Abstract
Bone morphogenetic proteins (BMPs) are potent morphogens that activate transcriptional programs for lineage determination. How BMP induction of a phenotype is coordinated with microRNAs (miRNAs) that inhibit biological pathways to control cell differentiation, remains unknown. Here, we show by profiling miRNAs during BMP2 induced osteogenesis of C2C12 mesenchymal cells, that 22 of 25 miRNAs which significantly changed in response to BMP2 are down-regulated. These miRNAs are each predicted to target components of multiple osteogenic pathways. We characterize two representative miRNAs and show that miR-133 directly targets Runx2, an early BMP response gene essential for bone formation, and miR-135 targets Smad5, a key transducer of the BMP2 osteogenic signal, controlled through their 3′UTR sequences. Both miRNAs functionally inhibit differentiation of osteoprogenitors by attenuating Runx2 and Smad5 pathways that synergistically contribute to bone formation. Although miR-133 is known to promote MEF-2-dependent myogenesis, we have identified a second complementary function to inhibit Runx2-mediated osteogenesis. Our key finding is that BMP2 controls bone cell determination by inducing miRNAs that target muscle genes but mainly by down-regulating multiple miRNAs that constitute an osteogenic program, thereby releasing from inhibition pathway components required for cell lineage commitment. Thus, our studies establish a mechanism for BMP morphogens to selectively induce a tissue-specific phenotype and suppress alternative lineages.
Keywords: C2C12 cells, miR-133, miR-135, osteoblast differentiation, Runx2
Skeletal development requires stringent control of a program for gene activation and suppression in response to physiological cues. The potent osteogenic bone morphogenetic proteins (BMPs) (BMP2/7) and canonical Wnt signaling activate skeletal-related genes for formation of cartilage and bone (1–3). BMPs regulate the differentiation of multiple cell types (4–6) but will also drive commitment of mesenchymal stem cells into a specific lineage. BMP2, for example, supports osteoblastogenesis (7, 8). There has been a principal focus on identifying the mechanisms by which only one cell phenotype is activated by BMP2. BMP2 intracellular receptor Smads, particularly Smad5, activates osteoblast-essential genes, including the transcription factors required for in vivo bone formation Runx2 (Cbfa1/AML3) and Osterix (9–11). The contribution of global mechanisms for suppression of alternative lineages is equally important for understanding tissue development and diseases.
MicroRNAs (miRNAs) have emerged as key negative regulators of diverse biological and pathological processes, including developmental timing, organogenesis, apoptosis, cell proliferation, and differentiation (reviewed in ref. 12) and in the control of tumorigenesis (13, 14). miRNAs are small (22-nt) endogenous noncoding RNAs that anneal to the 3′ UTR of target mRNAs to mediate inhibition of translation and lower protein levels. It remains to be established how specific miRNAs contribute to regulate the onset of a tissue-specific phenotype in response to a multifunctional morphogen.
Using BMP2-induced osteogenesis from premyoblast mesenchymal cells, miRNA profiling during the initial stages of osteoblast phenotype development showed up-regulation of a limited cohort of miRNAs that have predicted targets for muscle differentiation. Twenty-two miRNAs were down-regulated in response to the BMP2 signal. All of these miRNAs had related and overlapping predicted targets that are positive regulators of bone formation, including mediators of the Wnt, BMP, and FGF signaling pathways, and transcriptional regulators of osteogenesis. Here, we show miR-133, essential for myogenesis (reviewed in ref. 15), has a complementary role in suppressing osteogenesis by directly targeting Runx2, and miR-135 directly targets Smad5, the intracellular BMP2 receptor for osteoblast differentiation (10). Both Runx2 and Smad1/5 are essential for osteogenesis and synergize for activation of bone-specific genes (10, 11). Thus, our studies support a previously uncharacterized function for morphogens to promote cell lineage determination through regulation of miRNAs by both suppressing alternative phenotypes and releasing from inhibition the targets of miRNAs required for differentiation of a tissue-specific phenotype.
Results
Osteogenesis-Related miRNAs Are Down-Regulated During BMP2-Mediated C2C12 Osteogenic Differentiation.
