Abstract
The emergence of resistance during multidrug chemotherapy impedes the treatment of many human diseases, including malaria, TB, HIV, and cancer. Although certain combination therapies have long been known to be more effective in curing patients than single drugs, the impact of such treatments on the evolution of drug resistance is unclear. In particular, very little is known about how the evolution of resistance is affected by the nature of the interactions—synergy or antagonism—between drugs. Here we directly measure the effect of various inhibitory and subinhibitory drug combinations on the rate of adaptation. We develop an automated assay for monitoring the parallel evolution of hundreds of Escherchia coli populations in a two-dimensional grid of drug gradients over many generations. We find a correlation between synergy and the rate of adaptation, whereby evolution in more synergistic drug combinations, typically preferred in clinical settings, is faster than evolution in antagonistic combinations. We also find that resistance to some synergistic combinations evolves faster than resistance to individual drugs. The accelerated evolution may be due to a larger selective advantage for resistance mutations in synergistic treatments. We describe a simple geometric model in which mutations conferring resistance to one drug of a synergistic pair prevent not only the inhibitory effect of that drug but also its enhancing effect on the other drug. Future study of the profound impact that synergy and other drug-pair properties can have on the rate of adaptation may suggest new treatment strategies for combating the spread of antibiotic resistance.
Keywords: adaptation, antagonism, synergy, antibiotics, antibiotic resistance
Challenged by rapid emergence of drug-resistant pathogens and limited supply of new antibiotics, clinicians increasingly rely on multidrug treatments to combat infections (1–7). When drugs are applied together the effect of a drug can depend on the presence or absence of the other drug. Such interactions between drugs are classified as additive, synergistic, or antagonistic depending on whether their combined effect on bacterial growth is equal to, greater than, or less than expected based on the inhibitory abilities of the individual drugs (8, 9). Two main goals of drug treatment are stopping bacterial growth and preventing the evolution of drug resistance (5, 10–12). Although synergistically interacting drugs are often favored because of their greater combined ability to inhibit growth (13), little direct evidence for their ability to suppress the evolution of resistance exists, and some studies even suggest the contrary (14–18).
Examining the evolution of resistance in the context of a simple geometric model of drug–drug fitness landscapes, we find that mutations conferring full or partial resistance to one of the individual drugs may be more beneficial to bacteria in synergistic than in antagonistic drug treatments [Fig. 1; the situation for mutations conferring simultaneous resistance to both drugs is illustrated in supporting information (SI) Fig. S1]. This deduction is based on the simplifying assumption, demonstrated in previous work, that mutations conferring resistance to a single drug are effectively equivalent to a reduction in that drug's concentration (16, 17, 19, 20). When drugs amplify each other's effects (synergy) (9), this effective reduction in the concentration of one of the drugs not only relieves the effect associated with that drug but also reduces its enhancing influence on the other drug. In contrast, when drugs partially inhibit one another (antagonism), resistance mutations that remove some of the effect of one of the drugs will actually reveal the previously suppressed effect of the other drug (Fig. 1). This scenario has recently been experimentally observed for a hyperantagonistic drug pair in which horizontally transferred alleles that confer resistance to one of the drugs can actually be deleterious in the combined drug environment (17). This intuition applies whether the starting concentration is above or below an organism's minimal inhibitory concentration (MIC). Such expected differences in the selective advantage of resistant mutants in two-drug environments suggest the hypothesis that the rate of adaptation, the speed with which lineages carrying such mutations spread within evolving populations, might be greater for synergistic than for antagonistic drug combinations.
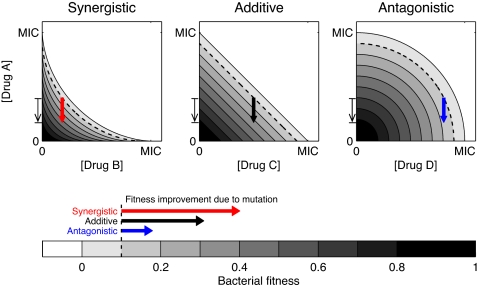
Fig. 1.
