even at the highest pulse strength, where the mechanism is EX1 and exchange is pH independent and limited by kop (20), the proton occupancy after 6 ms of refolding time does not reach its limiting value of 0.14 (determined by the proportion of residual H2O in the refolding buffer). The amide protons in the C and E helices belong to group W.
Fig. 1.
Representative plots of amide proton occupancy in partially refolded apomyoglobin as the 3.6-ms labeling pulse pH is varied from 7.0 to 10.7. (A) V10 (A helix), representative of the highly protected residues (class S). (B) A143 (H helix), representative of moderately protected residues (class M). (C) V66 (E helix), representative of weakly protected residues (class W). Red circles and blue triangles represent the data obtained at tf of 0.4 and 6 ms, respectively. The red lines and blue lines represent the fits to the 0.4- and 6-ms data, respectively, obtained by using Eqs. 2–5. The red and blue lines are coincident in A and B. The dotted lines show the expected curves for unprotected amides at tp of 3.6 ms.
Analysis of HD Exchange Data.
Amide protons in group S exhibit very similar exchange profiles at folding times of 6 and 0.4 ms, indicating that they are already fully protected within 0.4 ms of initiating refolding (Fig. 1A). With the sole exception of I30 (B helix), all of these amides belong to residues in the A and G helices. In contrast, the 0.4- and 6-ms refolding curves are not coincident for many of the amide protons belonging to groups M and W, an indication that the local folding process that results in the protection of these amides is continuing on a time scale of 0.4–6 ms. For each residue, the midpoint of the sigmoid transition is at nearly the same pH for the two refolding curves but there is a significant decrease in the plateau value of the proton occupancy at high pulse pH for the data at 0.4 ms relative to that at 6-ms refolding time (Fig. 1C and Fig. S1). This decrease can be attributed to an increased population of the NHopen state at 0.4-ms refolding time, rather than to differences in kcl and kop values, which would shift the pH midpoint of the sigmoid curve (20). In other words, many amide protons remain largely in the open conformation in the ensemble of compact states formed after 0.4 ms, and additional compaction or folding events reduce the fraction of NHopen after 6 ms of refolding. Amide protons located near the ends of the B helix and in the C and E helices exhibit this behavior, showing that these regions have not yet relaxed into stable hydrogen-bonded conformations in the compact states formed 0.4 ms after initiation of refolding.
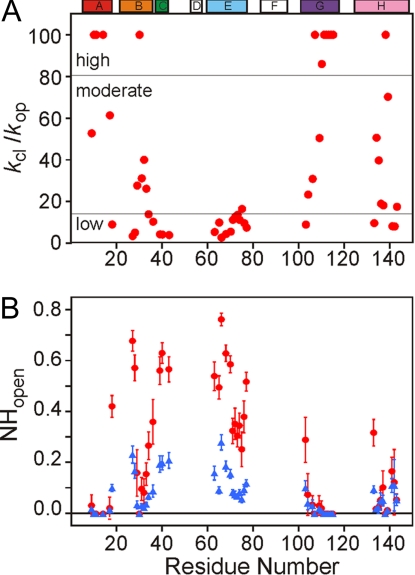
For each residue, the pH-dependent proton occupancies at 0.4- and 6-ms refolding times were fitted simultaneously to the Hvidt equations (18,19) (Eqs. 2 and 3 and Materials and Methods), assuming that the values of kop and kcl are the same at both refolding times. These equations were derived without assuming either EX1 or EX2 regimes (18). Eq. 4 was used to describe the time-dependent changes in [NHopen], assuming that all amide protons are in the open form at the moment of initiation of the refolding reaction. Examples of fitted curves are shown in Fig. 1 as solid lines. All of the exchange profiles can be fitted by the above equations, with the exception of amides in group S, which show only limited dependence on pH. The values of the stability (kcl/kop) and [NHopen] obtained from the data fitting are shown as a function of residue number in Fig. 2 and are listed in Table S1. For amides in group S, only lower limits can be placed on values of kcl/kop (>100) and kcl (more than ≈40,000 s−1) (see SI Text). Moderately protected amides, with kcl/kop between 20 and 100, are found toward the C terminus of the B helix and at the N-termini of the G and H helices (Fig. 2A). Weakly protected amides were identified not only in the C and E helices and CD loop, but also in the terminal regions of the helices that were otherwise highly or moderately protected. The relative stability of each amide in the kinetic intermediate ensemble provides a clue to the nature of the core structure and its mechanism of formation.