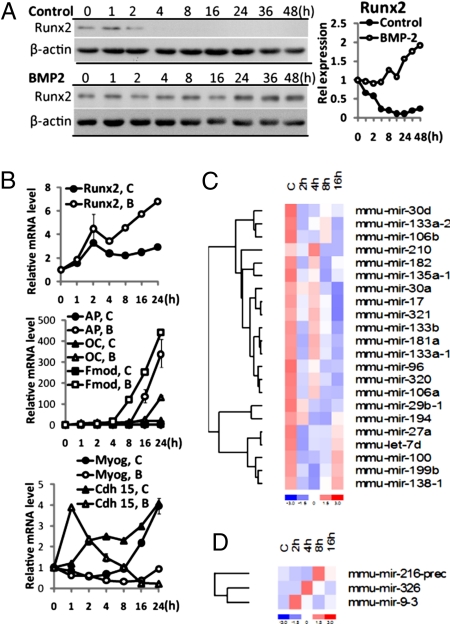
To detect miRNAs related to the osteogenic differentiation program, miRNA profiling (16) was performed by using total RNA from BMP2-treated C2C12 cells that are induced into osteoblasts by 8 h based on expression of muscle and bone phenotypic genes (Fig. 1 A and B). In the control group, Runx2 mRNA and protein rapidly decline during myoblast differentiation. In response to BMP2 treatment, Runx2 mRNA continually increased to 6-fold in mRNA and 2-fold in protein together with osteoblast markers that include alkaline phosphatase and osteocalcin. For the array studies, RNA from control and BMP2-treated samples were collected at 2, 4, 8, and 16 h. In two separate experiments under the same conditions, 22 of the 25 miRNAs that exhibited maximal change in response to BMP2 were down-regulated in the C2C12 samples that underwent osteoblast differentiation (Fig. 1C). Table 1 shows selected gene targets from the top 7% of potential targets listed in the miRanda, TargetScan, or PicTar databases, which play positive roles in osteoblast differentiation. These predicted targets included bone-related regulatory proteins Runx2 (9), Msx2 (17), Dlx3 (18), SMAD1 and SMAD5 (19); members of the Wnt/β-catenin pathway (WNT, CTNNB1) (3); BMPs and their receptors (BMP2, BMPR2, BMPR1A); JAK/STAT signaling components, including JAK2, STAT3, and STAT6; and MAPK (ERK1, ERK2, MAPK14) signaling pathways, which have all been reported to promote osteogenesis (20, 21). Only a limited number of miRNAs were up-regulated, and these miRs targeted putative myoblast related genes (22) (Fig. 1D, Table 1). Thus, BMP2 induced miRs to suppress muscle differentiation but principally inhibited miRNAs which targeted mRNAs that contribute to induction of osteoblastogenesis.
Fig. 1.
A program of miRNAs is expressed during C2C12 osteogenic differentiation. (A and B) Expression profile of osteoblastic and myogenic markers during C2C12 differentiation. (A) Western blot of Runx2 and β-actin (as control) expression in control and BMP2-treated groups. Right shows Runx2 densitometry quantitation normalized to β-actin. (B) mRNA levels (Q-PCR normalized to GAPDH) of osteoblastic markers Runx2, alkaline phosphatase (AP), osteocalcin (OC), and fibromodulin (Fmod), and muscle related genes myogenin (Myog) and cadherin 15 (Cdh 15) during the time course experiments used for miRNA profiling. Relative levels are expressed setting 0 time to 1 (B). (C and D) BMP2 down-regulated (C) and up-regulated (D) miRNAs from miRNA profiling data combined from two independent experiments. For presentation of hierarchical clustered gene, dChip software was used. See Materials and Methods for normalization of each miRNA data point.
Table 1.
Putative targets of significantly changed microRNAs in C2C12 osteogenic differentiation
| miRNA | Putative targets |
|---|---|
| mir-106a | BMP2, BMPR2, FZD1, FZD4, FZD7, SMAD5, STAT3, TGFBR2 |
| mir-106b | BMP2, FZD1, FZD4, FZD7, SMAD5, STAT3 |
| mir-133a | FGFR1, ID3, RUNX2, TCF7, TGFBR2 |
| mir-133b | FGFR1, RUNX2, TCF7 |
| mir-135a | BMPR1A, BMPR2, JAK2, MSX2, SMAD5, STAT6 |
| mir-138 | JUND, SMAD3, TCF7, TGFBR3 |
| mir-17–5p | BMP2, BMPR2, JAK1, SMAD5, STAT3,TGFBR2 |
| mir-181a | BMPR2, FOS, MAPK1, TGFBR1 |
| mir-182 | ACTR2, FZD3, SFRP1, SMAD1 |
| mir-27a | BMPR1A, BMPR2, DKK2, ID2, JUN, MAPK14, SMAD2, TGFBR1, WNT3A |
| mir-29b | COL1A1, COL1A2, COL2A1, COL4A2, DLX5, FOS, TGFB3 |
| mir-30–3p | FZD1, FZD7, JUN, SFRP-1, SMAD2, STAT1 |
| mir-320 | BMPR1A, CTNNB1, DLX3, JAK1, JAK2, MAPK1 |
| mir-96 | FZD3S, LRP6, MAD1, TGFBR1 |
| mir-216 | MEF2D |
| mir-326 | MEF2D, MYO1C, MYO1D |
| mir-9 | MYOD1, MYO1C, MYO1D, MYH1, MTPN |
miR-133 and miR-135 Expression Profiles During Bone and Muscle Phenotype Development.