A simple geometric model shows that a mutation conferring resistance to a single drug is most advantageous in a synergistic drug combination. Shown are isoboles, or lines of equal bacterial growth rate, in the plane of concentrations of drugs A with either drug B (where B interacts with drug A synergistically), drug C (additively), or drug D (antagonistically). The arrows shown on the isobolograms for the three types of interaction all correspond to the exact same mutation (indicated by a thin arrow along the axis of drug A's concentration), which confers partial resistance to drug A by reducing the effective concentration of drug A felt by the resistant mutant. The three arrows' origins represent environments that have the same initial concentration of drug A and the same fitness inhibition (10%, dotted line). Although the mutation changes the effective concentration of drug A by the same amount in all environments, the fitness gain conferred by the mutation is greatest in the synergistic case (it crosses more fitness contour lines).
To experimentally explore the relation between drug interaction and the rate of adaptation, we compared a drug pair that exhibits strong antagonism [doxycycline (DOX), a tetracycline antibiotic, and ciprofloxacin (CIP), a fluoroquinolone] to another pair showing strong synergy [DOX and a macrolide antibiotic, erythromycin (ERY)] (5, 17, 21, 22). For each pair, drugs were mixed in a two-dimensional array of wells in which the concentration of each of the single drugs varied from zero to above its MIC. Populations derived from clonal expansion of a single wild-type, drug-sensitive E. coli bacterium were introduced to all wells and propagated through daily serial transfers (see Materials and Methods) (23–25). The rates at which bacterial populations developed residual resistance to the various drug combinations were measured by tracking the evolution of these populations over ≈170 generations (Fig. 2). The degree of antibiotic resistance acquired by each population is manifest in the increase of its growth rate over time (The changes in MIC that accompanied these accelerations of growth were relatively mild, smaller than a factor of 2 in some cases and up to an 8-fold increase in others; data not shown). From daily growth curves (Fig. 2A) we estimated how the growth rate changed for each population as it evolved (Fig. 2B). Typically, we see a saturation curve with a relatively fast initial increase in fitness followed by a plateau with little or no additional adaptation in later times. We combined the increase in growth rate of each population during evolution, Δr, and the time it took the population to reach the half-way mark of that increase, tadapt, to define the rate of adaptation, α ≡ (Δr/2)/tadapt (Fig. 2B). This adaptation rate shows large variability among the different drug treatments, reflecting variability in both initial growth rates and adaptation times (Fig. 2C). In contrast, much less variability was manifest in the final growth rates attained by the populations, and by the end of the experiment most of the populations were growing at a rate similar to that in the drug-free environment (Fig. 2C).
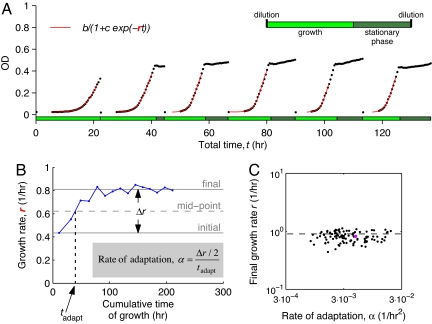
Fig. 2.
Parallel quantitative measurement of the rate of adaptation in multidrug environments. (A) Example of growth in one particular dosage of ciprofloxacin (CIP) and doxycycline (DOX), showing measurements of optical density as a function of time (OD, black dots). Cells are propagated in media containing the drugs through daily serial transfers over 15 days (only the first 6 days are shown), resulting in alternating periods of growth and stationary phase (Inset). Best fit of the logistic growth curve (red lines; indicated equation) defines the growth rate (r) for this population in each day. The measurement error in the growth rates was estimated to be ≈0.06/h (see Materials and Methods and Fig. S3). (B) Data points corresponding to daily growth rates show how the fitness in the population from A increases over time. Time is measured in hours of growth (time in stationary phase is excluded). The total increase in growth rate of the population is denoted by Δr, and the adaptation time, tadapt, is defined as the time at which the population crosses the midpoint between its initial and final growth rates (see Materials and Methods for more details). Measured Δr and tadapt are used for determining the rate of adaptation of each population (α, equation shown). (C) Scatter plot of rates of adaptation and final growth rates for CIP-DOX and ERY-DOX. The point highlighted in magenta corresponds to the population shown in A and B. Most populations that evolve recover the growth rate in a drug-free environment (dashed horizontal line).