Fig. 2.
Plots of stability and [NHopen] as a function of residue number. (A) The estimated stability (kcl/kop) for each amide. For highly protected amides with pH-independent proton occupancies, kcl/kop values are assigned a lower limit of 100. (B) [NHopen] values, estimated from kcl, kop, and tf. The red circles and blue triangles represent [NHopen] at tf of 0.4 and 6 ms, respectively. [NHopen] for highly protected amides is assigned a limiting value of 0. The location of the helices in the holomyoglobin structure is indicated at the top.
The [NHopen] values in different regions of the protein (Fig. 2B) give additional insights that can be interpreted in a time-dependent way. Amide protons in the center of the A, G, and H helices and at I30 in the B helix are strongly protected and have extremely small [NHopen] values at both 0.4- and 6-ms refolding times; no decrease in [NHopen] is observed at the longer folding time. In contrast, most amides in the B, C, and E helices possess much larger values of [NHopen] at 0.4 ms, which decrease significantly after 6 ms of refolding time. The fractional [NHopen] values at 0.4 ms indicate that these amides are involved in fluctuating and heterogeneous conformations in the intermediate ensemble formed after 0.4 ms of refolding. Stabilization of secondary structure and reduction of heterogeneity occurs in these regions as folding progresses in the time range between 0.4 and 6 ms. The amides of several terminal residues of the helices of the AGH folding core (E18 in A helix, Y103 in G helix, and K133 in H helix) also display time-dependent [NHopen] values, reflecting end-fraying of the helices.
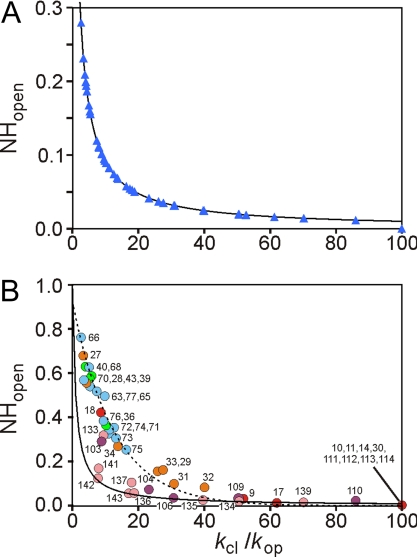
The difference in behavior of the various amides at 0.4- and 6-ms refolding time is emphasized by Fig. 3. For each amide, [NHopen], which is 1.0 in the unfolded state, starts to relax after the initiation of the refolding reaction to the equilibrium value defined by the respective kop and kcl rates. Limiting equilibrium values of [NHopen] can be calculated by using Eq. 4 for infinitely long refolding time, where the second term of the equation becomes zero and [NHopen] reduces to kop/(kop + kcl). Values of [NHopen] obtained by fitting the experimental data at 6-ms refolding time (Fig. 3, blue triangles) coincide with the calculated equilibrium values of [NHopen] (Fig. 3, solid curve), showing that all amides have achieved equilibrium between the open and closed states at this time (Fig. 3A). In contrast, at 0.4-ms refolding time, only the amides located in the helix AGH folding core (plus I30) have reached their equilibrium values of [NHopen] (Fig. 3B). For amides in the B, C, and E helix regions, [NHopen] values are much larger than the limiting equilibrium values, again indicating that these regions of the protein remain in a fluctuating and heterogeneous state in the folding intermediate ensemble formed after 0.4 ms. For amides in the B, C, and E helices, the kop values are all ≈200 s−1 (Table S1): their experimental [NHopen] values can be fitted by Eq. 4 by using kop = 200 s−1 and tf = 0.4 ms (broken line in Fig. 3B).
Fig. 3.
Equilibration of amide proton exchange at 0.4 and 6 ms. (A) [NHopen] versus kcl/kop for a refolding time of 6 ms. Experimental data points are shown as blue triangles. The solid line depicts equilibrium values of [NHopen] at long refolding time, calculated by using the equation [NHopen] = kop/(kop + kcl) derived from Eq. 4. (B) Plot of [NHopen] versus kcl/kop for a refolding time of 0.4 ms. Data points are colored according to their location in the apomyoglobin helices, as depicted in Fig. 2. The highly protected amides are assigned values of kcl/kop = 100 and [NHopen] = 0. The solid line depicts the equilibrium values of [NHopen], and the broken line was calculated by using Eq. 4 with kop = 200 s−1 and tf = 0.4 ms.