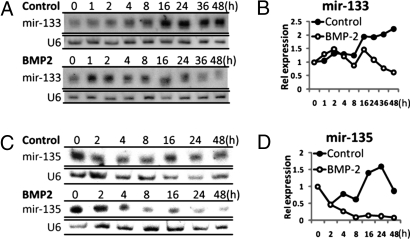
To provide evidence that BMP2 inhibition of specific miRs supports progression of osteoblast differentiation, we functionally characterized two down-regulated miRNAs, miR-133 and miR-135, which share several predicted targets with down-regulated miRs (Fig. 1C, Table 1). miR-133 was predicted to target osteogenic transcription factors, whereas miR-135 was predicted to target many components of BMP signaling. Precursors of miR-133 (mmu-miR-133a-1, mmu-miR-133a-2 and mmu-miR-133b) and miR-135 (mmu-miR-135a-1) were robustly down-regulated (>2-fold) by BMP2 treatment during osteoblast differentiation and increased during the time course in the absence of BMP2 in myoblasts [supporting information (SI) Fig. S1]. We validated the decreased expression of mature miR-133 and miR-135 during BMP2-induced osteogenic differentiation from a replicate study during 48 h (Fig. 2). RNA blot analysis shows a significant up-regulation of miR-133 (Fig. 2 A and B) and miR-135 (Fig. 2 C and D) throughout myoblast differentiation. These miRNAs were abruptly down-regulated early during BMP2 commitment to the osteoblast phenotype. Consistent with loss of miR-133 and miR-135 in C2C12-differentiated osteoblasts, we observed that miR-133 is not expressed in committed and differentiating MC3T3 osteoblasts, and miR-135 is found at very low levels at the preosteoblast stage (data not shown). These findings suggest that miR-133 and miR-135 actively inhibit the osteogenic phenotype in pluripotent cells and during muscle differentiation by targeting essential osteogenic factors. Thus, for osteogenesis to proceed, there is a requirement for down-regulation of miR-133 and miR-135 and other miRNAs in the dataset responsive to BMP2 to release the osteogenic factors from miRNA inhibition.
Fig. 2.
miR-133 and miR-135 expression during BMP2-induced C2C12 osteogenic differentiation. Validation of the array data shown for miR-133 (A and B) and miR-135 (C and D) was performed with RNA from replicate time course up to 48 h. Northern blot analysis of miR-133 and miR-135 expression in control or BMP2 treatment is shown (A and C). U6 snRNA was used as a loading control and for normalization after densitometric quantitation plotted in B and D.
miR-133 Directly Targets Runx2, and miR-135 Directly Targets Smad5.
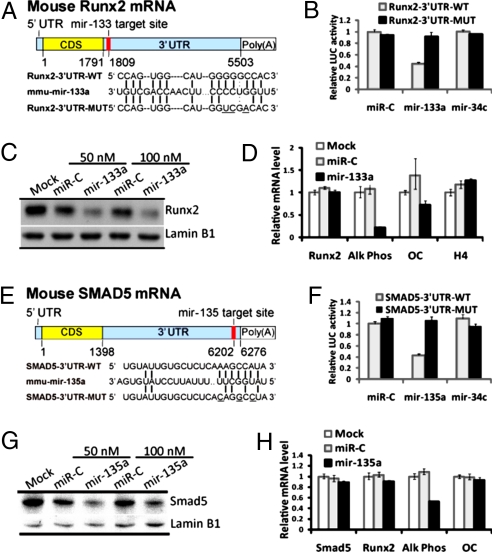
The putative targets of miR-133 (Runx2) and miR-135 (Smad5) reside in parallel but interconnected pathways (9, 11). Smad5, an intracellular BMP2 coreceptor, was selected as the target for validation based on its role in osteoblast differentiation and ability to form a coregulatory complex with the bone-essential transcription factor Runx2 (10, 11). One putative miR-133-binding site is located in the Runx2 3′UTR (Fig. 3A), and one miR-135 putative target site is contained in the mouse Smad5 mRNA 3′UTR (Fig. 3E). We investigated whether these miRs regulate their predicted targets by testing miR-133 activity on Runx2 (Fig. 3 A–D) and miR-135 activity on Smad5 (Fig. 3 E–H). These studies were performed by using a reporter plasmid in which the wild-type or mutated putative 3′UTR coding sequences for Runx2-binding sites (miR-133 “seed region”) or those for Smad5 (miR-135 “seed region”) are cloned into the 3′ UTR of the luciferase gene (shown in Fig. 3 A and E). We find miR-133 ectopic expression significantly repressed luciferase activity of the wild-type Runx2 3′UTR reporter plasmids but not that of the mutated-Runx2 3′UTR reporters (Fig. 3B; Fig. S2). Functional activity of miR-133 was specific because neither the miRNA-control (miR-C) nor miR-34c (which does not target Runx2) affected wild-type and mutant constructs, which validated that miR-133 directly regulates Runx2 mRNA through its 3′UTR. A similar functional luciferase activity assay showed that ectopic expression of miR-135a significantly repressed the luciferase activity of wild-type Smad5 3′UTR reporter plasmids, but had no effect on mutated Smad5 3′UTR reporters (Fig. 3F; Fig. S2). Specificity was confirmed by two types of control miRNAs, which had no effect. In complementary studies, we transfected anti-miR-133 and anti-miR-135 in C2C12 cells, which express these miRs. We observed a 2-fold increased reporter activity of the Runx2 and Smad5 3′UTR-luc reporters and a 2-fold decrease by miRs (Fig. S2). Thus, by overexpression of the miRs and anti-miRs on Runx2 and Smad5 3′UTR wild-type and mutant sequences in both osteoblasts and premyoblasts, we show these genes are direct targets of miR-133 and miR-135, respectively.