Results
Our data show that drug combinations have a strong effect on the evolution of drug resistance (Fig. 3). Different drug pairs, however, have profoundly different impacts on the rate of adaptation. For ERY-DOX, which is a strongly synergistic drug pair, we see accelerated adaptation when the drugs are used in combination (center of the 2-D drug grid, arrow in Fig. 3C) relative to drug treatments involving either of the drugs alone (the two edges lying on the drug concentration axes). For the antagonistic drug pair CIP-DOX, such an acceleration is absent in the combinations that we tested. Moreover, the opposite effect—a depression of the rate of adaptation relative to the single-drug environments—is apparent for some CIP-DOX combinations (Fig. 3D). Related to these observations is the fact that resistance to the synergistic drug pair evolves rapidly, not only compared to each drug separately, but also compared to the antagonistic pair. This link between the way drugs interact and the rate of adaptation is consistent with the hypothesis described in Fig. 1, and further exploration of this relation was achieved by the additional experiments and analyses described below.
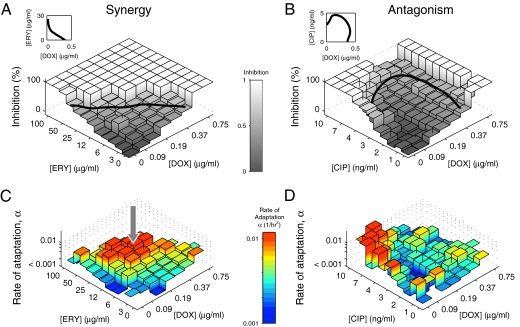
Fig. 3.
Different drug pairs vary profoundly in their impact on the rate of adaptation. (A and B) For two pairs of drugs (A, synergistic ERY-DOX; B, antagonistic CIP-DOX), the initial level of inhibition is shown for a matrix of concentrations of the two drugs. The level of inhibition is defined as 1 − r/r0, where r is the growth rate of the population in the presence of antibiotics and r0 is the drug-free growth rate. The solid line corresponds to the line of 50% inhibition and is also shown on a linear scale in the insets (see Fig. 1 for comparison). (C and D) The rate of adaptation for the drug combinations shown in A and B. The arrow in C points to a region of drug concentrations where the rate of adaptation for the synergistic ERY-DOX combination is accelerated relative to the single-drug treatments. This acceleration is surprising because the more expected outcome of combining drugs is for adaptation to slow down.
We expanded on the comparison of strongly synergistic and strongly antagonistic drug combinations by including two additional drug pairs in our study—CIP with the aminoglycoside amikacin (AMI) and AMI with DOX—which interact synergistically at some dose combinations but antagonistically at others. In addition, we used the fact that for any drug pair, including strongly synergistic or antagonistic ones, the strength of interactions varies for different drug ratios and dosages (26). Therefore, for all drug pairs, for each combination of concentrations of two drugs, we measured the degree of synergy (S, defined in Materials and Methods and in Fig. 4), which ranges from +1 for strongly synergistic interactions to −1 for strongly antagonistic ones, and compared it with the rate of adaptation (Fig. 4; see raw data in Fig. S2). We find a positive correlation between synergy and the rate of adaptation (ρ = 0.683, P < 0.001). Two aspects of the data contribute to this correlation: the correlation among the means of the four drug pairs and the correlation within each pair. To isolate the latter, we calculated the partial correlation between synergy and the rate of adaptation when controlling for drug-pair membership, which yields ρ = 0.533, P < 0.001 (see SI Text). Another factor that is likely to influence the rate of adaptation is the initial inhibition of growth of the ancestral strain in each environment. Again, we have verified that the observed correlation between adaptation rate and synergy remains when accounting for the confounding factor of initial inhibition (see SI Text). Indeed, the partial correlation between rate of adaptation and degree of synergy when controlling for initial inhibition supports the same conclusion as before (P < 0.001), namely, that synergy promotes faster emergence of drug resistance.
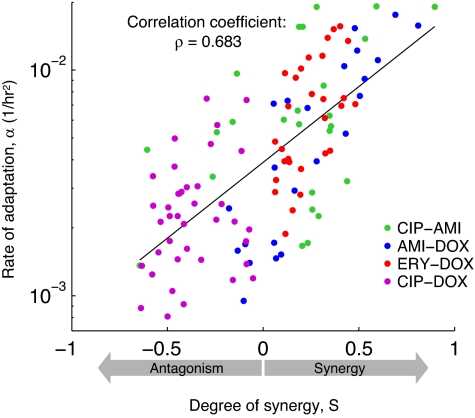
Fig. 4.