Discussion
Formation of the Folding Core Is Associated with Initial Hydrophobic Collapse.
Conventional stopped-flow and quench-flow hydrogen exchange experiments on refolding of apomyoglobin at 5°C revealed formation of a helical intermediate within the 6-ms dead time of the instrument (11, 12). The present studies extend kinetic pulse-labeling experiments on apomyoglobin refolding into the submillisecond time regime and provide insights into the early folding events. In these experiments, most of the observable amide protons in the A, G, and H helix regions are significantly protected from exchange even at the shortest (0.4 ms) refolding time (Figs. 2A and 3B). Many amides in the A and G helix regions show little tendency to exchange even with the highest pH labeling pulse. Some amides in the middle of G (residues 106, 109, and 110) and those in the middle of H (residues 134–137 and 139) are more labile but still appear to have reached their maximal protection in the intermediate formed after 0.4 ms. These data show that the AGH core is fully formed after 0.4 ms of refolding time, although not all amides are equally well protected from exchange. With the exception of residue I30, and to a lesser extent R31 and L32, which have small values of [NHopen] (<0.1), amides in the B, C, and E helices are not strongly protected in the 0.4-ms intermediate, suggesting that stable helical structure has not yet formed.
Although stable helical structure is a prerequisite for the observation of amide proton protection during folding, it is not the major driving force in the folding process. Studies of isolated peptide fragments of apomyoglobin indicate that the H helix possesses the highest propensity for spontaneous formation of helical structure in aqueous solution, whereas the fragments corresponding to the G helix possess poor helix propensity in isolation (24, 25). Both the G and A helices show extensive helix formation in longer peptides where interhelix contacts are possible (26). These data clearly show the importance of tertiary interactions for folding of the A and G helices. The A, B, G, and H helix regions are also the sites of transient long-range contacts formed in the acid-unfolded state of apomyoglobin at equilibrium, which have been directly related to the local hydrophobic nature of the sequence (27–29). Because hydrophobic collapse of the apomyoglobin polypeptide chain and formation of the first tertiary contacts occurs within 300 μs (1, 4, 30), our observation that the A, G, and H helices contain the most highly protected amides at 0.4 ms is consistent with a model in which collapse and coalescence of local hydrophobic clusters in these regions drives formation and stabilization of helical secondary structure. A strong propensity for spontaneous helix formation is not a prerequisite for efficient folding, as shown clearly by previous folding studies of H helix-destabilizing apomyoglobin mutations (31).
It is notable that, although amides in the middle of the H helix are well protected after 0.4 ms and show no decrease in [NHopen] values after 6 ms of refolding time, they are in general less well protected than amides in the A and G helices (Fig. 2). This finding contrasts with the native apoprotein and the pH 4 equilibrium molten globule intermediate, where the protection factors for exchange of amides in the H helix are comparable to those of the A and G helices (10).
Docking of B and E Helices to the Folding Core.
Five amides in the B helix (residues 29, 30, 31, 32, and 33) have relatively small [NHopen] (<0.16) and are moderately protected at 0.4-ms refolding time. These residues contact the C/D region, the N-terminal region of the E helix, and the middle of the G helix in the native state. Of these regions, only the G helix contains amides that show high protection and low [NHopen], which implies that the primary contact for the B helix in the intermediate ensemble is to helix G. These observations are consistent with previous studies of the L32A mutant (32), in which the substitution caused a reduction in the observed proton occupancies at G-helix residues I107 and E109, which contact L32 in the folded state. We therefore suggest that, at 0.4-ms refolding time, the region between residues 29–33 of the B helix is sampling contacts with the fully structured G helix; the interactions with G and the helical structure of B become further stabilized as folding proceeds in the time domain from 0.4 to 6 ms, as evidenced by the decrease in [NHopen] observed for the B-helix residues over this time period.