Fig. 3.
miR-133 and miR-135 functional activity on target genes. (A) Schematic of miR-133 putative target site in mouse Runx2 3′UTR and alignment of miR-133a with wild-type (WT) and mutant (MUT) 3′UTR region of Runx2 showing complementary pairing. The 3 mutated nucleotides are underlined. (B) MC3T3 cells were cotransfected with the luciferase reporters carrying wild-type Runx2 3′UTR or mutated Runx2 3′UTR, phRL-null (Renilla plasmid) and 100 nM RNA oligonucleotides of miR-Control (miR-C), the miR-133a or miR-34c as an irrelevant control. Effects of mir-133a and control miRNAs on the reporter constructs were shown after 36 h. The ratio of reporter (Firefly) to control phRL-null plasmid (Renilla) in relative luminescence units was plotted. Error bars represent the standard error for n = 3. (C) miR-133 directly targets and regulates Runx2 and inhibits osteoblastogenesis. MC3T3 osteoblast cells were transfected with miR-133a, miRNA-Control or transfection reagent only (Mock) at the indicated concentrations. Western blots for Runx2 and Lamin B1 (as control) were performed on total cell lysates collected at 48 h. (D) Quantitative mRNA levels (normalized by GAPDH) by Q-PCR for Runx2, alkaline phosphatase (Alk Phos), osteocalcin (OC) and histone H4 in 100 nM oligo transfection. Values represent mean ± SD of n = 3 from two independent experiments. (E) Schematic of the miR-135 putative target site in the mouse Smad5 mRNA 3′UTR and alignment of miR-135a with wild-type (WT) and mutant (MUT) 3′ UTR region of Smad5. Complementary pairing between miR-135a and Smad5 is illustrated. Mutated 3′UTR nucleotides (n = 3) are underlined. (F) Functional activity of the luciferase reporter plasmid carrying wild-type or mutated Smad5 3′UTR was assessed as described above in B. Error bars represent the standard error for n = 3. (G) MC3T3 cells were transfected with miR-135a and miRNA-Control as described in C. (H) The mRNA levels of osteoblast marker genes after transfection of miR-Control and miR-135a were determined as described in D.
To address the hypothesis that miR-133 and miR-135 negatively regulated osteoblast differentiation by targeting key signal transduction factors, we introduced the miRNAs into mouse MC3T3 osteoblasts, which express abundant Runx2 protein (Fig. 3C, mock lane). Expression of miR-133 down-regulated endogenous Runx2 protein (Fig. 3C) with no effect on mRNA levels (Fig. 3D), consistent with the mechanism of miRNA regulation. Alkaline phosphatase and osteocalcin mRNA, which are Runx2 target genes, were also down-regulated, whereas histone H4/n mRNA reflecting cell proliferation was not affected in MC3T3 cells overexpressing miR-133 (Fig. 3D). When miR-135 was expressed in MC3T3 cells (Fig. 3G), Smad5 protein was significantly down-regulated which consequently reduced expression of early markers of osteoblast differentiation (Fig. 3H). Because of redundant Smad1 and Smad5 activities in osteoblasts, the effect of miR-135a (50% inhibition) was not as striking as miR-133 repression of Runx2 on osteoblastogenesis (80% inhibition). These observations further support the concept that multiple miRNAs are required for regulation of phenotype development. Taken together, our results indicate that miR-133 directly inhibits expression of Runx2 protein and miR-135 inhibits Smad5 protein and as a consequence together significantly decrease osteoblast differentiation.
miR-133 and miR-135 Inhibit BMP2-Induced Osteogenic Differentiation.