The degree of synergy and the rate of adaptation are positively correlated: The more synergistically the drugs interact, the faster the bacteria evolve resistance. Shown are data from all four drug pairs in our study (legend). Each dot represents the rate of adaptation for a given drug pair and concentrations versus the degree of synergy (S). The variability in rates of adaptation has two contributions: an error of approx 25% due to errors in measuring growth rates, and an inherent stochasticity of the mutation process (see Materials and Methods and Figs. S3 and S4). For a combination (x,y) of drug concentrations, S is measured as the deviation from the neutral expectation defined by Bliss independence: S = (fx0/f00)(f0y/f00) − fxy/f00, where fxy denotes wild-type growth rates when the concentration of one drug is x and that of the other is y. Following this definition, positive values of S corresponds to synergistic interactions and negative values to antagonistic ones. The solid line is the best fit to all 116 data points from all of the drug pairs.
Discussion
Our simple geometric model provides some insight into how synergistic drug interactions may lead to faster adaptation (Fig. 1). It should be noted that the same model also indicates the influence of another factor on the rate of adaptation: the availability of mutations that confer simultaneous resistance to both drugs (27, 28) (Fig. S1). It is certainly possible that the level of pleiotropy in mutational effects contributes to the observed differences in the rates of adaptation between drug pairs in our experiments. Indeed, drug pairs with similar mechanisms of action, such as ERY and DOX, could have increased pleiotropy. Conversely, pleiotropy could inhibit evolution if mutation conferring resistance to one drug reduces resistance to another. Further, the rates of mutation may itself vary with some of the drugs, especially with DNA synthesis inhibitors such as CIP. The contribution of these additional factors to the rate of adaptation and their possible correlation with drug synergy is, however, beyond the scope of this work.
We have shown that the rate at which laboratory populations of bacteria adapt to multidrug treatments is correlated with the degree of synergy between the drugs. This correlation implies that antagonistic drug combinations, although typically avoided in clinical settings, may be more effective than synergistic combinations in forestalling the development of antibiotic resistance. Antagonism between drugs may generate sign epistasis between single-resistant mutations, thereby limiting the mutational paths leading to adaptation (29). Further research is needed, however, to see how these results relate to clinical measurements of success in multidrug chemotherapies, whether with bacteria, viruses, or cancer cells. Success in such treatments is the consequence of a multitude of factors, including pharmacokinetics, heterogeneous drug concentrations, horizontal gene transfer, enhanced bactericidal activity, and microbial species and host interactions. Nevertheless, our findings suggest that the choice of drug combinations may involve a tradeoff between two important factors: immediate inhibition of growth and eventual delay in the evolution of resistance. Synergy, by definition, has the advantage that less drug achieves more inhibition or faster killing of wild-type cells. This enhanced inhibition may be crucial to the efficacy of some drug treatments, especially when it is possible to reach in vivo concentrations so high that resistant mutants cannot survive. On the other hand, when treatment dosages fluctuate close to the MIC, which is where antibiotic resistance is most likely to evolve (30), antagonistic drug pairs may be a more effective antimicrobial weapon in forestalling the emergence of resistance. Our work suggests that antagonistic drug combinations may form an important part of treatment strategies designed to achieve this goal.
Materials and Methods
Media and Strains.
All experiments were conducted in M9 Minimal Media (Bio101 #3035-012) supplemented with 0.2% glucose and 0.1% Casamino Acids. Drug solutions were made from powder stocks [doxycycline hyclate (Sigma D-9891); ciprofloxacin (Fluka 17850); erythromycin (Fluka 45673); and amikacin disulfate stock (Sigma A-1774)] and diluted in the growth media on a 2-D grid at indicated concentrations in 96-well plates, with 150 μl per well. To ensure constant drug conditions, all plates of a given drug pair were made from a single master deep-well plate and stored at −20°C (no measurable decay of drugs was observed during the experiment). The populations were founded from an individual colony of strains of fluorescently labeled MC4100 E. coli described previously (24). This strain was not preadapted to the growth conditions of these experiments.
Properties of Antibiotics Used.