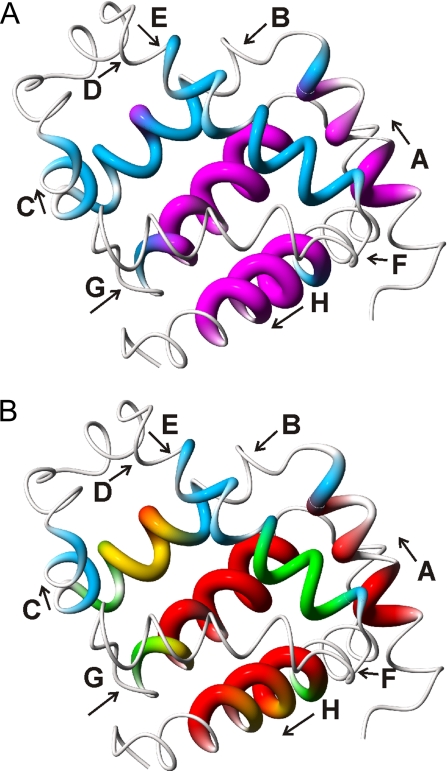
The E helix can be divided into two regions that demonstrate different docking dynamics to the AGH core domain. At the C terminus of the E helix, residues 71–76 possess moderate NHopen at 0.4 ms and low protection (average kcl/kop = 12 ± 2), whereas at the N terminus, residues 63–70 possess high NHopen and marginal protection (average kcl/kop = 5 ± 3). The C-terminal half of the E helix is located at the junction of the A, G, and H helices in the folded protein (Fig. 4), providing a possible reason for its increased stability in the intermediate state. In addition, the marginal stability of the N-terminal half of the E helix, which contacts the B helix in the native structure, suggests inefficient packing between the E and B helices in the intermediate. These results imply that the docking of the B and E helices to the core occurs independently and that interactions between the B and E helices are not established during this time period. It follows that the formation of the correct contacts between the different helices that dock to the core might be an important kinetic bottleneck in the folding of apomyoglobin.
Fig. 4.
Location of the most rapidly protected amides. (A) Distribution of fully and nonequilibrated amides in the 0.4-ms intermediate ensemble. The structure of holomyoglobin (38) is represented as a tube of varying radius. Residues whose amides are fully equilibrated after 0.4 ms of refolding, i.e., for which [NHopen] has reached the equilibrium value, are depicted in magenta with increased tube radius. Residues for which [NHopen] has not reached the equilibrium values and which are fluctuating between folded and unfolded states are depicted by a thinner blue tube. Very thin tubes represent regions that do not exhibit exchange protection and probably remain unstructured in the folding intermediate. (B) Mapping of kcl values onto the holomyoglobin structure. Closing rates are depicted by variations in color and tube radius. Red, kcl > 9,500 s−1; yellow, 5,000 < kcl < 9,500 s−1; green, 2,500 < kcl < 5,000 s−1; blue, kcl < 2,500 s−1. This figure was prepared by using MolMol (39).
Cooperative Dynamics in the Intermediate Ensemble.
Analysis of kop and kcl and the time-dependent changes in [NHopen] yields profound insights into the structural heterogeneity and dynamic cooperativity of the intermediate ensemble. Our model assumes that the open-closed dynamics of different amides can be described by independent kop and kcl values. Exchange between open and closed states is fast enough (kop + kcl > 1,000 s−1) that the exchange process in the intermediate ensemble reaches equilibrium within the 6-ms refolding time, i.e., [NHopen] is equal to kop/(kop + kcl) (Eq. 4). That the system has equilibrated within this refolding time is confirmed by Fig. 3A, which shows that the fitted values of [NHopen] at 6 ms are coincident with the calculated equilibrium values, i.e., all protected amides in the intermediate ensemble have reached equilibrium after 6 ms.
In contrast to the data at 6 ms, the plot of [NHopen] versus kcl/kop for the data at 0.4 ms (Fig. 3B) shows that the amides fall into two groups. First, the amides in the AGH folding core are fully equilibrated even after 0.4 ms of refolding, and their [NHopen] values are identical to those of the equilibrated state at 6 ms. Amides in these regions are highly protected from exchange and kcl values are large, i.e., folding is very rapid. Second, the amides in the B and E helices possess [NHopen] values that are significantly shifted from the equilibrated values; for these amides, kop + kcl is not fast enough for equilibrium to be reached within the 0.4-ms refolding time. The B-helix amides cluster around kcl/kop of ≈20–40, and the amides in the N-terminal and C-terminal halves of the E helix cluster around values of 5 and 10, respectively. The distribution of [NHopen] values for amides in these helices can be fitted reasonably well by Eq. 4 by using kop of 200 s−1 and tf of 0.4 ms (Fig. 3B). Amides in the C helix also belong to this group. The location of the equilibrated and nonequilibrated amides mapped onto the holomyoglobin structure is shown in Fig. 4A.