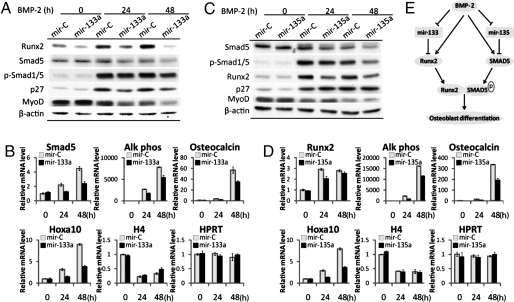
To determine whether miR-133 and miR-135 are directly coupled to BMP2-induced C2C12 osteoblastogenesis through their targets, C2C12 cells were treated with BMP2 for 24 and 48 h after expressing miR-133 for 12 h. As expected, Runx2 protein level is enhanced by BMP2 in cells transfected with miR-Control (Fig. 4 A and C). However, miR-133 expression restrained the increase of Runx2 protein (Fig. 4A) but not Runx2 mRNA after BMP2 treatment (data not shown). MiR-133a ectopic expression affected BMP2 signaling as both Smad5 and p-Smad1/5 protein and Smad5 mRNA decreased in control cells and the increase in response to BMP2 was further decreased by miR-133 expression (Fig. 4 A and B). The cell cycle regulator p27KIP1, which promotes osteoblast differentiation (23), is also up-regulated by BMP2 and inhibited by miR-133. MyoD protein is present in controls, and expression is decreased by BMP2 treatment at 24 h, again supporting the reciprocal relationship between muscle and osteoblast differentiation. The miR-133 block Runx2 protein translation. Thus, the BMP2 mediated down-regulation of miR-133 becomes essential for induction of Runx2 and osteogenic BMP2 signaling. This finding was confirmed by observing that miR-133a overexpression decreased the markers of osteoblast differentiation, including alkaline phosphatase, osteocalcin, and Hoxa10, which were all induced by BMP2, and this induction was prevented by miR-133 (Fig. 4B). However, H4/n mRNA, which is coupled to DNA synthesis, was not affected, nor was the HPRT gene control. Thus, the observed response of p27 to miR-133 supports a role for miR-133 in differentiation directly and not by affecting changes in proliferation or as a nonspecific effect. Taken together, our results suggest that miR-133 functions as a negative regulator of osteogenic differentiation by repressing Runx2 and thereby inhibiting expression of osteoblast-related genes.
Fig. 4.
miR-133 and miR-135 inhibit BMP2-induced C2C12 osteogenic differentiation. (A) miR-133a overexpression restrained BMP2-induced Runx2, p-Smad1/5, and p27 protein increase. C2C12 cells transfected with 100 nM miR-133a and miRNA-Control (miR-C) for 12 h, then cultured in 0.25% albumin serum-free medium, which contains 300 ng/ml BMP2. Western blots for indicated proteins were performed on total cell lysates as shown. (B) miR-133 inhibits osteoblast markers Smad5, alkaline phosphatase (Alk Phos), Osteocalcin, and Hoxa10 mRNA (Q-PCR normalized by GAPDH), but has no effect on H4 and HPRT (hypoxanthine guanine phosphoribosyl transferase 1). (C) mir-135 down-regulates the BMP2 transducer Smad5. C2C12 cells transfected with 100 nM miR-135a were harvested and for consequences on the indicated proteins under conditions described in A. Western blots for Runx2, Smad5, p-Smad1/5, p27, MyoD, and β-actin (as control) were performed on total cell lysates as shown. (D) miR-135 decreases osteogenesis. mRNA levels of osteoblast markers Runx2, alkaline phosphatase (Alk Phos), Osteocalcin and Hoxa10, and H4 and HPRT were detected as described in B. (E) Model of miR-133 and miR-135-mediated regulation of osteoblast differentiation. MiR-133 and miR-135 targeted osteogenic factors Runx2 and Smad5, respectively. BMP2 treatment down-regulates expression of miR-133 and miR-135 and releases expression of Runx2 and Smad5 to promote osteoblast differentiation.
Using the strategy described for miR-133, we tested the functional activity of miR-135 during BMP2-induced C2C12 osteogenic differentiation (Fig. 4 C and D). Western blot results showed that Smad5 protein (Fig. 4C) but not RNA (data not shown) was down-regulated in cells ectopically expressing miR-135. P-Smad1/5 and Runx2 protein were enhanced after BMP2 treatment of the control cells, but this increase was restrained by miR-135a expression (Fig. 4C). Consequently, osteoblast markers of differentiation (Alk Phos, Osteocalcin, and Hoxa10) (Fig. 4D) were inhibited by miR-135. The cell cycle regulator p27 was not affected by miR-135a overexpression (Fig. 4C), suggesting that miR-135 does not have a direct effect in the transition from proliferation to differentiation controlled by p27 (23). These results demonstrate that miR-135a, like miR-133, inhibited C2C12 osteogenic differentiation, and that down-regulation of miR-135 even after BMP2 treatment permits BMP2 signaling through the receptor-SMAD5 target. Together, the functional characterization of two representative BMP2 down-regulated miRNAs, miR-135 and miR-133, demonstrate a previously undescribed pathway for regulation of induced osteoblastogenesis, which is illustrated in Fig. 4E.