The four antibiotics used in the experiments, DOX, ERY, CIP, and AMI, represent a range of action mechanisms (17). DOX is a tetracycline antibiotic used to treat both Gram-negative and Gram-positive infections; it inhibits protein synthesis by blocking aminoacyl–tRNA binding at the A-site in the 30S ribosomal subunit (22). ERY is a macrolide that binds the 23S rRNA molecule in the 50S subunit and also inhibits protein synthesis (5). CIP is a fluoroquinolone, a derivative of nalidixic acid, and is a broad-spectrum antibiotic that inhibits the action of DNA gyrase in both Gram-positive and Gram-negative bacteria (5). AMI is an aminoglycoside antibiotic and works by inhibiting protein synthesis (5).
Parallel Evolution.
Cultures were propagated in parallel for >150 generations with daily serial transfers. OD curves were measured in a fully automated robotic system (Staccato Sciclone Cell Station, Caliper LifeSciences), including a microplate shaker (Liconic LPX40), and a plate reader (Victor3, Perkin-Elmer). The equipment was maintained in an environmental room at constant temperature (30°C) and relative humidity (70%). One initial OD measurement [absorbance at 600 nm (OD600)] was taken at the beginning of each day followed by a 5-h gap and then measurements every 24 min. Except for read periods, plates were shaken continuously at ≈1000 rpm. At the end of each day, plates were removed from the robotic systems and cells were transferred from each population to freshly thawed antibiotic gradient plates by using a 96-pin replicator (VP409). Volume transferred by the pins was measured by using flourescein solution and was found to be 0.04 μl per pin, with a variability of 20% between pins. These volumes correspond to an ≈3,300-fold dilution or ≈11.7 generations of binary fission.
Determination of Adaptation Rate.
We estimated the growth rate, r [as well as the three parameters for the baseline, a, yield, a + b, and lag time, ln(c)/r], for each population on each day by the best fit of the function OD(t) = a + b/[1 + cexp(−r·t)] to the OD measurements taken during that day. It is also possible to extract growth rates by fitting a linear function to log-transformed and background-subtracted OD measurements during exponential phase, but for our data this alternative method leads to a larger error than the method that we employ (Fig. S3). Because a spreading mutant population will keep growing until the resources are exhausted (stationary phase) or until the time of dilution at the end of the day, we measure time in terms of hours spent in growth. For each population we keep track of the time (in hours) spent in growth (and not in stationary phase) since the beginning of the experiment. Let t(i) be the time spent in growth at the end of day i of the experiment. Then the growth rate calculated from the OD measurements of day i is interpreted as the growth rate of the population, r(T), at time T = t(i − 1) + [t(i) − t(i − 1)]/2. The measurements of r versus T for all populations are shown in Fig. S2. All of the populations that survived the antibiotic treatment but were initially inhibited exhibited an increase in their growth rate. The growth rate measured in the 1st day of the experiment was defined as the initial growth rate. The final growth rate was calculated as the average growth rate over the last 6 days of the experiment. From these we calculated the total increase in growth rate, Δr, as the difference between calculated final and initial growth rates. We focused on all populations for which Δr > 0.2 1/h, which is above the estimated error in growth rate measurement (this filter was applied for calculating correlations but not for plotting the data shown in Fig. 3). The time of adaptation for each of these populations, tadapt (Fig. 2B), is defined as the interpolated time at which the growth-rate improvement of the population reached half its maximum value, and the rate of adaptation is defined as α ≡ (Δr/2)/tadapt. Defining tadapt as the time to traverse a different fraction of the fitness increase, for example ¼ or ¾ rather than ½, leads only to small changes in the correlations and partial correlations between synergy and the rate of adaptation and does not change their statistical significance. The results are also essentially unchanged if one employs a different definition of the rate of adaptation, namely α ≡ 1/tadapt, a definition that focuses only on adaptation times.
Estimation of Measurement Errors in Growth Rates and Rates of Adaptation.
We used replicate measurements to assess the error in the measurement of growth rates. On the 1st day of the evolutionary experiment, when all bacterial populations were primarily composed of the ancestral strain, we compared the growth rates for pairs of wells that contained exactly the same drug concentrations. These wells were: four drug-free wells, one from each plate; three columns containing identical gradients of concentrations of DOX alone from the AMI-DOX, ERY-DOX, and CIP-DOX plates; two rows containing only CIP from the CIP-AMI and CIP-DOX plates; and wells in the column containing AMI alone in the CIP-AMI plate that correspond to wells with the same concentrations of AMI in the AMI-DOX plate. The error was inferred from the standard deviation of the differences between replicates and was found to be ≈0.06/h (Fig. S3).