With the exception of the H helix, kop values lie in a fairly narrow range (≈200–400 s−1). The fact that kop values are similar for most residues implies that H/D exchange occurs through a cooperative, global opening process that involves unfolding of the intermediate. Opening rates in the H helix, especially toward the C terminus, are significantly faster and suggest fraying of the helix. In contrast, considerable variation is seen in closing rates (kcl), which provides important insights into the folding process. For residues in the G and H helices that display pH-dependent proton occupancies, folding is very fast and values of kcl are mostly >10,000 s−1. Although it is not possible to fit kinetic parameters for highly protected residues in helices A and G whose proton occupancies show no pH dependence, a lower limit of ≈40,000 s−1 can be placed on the kcl rates. Closing rates near the ends of the A, G, and H helices are significantly slower and reflect the decreased stability of the helix terminals in the conformational ensemble of the intermediate.
The average value of kcl in the central region of the B helix is ≈5,000 s−1, suggesting that this region folds more slowly than the AGH core. The similar values of kcl and kop for these residues argue that both folding and unfolding of the central region of the B helix is cooperative. The N- and C-terminal halves of the E helix have the same kop rates but display significantly different values of kcl; for residues 63–70, the average kcl is ≈1,400 s−1, whereas the average kcl for residues 71–76 is ≈3,000 s−1. These results suggest that the E helix comprises two independent subdomains, and that folding and unfolding of each subdomain occurs cooperatively. It is also of note that kcl values at the N terminus of the B helix (residues D27 and I28) are the same as the N-terminal residues of helix E, suggesting that these two regions, which are in contact in the native myoglobin structure, fold at the same rate. The kcl values are mapped to the myoglobin structure in Fig. 4B.
Apomyoglobin Folding Mechanism.
Based on the measured kinetic parameters, we propose a hierarchical model for folding of apomyoglobin. The A, G, and H helix regions undergo rapid collapse and fast formation of helical structure (red regions in Fig. 4B). The B, C, and E helices then dock onto the AGH core domain and fold in the following order: B helix (Fig. 4B, yellow), C-terminal subdomain of the E helix (Fig. 4B, green), and N-terminal subdomain of the E helix and C helix (Fig. 4B, blue). This interpretation is consistent with previous apomyoglobin folding schemes involving two sequential on pathway intermediates separated by an energy barrier (1, 33, 34). Jamin and Baldwin (33) identified two intermediates, designated Ia and Ib, that exist in equilibrium in the pH 4.2 molten globule state of sperm whale apomyoglobin and are populated transiently during kinetic refolding (U ↔ Ia ↔ Ib ↔ N). Fast kinetic refolding experiments with horse apomyoglobin revealed formation of an initial collapsed intermediate, I1, within 0.3 ms, which converts to a second intermediate, I2, with a time constant of 5 ms (1). It appears that I1 and I2 correspond to the Ia and Ib intermediates, respectively, of the sperm whale protein (1, 34). The two intermediates are equally compact (Rg, 23.7 Å) but differ in helical content (30% and 44% helix for I1 and I2, respectively). The initial intermediate, Ia/I1, likely corresponds to the ensemble of heterogeneous conformations observed in the present work after 0.4 ms of refolding, containing helical structure in the A, G, and H regions. The second intermediate, Ib/I2, would then correspond to the equilibrated state formed after 6 ms, in which additional helical structure has become stabilized in the B-, C-, and E-helix regions. The increase in helicity between I1 and I2 revealed by CD (1) is quite consistent with the present pulse-labeling experiments, which show increased exchange protection between 0.4 and 6 ms for regions that encompass 17–18 residues in the B, C, and E helices. Although the present data suggest that different domains of the B and E helices fold at different rates, these processes would be difficult to resolve by the low-resolution CD and fluorescence methods used in the previous kinetic measurements, which detected only the complete folding of both the B and E helices as a single kinetic event. Further studies on the opening and closing dynamics of the individual amides in helices B and H and on the heterogeneity of the intermediate ensemble are required to understand the kinetic cooperativity and heterogeneity in apomyoglobin folding.
Materials and Methods
Rapid Solution Mixer.