Discussion
Our studies have identified a global mechanism by which miRNAs control BMP induction of cell lineage determination as illustrated by BMP2 initiating a selective program of osteoblast differentiation. The mechanism involves BMP2 down-regulation of multiple miRNAs that inhibit translation of bone-forming genes that include essential transcription factors and developmental signaling molecules and their receptors that are required for the complex process of osteoblastogenesis. Through BMP2-inhibited expression of numerous miRNAs with overlapping signal transduction pathway-related components, an osteogenic program is released from repression to drive tissue specific differentiation.
During BMP2-induced osteogenic differentiation of the mesenchymal C2C12 cells, our microarray analysis revealed, first, three up-regulated miRNAs that inhibit myogenesis but 22 miRNAs significantly down-regulated during osteogenic differentiation from ≈318 mouse miRNAs on the array; second, the down-regulated miRNAs were each predicted to target mainly regulatory factors that function to promote osteoblast differentiation; third, the different down-regulated miRNAs shared many common targets, suggesting that this set of BMP2 down-regulated miRNAs constitute a program of miRs required to suppress osteogenesis. Hence, BMP2 osteogenic activities not only involve transcription of bone-promoting factors through Runx2, Smad and MAPK signaling, but BMP2 is also releasing a protein translational block in osteogenic signaling factors required for bone formation. In support of the concept that miRs play a key role in osteogenesis, a recent miRNA profiling study of calvarium, muscle and limb tissues (24) shows the presence of several miRs that we have identified are responsible to BMP2.
miRNAs mediate translational inhibition and alter the levels of critical regulators of biological pathways, thereby providing a mechanism for spatiotemporal control of developmental and homeostatic events across a wide range of plants and animals (25). The developmental morphogen BMP2 regulates growth and differentiation of distinct cell populations, e.g., in intestinal crypts, keratinocytes, osteocyte, dermal papilla cells, and skeletal cells (4–7). Here, we have shown that BMP2 can commit mesenchymal cells to the osteoblast lineage and inhibit myoblast differentiation by both increasing miRNAs to inhibit one cell type and down-regulating other miRNAs that will allow progression of a specific cell phenotype.
The muscle-specific miRNAs, miR-1 and miR-133, were studied in C2C12 cells and shown to control the proliferation and differentiation of the muscle phenotype (26, 27). Our studies demonstrate miR-133 is a negative regulator of the bone-essential Runx2 transcription factor by directly targeting the 3′UTR putative binding sequence for miR-133 “seed region,” important for miRNA–mRNA interactions via Watson–Crick base pairing (28). The binding of miR-133 to Runx2 inhibited translation of Runx2 and by decreasing cellular protein inhibited BMP2-induced C2C12 osteogenic differentiation. Notably, miR-133 and miR-181 in our profile (which we validated; Fig. S3), are muscle-related, contributing to functional activation of muscle-specific transcription factors for differentiation (22). Indeed, we show a complete absence of miR133 and very low levels of miR-135 in committed MC3T3 osteoblasts. Taken together, miR-133 represents a key mechanism for suppressing the osteoblast phenotype in nonosseous cells by its ability to target and negatively regulate Runx2 and likely several osteogenic factors (Table 1) and be responsive to down-regulation by BMP2.
miR-135a, also down-regulated by BMP2, is predicted to target Smad5, a BMP intracellular receptor essential for osteogenesis. A reduction in activated p-Smad and Runx2 expression occurs, but the reduced Runx2 was not a consequence of direct miR-135 targeting to Runx2 3′ UTR (data not shown). Decreased Runx2 levels and functional activity can occur, resulting from decreased phosphorylated Smad1/5, which is necessary for Runx2–Smad5 interactions (29). Cooperation between Runx2 and sustained p-Smad5 is clearly established for osteoblast-specific gene expression (10, 11). In addition, the BMP-Smad pathway contributes to induced Runx2 expression through induction of homeodomain transcription factors (30). Hence, reduced Smad5 protein by miR-135 decreases Runx2 expression, coregulatory protein interactions, and transactivation functions that result in inhibited expression of many osteoblast-related genes that synthesize the bone matrix (9). Thus, miR-135 has a broad range of inhibitory effects on osteoblastogenesis by targeting Smad5 and its down-regulation by BMP2 facilitates activation of the required Smad5 osteogenic signals for bone formation (10).