We then used the estimated error in growth rate measurements to assess their contribution to uncertainty in the rates of adaptation. For each growth-rates trajectory, 100 randomly modified trajectories were generated by adding to each daily measured growth rate an independent sample from a normal distribution with a mean of zero and a standard deviation of 0.06/h. The rate of adaptation was then calculated separately from each of these trajectories, providing a distribution of values typically around the measured rate of adaptation, α. The standard deviation of this distribution is interpreted as the error in α that is caused by the error in measuring growth rates. We found the error in α to be roughly proportional to α, corresponding to an average multiplicative error of 25%.
To estimate the role of inherent biological variability in the adaptive process between wells, we compared rates of adaptation between identical wells (the subset of the replicate well-pairs listed above for which a rate of adaptation was determined). In some cases, differences in rates of adaptation between replicate experiments were observed that were significantly larger than the measurement error, indicating the presence of underlying mutational events that led to markedly different adaptive trajectories (Fig. S4).
Definition of Degree of Synergy.
Antibiotic interactions–synergy or antagonism—are defined as deviations from neutral or additive expectations (8). We define neutrality by using Bliss independence (9), where the expected fitness of a bacterium in the presence of two antibiotics is the product of its fitness in the presence of each of the antibiotics separately:
where fxy/f00 is the fitness of the bacteria in the two-drug environment relative to its drug-free fitness. Note that when two drugs show Bliss independence, the fitness advantage of a single-drug resistance mutant does not depend on the presence or absence of the other drug. Synergy is defined as the difference between the neutral expectation and the actual measured value of the bacterial fitness under antibiotic combinations:
The value of S is calculated based on inhibition of the wild-type in the single and double perturbed environments, as measured by the growth rates in the 1st day of the experiment. Synergy is not defined when the growth rate is zero in the combined drug environment and in at least one of the single drug environments.
Supplementary Material
Acknowledgments.
We thank E. Angelino, T. Bollenbach, R. Chait, A. DeLuna, M. Ernebjerg, M. Kirschner, M. Lipsitch, C. Marx, J. Michel, R. Milo, M. Ruvolo, R. Moellering, K. Vestigian, D. Weinreich, and P. Yeh for helpful discussions and comments on the manuscript. This work was supported in part by National Institutes of Health Grant R01GM081617 (to R.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805965105/DCSupplemental.
References
- 1.Daniels M, Hill AB. Chemotherapy of pulmonary tuberculosis in young adults - An analysis of the combined results of 3 Medical Research Council trials. Brit Med J. 1952;1:1162–1168. doi: 10.1136/bmj.1.4769.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klastersky J, Cappel R, Daneau D. Clinical significance of in-vitro synergism between antibiotics in gram-negative infections. Antimicrob Agents Chemother. 1972;2:470–475. doi: 10.1128/aac.2.6.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies J, Wright GD. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997;5:234–240. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 4.Smith RD, Coast J. Antimicrobial resistance: A global response. Bull World Health Organ. 2002;80:126–133. [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh C. Antibiotics: Actions, Origins, Resistance. Washington, DC: American Society for Microbiology Press; 2003. [Google Scholar]
- 6.Levy SB, Marshall B. Antibacterial resistance worldwide: Causes, challenges and responses. Nature Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 7.Norrby SR, Nord CE, Finch R. Lack of development of new antimicrobial drugs: A potential serious threat to public health. Lancet Infect Dis. 2005;5:115–119. doi: 10.1016/S1473-3099(05)01283-1. [DOI] [PubMed] [Google Scholar]
- 8.Berenbaum MC. What is synergy. Pharmacol Rev. 1989;41:93–141. [PubMed] [Google Scholar]