For the pulse-labeling experiments, a series of three consecutive timed mixing steps is required: initiation of the refolding reaction, H/D exchange, and quench. A schematic drawing of the equipment, modified from the previous versions (2, 21, 22), is shown in Fig. S2. Usage and calibration of the mixer are described in SI Text.
H/D Exchange, pH Pulse Labeling for Apomyoglobin.
15N-labeled sperm whale apomyoglobin was produced in Escherichia coli BL21-DE3 cells and purified as described (35). A solution containing the fully unfolded apomyoglobin (≈80 μM) at pH 2.2 in HCl/H2O was placed in syringe P (Fig. S2). Refolding and H/D exchange was performed at room temperature. The refolding reaction was initiated by a 6-fold dilution of the acid-unfolded apomyoglobin with either 60 mM acetate buffer or 30 mM citrate buffer in 2H2O delivered from syringe Q, to a final pH* (measured pH, uncorrected for the deuterium isotope effect) of 5.8. After refolding times (tf) of 0.4 or 6 ms, the H/D exchange reaction was initiated by a 1/6-fold dilution of the sample solution with 2H2O buffer containing 350-mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) delivered from syringe R at the desired pH* from 7 to 10.7 to enhance exchange of unprotected amide protons to deuterons. The H/D exchange labeling was allowed to proceed for 3.6 ms (tp), and the pH* was then rapidly lowered to 5.7 by mixing with 250 mM citrate buffer in 2H2O, precooled on ice. To avoid further undesired H/D exchange, the samples were kept on ice or at 4°C after the final mixing.
Because holomyoglobin is more stable to exchange than apomyoglobin, heme was added to the exchange-labeled apomyoglobin and the CO complex was formed by a modification of previous procedures (12). The pulse-labeled samples were mixed with a 6-fold molar excess of hemin (Sigma) in a 2H2O buffer containing 250 mM Hepes and 0.1-mg/ml KCN adjusted to pH* 8.2. After a 5-s delay to allow heme insertion and isomerization, the pH* was decreased to 6.0 by addition of 700 mM citrate buffer in 2H2O. The holomyoglobin was concentrated by using a centrifugal filter (Amicon Ultra-15; Millipore) and the buffer was exchanged to 50 mM phosphate in 2H2O at pH* 5.6. The holomyoglobin was reduced by adding sodium dithionite to a final concentration of 1.3 mg/ml and purged with carbon monoxide to prepare the CO-bound complex. Excess dithionite was removed under a CO atmosphere by using a Sephadex G15 column equilibrated with 50 mM phosphate buffer in 2H2O at pH* 5.6. The eluate was collected directly in an NMR tube purged with CO.
NMR Spectroscopy.
15N-1H heteronuclear single quantum coherence (HSQC) spectra were recorded by using a Bruker DRX600 spectrometer at a probe temperature of 35°C. The spectral widths were adjusted to 9,615 Hz and 2,000 points in the 1H dimension and 1,538 Hz and 128 points in the 15N dimension. Proton occupancies were determined from HSQC cross-peak intensities (see SI Text).
Data Analysis.
Amide hydrogen exchange was interpreted based on Scheme 1, in which a given amide can be in a protected state that is nonexchangeable (NHclosed), an open state from which exchange occurs (NHopen), or in the fully exchanged state (NDexchanged) (18, 19, 36). kop and kcl are the local opening and local closing rates, and kex is the exchange rate of the unprotected amide. The latter depends on a variety of conditions and varies with pH as described in Eq. 1 (23, 37):
where kexpH7 is the intrinsic exchange rate of the unprotected amide at pH 7. Because kex increases with increasing pH, the fraction of protonated amide (fH) varies sigmoidally with labeling pulse pH, as expressed by Eq. 2 (20):
 |
where
 |
[NHopen]tf is the population of the open state at refolding time tf. In practice, we observe a proton occupancy (Hobs), which is the sum of fH and the remaining fraction of H2O after the second mixing (= 1/7 for the 7-fold dilution with refolding buffer and pulse labeling buffer in our experimental protocol).
The Hobs values as a function of pH were fitted to the above equations by using the program IGOR Pro (Wavemetrics).
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grant DK34909 (to P.E.W.). T.U. was supported by a fellowship from the Japan Society for the Promotion of Science to Young Scientists, and S.A. was supported by PRESTO, the Japan Science and Technology Agency.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804033105/DCSupplemental.