In this study, we have identified osteoblast-related miRNAs that are critical for regulation of the osteoblast phenotype. Our data indicate that these BMP2-repressible “osteomiRs” attenuate common osteogenic pathways, and that some miRNAs, like mir-133, have dual functions by promoting myogenesis and actively inhibiting osteogenesis. In addition BMP2 up-regulates miRNAs that actively inhibit muscle differentiation. The myogenic and osteogenic miRNAs may compete at early stages of lineage commitment when cells still have considerable plasticity to differentiate to distinct lineages until a morphogen regulates a set of miRNA for commitment to one lineage. An important implication of our studies is that multiple miRNAs are needed to maintain suppression of one phenotype at the expense of another phenotype. Using the model of osteoblast differentiation, we have provided direct functional evidence for the powerful control of lineage direction by BMP2 through miRNAs.
To conclude, after BMP2 treatment of nonosseous cells, down-regulation of multiple miRNAs with common target genes that are mediators of many signaling pathways required for bone formation, identifies a mechanism for rapid induction of the osteoblast phenotype. These studies have clinical relevance in that anti-miRNA therapies directed to inhibit miR-133 and miR-135 could increase allocation of mesenchymal stem cells into the osteogenic lineage for rebuilding the skeleton.
Materials and Methods
Cell Culture
Mouse premyogenic C2C12 cells were maintained in regular growth medium (DMEM, 10% FBS and antibiotics) and differentiate in low serum conditions (7). Osteogenesis was induced by a detailed protocol (8). Cells were cultured in 0.25% BSA containing media for 24 h then treated with 300 ng/ml recombinant human BMP2 (Wyeth Research Genetics Institute) or vehicle (as control) and harvested at the indicated times. The MC3T3 osteoprogenitor cell line established from mouse calvaria (31) were maintained as described in ref. 30.
Protein and mRNA Analyses.
Western blot.
Procedures were described in ref. 32. Primary antibodies used are shown in Table S1. The signal was detected by using a chemiluminescence detection kit (PerkinElmer).
Real-time PCR.
RNAs were analyzed by quantitative RT-PCR as described (30, 32). Briefly, TRIzol reagent (Invitrogen) was used for RNA isolation and purified with the DNA-free RNA kit (Zymo Research). Purified total RNA was used to make cDNA by reverse transcription reaction with oligo(dT) primers (Applied Biosystems). Real-time PCR was performed with SYBR green reagents (Bio-Rad). mRNA levels were normalized to GAPDH (32). Primers used for amplification are listed in Table S2.
miRNA Microarray and Data Analysis.
Microarray procedure and data analysis was performed at Ohio State University as described in ref. 16. Total RNA (5 μg) from each control or BMP2-treated C2C12 groups at selected times (2, 4, 8, and 16 h) were labeled with biotin and hybridized in triplicate by using miRNA microarray chips containing >634 probes corresponding to 316 human and 318 mouse miRNA genes. Detection and quantification of hybridization signals are detailed elsewhere (16). For each miRNA, the relative fold change in each time point was equal to the value of fluorescence in the BMP2 treatment group normalized by the value of fluorescence in controls. The average fold changes of two separate experiments were hierarchically clustered by using dChip software (33).
Northern Blot.
Two independent C2C12 time courses were examined (up to 48 h) for validation of miRNA profiling studies. Total RNA was separated on a 12% acrylamide/urea gel and transferred onto Hybond-XL membranes (Amersham Biosciences). [γ-32P]ATP 5′-end-labeled oligonucleotide probes corresponding to mature miR-133 or miR-135 were used to hybridize RNA blots in Rapid-Hyb Buffer (Amersham Biosciences) following the manufacturer's instructions. The blots were reprobed for U6 RNA to control for equal loading. Band intensity was measured by using AlphaImager software (Alpha Innotech) followed by exposure to x-ray film (Kodak), and the relative expression of the specific miRNA samples was normalized to U6 snRNA and plotted as fold change setting 0 time as 1. All probes are listed in Table S3.
DNA Constructs.
For functional analyses of miR-133 and miR-135, segments of the mRNA 3′UTR sequences of the Runx2 and Smad5 genes were PCR-amplified from mouse genomic DNA by using the sense and antisense primers. This product was then subcloned into the XbaI-FseI site immediately downstream of the stop codon in the pGL3-Control Firefly Luciferase reporter vector (Promega). Sequences of these and mutagenic primers for each target gene are listed in Table S4. The mutated sites were confirmed by digestion of plasmid with SalI (for miR-133) and with StuI (for miR-135), and plasmid DNA controls were sequenced to ensure authenticity.
Transfection Assay.