- 9.Greco WR, Bravo G, Parsons JC. The search for synergy - A critical-review from a response-surface perspective. Pharmacol Rev. 1995;47:331–385. [PubMed] [Google Scholar]
- 10.Bliziotis IA, Samonis G, Vardakas KZ, Chrysanthopoulou S, Falagas ME. Effect of aminoglycoside and beta-lactam combination therapy versus beta-lactam monotherapy on the emergence of antimicrobial resistance: A meta-analysis of randomized, controlled trials. Clin Infect Dis. 2005;41:149–158. doi: 10.1086/430912. [DOI] [PubMed] [Google Scholar]
- 11.Sweany HC, Dunbar FP, Wood E. The problem of bacterial resistance in antimicrobial therapy of tuberculosis. Dis Chest. 1955;28:260–274. doi: 10.1378/chest.28.3.260. [DOI] [PubMed] [Google Scholar]
- 12.Eggimann P, Revelly JP. Should antibiotic combinations be used to treat ventilator-associated pneumonia? Semin Respir Crit Care Med. 2006;27:68–81. doi: 10.1055/s-2006-933675. [DOI] [PubMed] [Google Scholar]
- 13.Pillai SK, Moellering RC, Eliopoulos GM. In: Antibiotics in Laboratory Medicine. Lorian V, editor. Philadelphia: Lippincott Williams & Wilkins; 2005. pp. 365–440. [Google Scholar]
- 14.Daschner FD. Combination of bacteriostatic and bactericidal drugs - Lack of significant invitro antagonism between penicillin, cephalothin, and rolitetracycline. Antimicrob Agents Chemother. 1976;10:802–808. doi: 10.1128/aac.10.5.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein M, Schorr SE. The role of bacterial resistance in antibiotic synergism and antagonism. J Bacteriol. 1953;65:454–465. doi: 10.1128/jb.65.4.454-465.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipsitch M, Levin BR. The population dynamics of antimicrobial chemotherapy. Antimicrob Agents Chemother. 1997;41:363–373. doi: 10.1128/aac.41.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chait R, Craney A, Kishony R. Antibiotic interactions that select against resistance. Nature. 2007;446:668–671. doi: 10.1038/nature05685. [DOI] [PubMed] [Google Scholar]
- 18.Jawetz E. Infectious diseases - Problems of antimicrobial therapy. Annu Rev Med. 1954;5:1–26. doi: 10.1146/annurev.me.05.020154.000245. [DOI] [PubMed] [Google Scholar]
- 19.Gunnison JB, Shevky MC, Bruff JA, Coleman VR, Jawetz E. Studies on antibiotic synergism and antagonism - The effect invitro of combinations of antibiotics on bacteria of varying resistance to single antibiotics. J Bacteriol. 1953;66:150–158. doi: 10.1128/jb.66.2.150-158.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez-Martinez JM, et al. Mutant prevention concentrations of fluoroquinolones for Enterobacteriaceae expressing the plasmid-carried quinolone resistance determinant qnrA1. Antimicrob Agents Chemother. 2007;51:2236–2239. doi: 10.1128/AAC.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeh P, Tschumi AI, Kishony R. Functional classification of drugs by properties of their pairwise interactions. Nat Genet. 2006;38:489–494. doi: 10.1038/ng1755. [DOI] [PubMed] [Google Scholar]
- 22.Chopra I, Roberts M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elena SF, Lenski RE. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 24.Hegreness M, Shoresh N, Hartl D, Kishony R. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science. 2006;311:1615–1617. doi: 10.1126/science.1122469. [DOI] [PubMed] [Google Scholar]
- 25.DeLuna A, et al. Exposing the fitness contribution of duplicated genes. Nat Genet. 2008;40:676–681. doi: 10.1038/ng.123. [DOI] [PubMed] [Google Scholar]
- 26.Berenbaum MC, Yu VL, Felegie TP. Synergy with double and triple antibiotic combinations compared. J Antimicrob Chemother. 1983;12:555–563. doi: 10.1093/jac/12.6.555. [DOI] [PubMed] [Google Scholar]
- 27.Chao L. Unusual interaction between target of nalidixic-acid and novobiocin. Nature. 1978;271:385–386. doi: 10.1038/271385a0. [DOI] [PubMed] [Google Scholar]
- 28.Szybalski W. Genetic studies on microbial cross resistance to toxic agents. 4. Cross resistance of bacillus-megaterium to 44 antimicrobial drugs. Appl Microbiol. 1954;2:57–63. doi: 10.1128/am.2.2.57-63.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 30.Drlica K. The mutant selection window and antimicrobial resistance. J Antimicrob Chemother. 2003;52:11–17. doi: 10.1093/jac/dkg269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.