References
- 1.Uzawa T, et al. Collapse and search dynamics of apomyoglobin folding revealed by submillisecond observations of α-helical content and compactness. Proc Natl Acad Sci USA. 2004;101:1171–1176. doi: 10.1073/pnas.0305376101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uzawa T, et al. Time-resolved small-angle X-ray scattering investigation of the folding dynamics of heme oxygenase: Implication of the scaling relationship for the submillisecond intermediates of protein folding. J Mol Biol. 2006;357:997–1008. doi: 10.1016/j.jmb.2005.12.089. [DOI] [PubMed] [Google Scholar]
- 3.Arai M, et al. Microsecond hydrophobic collapse in the folding of Escherichia coli dihydrofolate reductase, an α/β-type protein. J Mol Biol. 2007;368:219–229. doi: 10.1016/j.jmb.2007.01.085. [DOI] [PubMed] [Google Scholar]
- 4.Lapidus LJ, et al. Protein hydrophobic collapse and early folding steps observed in a microfluidic mixer. Biophys J. 2007;93:218–224. doi: 10.1529/biophysj.106.103077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Englander SW. Protein folding intermediates and pathways studied by hydrogen exchange. Annu Rev Biophys Biomol Struct. 2000;29:213–238. doi: 10.1146/annurev.biophys.29.1.213. [DOI] [PubMed] [Google Scholar]
- 6.Bai Y. Protein folding pathways studied by pulsed- and native-state hydrogen exchange. Chem Rev. 2006;106:1757–1768. doi: 10.1021/cr040432i. [DOI] [PubMed] [Google Scholar]
- 7.Wright PE, Baldwin RL. In: Mechanisms of Protein Folding. Pain RH, editor. Oxford: Oxford Univ Press; 2000. pp. 309–329. [Google Scholar]
- 8.Eliezer D, Wright PE. Is apomyoglobin a molten globule? Structural characterization by NMR. J Mol Biol. 1996;263:531–538. doi: 10.1006/jmbi.1996.0596. [DOI] [PubMed] [Google Scholar]
- 9.Lecomte JT, Sukits SF, Bhattacharjya S, Falzone CJ. Conformational properties of native sperm whale apomyoglobin in solution. Protein Sci. 1999;8:1484–1491. doi: 10.1110/ps.8.7.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimura C, Dyson HJ, Wright PE. Enhanced picture of protein-folding intermediates by using organic solvents in H/D exchange and quench-flow experiments. Proc Natl Acad Sci USA. 2005;102:4765–4770. doi: 10.1073/pnas.0409538102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jennings PA, Wright PE. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science. 1993;262:892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura C, Dyson HJ, Wright PE. The apomyoglobin folding pathway revisited: Structural heterogeneity in the kinetic burst phase intermediate. J Mol Biol. 2002;322:483–489. doi: 10.1016/s0022-2836(02)00810-0. [DOI] [PubMed] [Google Scholar]
- 13.Nishimura C, Dyson HJ, Wright PE. Identification of native and nonnative structure in kinetic folding intermediates of apomyoglobin. J Mol Biol. 2006;355:139–156. doi: 10.1016/j.jmb.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 14.Tsui V, et al. Quench-flow experiments combined with mass spectrometry show apomyoglobin folds through an obligatory intermediate. Protein Sci. 1999;8:45–49. doi: 10.1110/ps.8.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughson FM, Wright PE, Baldwin RL. Structural characterization of a partly folded apomyoglobin intermediate. Science. 1990;249:1544–1548. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- 16.Eliezer D, Chung J, Dyson HJ, Wright PE. Native and nonnative structure and dynamics in the pH 4 intermediate of apomyoglobin. Biochemistry. 2000;39:2894–2901. doi: 10.1021/bi992545f. [DOI] [PubMed] [Google Scholar]
- 17.Eliezer D, Yao J, Dyson HJ, Wright PE. Structural and dynamic characterization of partially folded states of myoglobin and implications for protein folding. Nat Struct Biol. 1998;5:148–155. doi: 10.1038/nsb0298-148. [DOI] [PubMed] [Google Scholar]
- 18.Hvidt A. A discussion of the pH dependence of the hydrogen-deuterium exchange of proteins. C R Trav Lab Carlsberg. 1964;34:299–317. [PubMed] [Google Scholar]