Double-stranded RNA oligos representing mature sequences that mimic endogenous miRNAs of miR-133a, miR-135a, and miRNA negative control #2 (obtained from Ambion) were transfected into MC3T3-E1 at 30–50% confluence at 50 or 100 nM concentration with Oligofectamine (Invitrogen). Cells were harvested 48 h after transfection for protein and mRNA analysis. miR-34c (irrelevant control), the miRNA inhibitors anti-miR-133a and anti-miR-135a, designed to inhibit endogenous miR-133a and miR-135a, respectively, and anti-miR negative control #1 were also obtained from Ambion. For luciferase activity analysis, miRNAs or anti-miRNAs and plasmid DNAs were cotransfected into cells with Lipofectamine 2000 (Invitrogen) for 36 h. The Dual-luciferase reporter assay system (Promega) was used to quantify luminescent signal by using a luminometer (Glomax, Promega). Each value from the firefly luciferase assay was normalized to the Renilla luciferase assay value from the cotransfected phRL-null vector (Promega).
Supplementary Material
Acknowledgments.
We thank Ying Zhang and Ricardo Medina for helpful discussions, Matthew Mandeville for cell culture support, and Judy Rask for editorial assistance. These studies were supported by National Institutes of Health Grants AR039588 (to G.S.S.), DE012528 (to J.B.L.), and P01CA81534 (to C.M.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804438105/DCSupplemental.
References
- 1.Wu X, Shi W, Cao X. Multiplicity of BMP signaling in skeletal development. Ann NY Acad Sci. 2007;1116:29–49. doi: 10.1196/annals.1402.053. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay A, et al. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodine PV, Komm BS. Wnt signaling and osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:33–39. doi: 10.1007/s11154-006-9002-4. [DOI] [PubMed] [Google Scholar]
- 4.Fukuda S, Taga T. Cell fate determination regulated by a transcriptional signal network in the developing mouse brain. Anat Sci Int. 2005;80:12–18. doi: 10.1111/j.1447-073x.2005.00097.x. [DOI] [PubMed] [Google Scholar]
- 5.Plikus MV, et al. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature. 2008;451:340–344. doi: 10.1038/nature06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rendl M, Polak L, Fuchs E. BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes Dev. 2008;22:543–557. doi: 10.1101/gad.1614408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katagiri T, et al. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 1994;127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balint E, et al. Phenotype discovery by gene expression profiling: Mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J Cell Biochem. 2003;89:401–426. doi: 10.1002/jcb.10515. [DOI] [PubMed] [Google Scholar]
- 9.Lian JB, et al. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- 10.Lee KS, et al. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between runx2 and smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20:8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Javed A, et al. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J Biol Chem. 2008;283:8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 13.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 14.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 15.van RE, Liu N, Olson EN. MicroRNAs flex their muscles. Trends Genet. 2008;24:159–166. doi: 10.1016/j.tig.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Liu CG, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng SL, Shao JS, Charlton-Kachigian N, Loewy AP, Towler DA. Msx2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. J Biol Chem. 2003;278:45969–45977. doi: 10.1074/jbc.M306972200. [DOI] [PubMed] [Google Scholar]
- 18.Hassan MQ, et al. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by Dlx3 and a homeodomain transcriptional network. J Biol Chem. 2006;281:40515–40526. doi: 10.1074/jbc.M604508200. [DOI] [PubMed] [Google Scholar]
- 19.Lee KS, Hong SH, Bae SC. Both the Smad and p38 MAPK pathways play a crucial role in Runx2 expression following induction by transforming growth factor-beta and bone morphogenetic protein. Oncogene. 2002;21:7156–7163. doi: 10.1038/sj.onc.1205937. [DOI] [PubMed] [Google Scholar]
- 20.Bellido T, Borba VZ, Roberson P, Manolagas SC. Activation of the Janus kinase/STAT (signal transducer and activator of transcription) signal transduction pathway by interleukin-6-type cytokines promotes osteoblast differentiation. Endocrinology. 1997;138:3666–3676. [Google Scholar]
- 21.Ge C, Xiao G, Jiang D, Franceschi RT. Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J Cell Biol. 2007;176:709–718. doi: 10.1083/jcb.200610046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Rooij E, Olson EN. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drissi H, et al. The cell cycle regulator p27kip1 contributes to growth and differentiation of osteoblasts. Cancer Res. 1999;59:3705–3711. [PubMed] [Google Scholar]
- 24.Kobayashi T, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci USA. 2008;105:1949–1954. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 29.Afzal F, et al. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J Cell Physiol. 2005;204:63–72. doi: 10.1002/jcp.20258. [DOI] [PubMed] [Google Scholar]
- 30.Hassan MQ, et al. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudo H, Kodama H-A, Amagai Y, Yamamoto S, Kasai S. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J Cell Biol. 1983;96:191–198. doi: 10.1083/jcb.96.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaur T, et al. Canonical WNT signaling promotes osteogenesis by directly stimulating RUNX2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 33.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.