- 19.Hvidt A, Nielsen SO. Hydrogen exchange in proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- 20.Krishna MM, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Akiyama S, Takahashi S, Ishimori K, Morishima I. Stepwise formation of α-helices during cytochrome c folding. Nat Struct Biol. 2000;7:514–520. doi: 10.1038/75932. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi S, Yeh SR, Das TK, Gottfried DS, Rousseau DL. Folding of cytochrome c initiated by submillisecond mixing. Nat Struct Biol. 1997;4:44–50. doi: 10.1038/nsb0197-44. [DOI] [PubMed] [Google Scholar]
- 23.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin H-C, et al. Peptide models of protein folding initiation sites. 3. The G-H helical hairpin of myoglobin. Biochemistry. 1993;32:6356–6364. doi: 10.1021/bi00076a008. [DOI] [PubMed] [Google Scholar]
- 25.Waltho JP, Feher VA, Merutka G, Dyson HJ, Wright PE. Peptide models of protein folding initiation sites. 1. Secondary structure formation by peptides corresponding to the G and H helices of myoglobin. Biochemistry. 1993;32:6337–6347. doi: 10.1021/bi00076a006. [DOI] [PubMed] [Google Scholar]
- 26.Reymond MT, Merutka G, Dyson HJ, Wright PE. Folding propensities of peptide fragments of myoglobin. Protein Sci. 1997;6:706–716. doi: 10.1002/pro.5560060320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lietzow MA, Jamin M, Dyson HJ, Wright PE. Mapping long-range contacts in a highly unfolded protein. J Mol Biol. 2002;322:655–662. doi: 10.1016/s0022-2836(02)00847-1. [DOI] [PubMed] [Google Scholar]
- 28.Yao J, Chung J, Eliezer D, Wright PE, Dyson HJ. NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding. Biochemistry. 2001;40:3561–3571. doi: 10.1021/bi002776i. [DOI] [PubMed] [Google Scholar]
- 29.Felitsky DJ, Lietzow MA, Dyson HJ, Wright PE. Modeling transient collapsed states of an unfolded protein to provide insights into early folding events. Proc Natl Acad Sci USA. 2008;105:6278–6283. doi: 10.1073/pnas.0710641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishiguchi S, Goto Y, Takahashi S. Solvation and desolvation dynamics in apomyoglobin folding monitored by time-resolved infrared spectroscopy. J Mol Biol. 2007;373:491–502. doi: 10.1016/j.jmb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Cavagnero S, Dyson HJ, Wright PE. Effect of H helix destabilizing mutations on the kinetic and equilibrium folding of apomyoglobin. J Mol Biol. 1999;285:269–282. doi: 10.1006/jmbi.1998.2273. [DOI] [PubMed] [Google Scholar]
- 32.Nishimura C, Wright PE, Dyson HJ. Role of the B helix in early folding events in apomyoglobin: Evidence from site-directed mutagenesis for native-like long-range interactions. J Mol Biol. 2003;334:293–307. doi: 10.1016/j.jmb.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 33.Jamin M, Baldwin RL. Two forms of the pH 4 folding intermediate of apomyoglobin. J Mol Biol. 1998;276:491–504. doi: 10.1006/jmbi.1997.1543. [DOI] [PubMed] [Google Scholar]
- 34.Weisbuch S, et al. Cooperative sub-millisecond folding kinetics of apomyoglobin pH 4 intermediate. Biochemistry. 2005;44:7013–7023. doi: 10.1021/bi047372v. [DOI] [PubMed] [Google Scholar]
- 35.Jennings PA, Stone MJ, Wright PE. Overexpression of myoglobin and assignment of the amide, Cα and Cβ resonances. J Biomol NMR. 1995;6:271–276. doi: 10.1007/BF00197808. [DOI] [PubMed] [Google Scholar]
- 36.Linderström-Lang K. Deuterium exchange between peptides and water. Chem Soc Spec Publ. 1955;2:1–20. [Google Scholar]
- 37.Connelly GP, Bai Y, Jeng M-F, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 38.Kuriyan J, Wilz S, Karplus M, Petsko GA. X-ray structure and refinement of carbon-monoxy (Fe II)-myoglobin at 1.5-Å resolution. J Mol Biol. 1986;192:133–154. doi: 10.1016/0022-2836(86)90470-5. [DOI] [PubMed] [Google Scholar]
- 39.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.