Abstract
Aire promotes T cell tolerance by inducing the expression of a broad swath of genes encoding peripheral tissue antigens (PTAs) in medullary epithelial cells (MECs) of the thymus. The exact mechanism used in inducing this ectopic transcription remains obscure. To address this issue, we generated transgenic mice expressing Aire in pancreatic islet β cells. Gene-expression profiling of such islets revealed that Aire can have a significant impact on transcription in these cells, mainly inducing, but also repressing, transcript levels in a manner comparable with its influence on MECs. The exact transcripts affected differed in MECs and β cells, with limited overlap between the two sets of Aire-modulated genes. We propose that Aire promotes ectopic gene expression by a generic mechanism that does not depend on any particular characteristics or transcription mechanisms operating in MECs, whereas the cellular environment does govern which genes are actually susceptible to Aire regulation.
Keywords: autoimmunity, tissue
The first checkpoint for T cell tolerance is enforced in the thymus. After positive selection in the thymic cortex, negative selection purges the T cell repertoire of self-reactive clones, through clonal deletion, inactivation, or deviation. Negative selection can occur in response to self-antigens presented in either the thymic cortex or medulla. The scope of self-antigens presented for negative selection is increased beyond those expressed ubiquitously within circulating hematopoietic cells or thymic stromal cells by ectopic expression in the thymic epithelium of a large number of genes, whose expression is normally restricted to differentiated tissues (peripheral tissue antigens [PTAs]) (1). Important insights into this process have been provided by the autoimmune pathology presented by patients, and later by knockout mice, defective in expression or function of the Aire protein (2–5). Aire was found to be responsible, at least in part, for ectopic PTA expression (4). In addition, Aire was shown to enhance the presentation of ectopically expressed antigens (6) and to promote the death of terminally differentiated medullary epithelial cells (MECs) that express it (7), a phenomenon that may promote cross-presentation (8). Thus, the absence of Aire creates a defect in the pool of tolerizing self-peptides and their presentation, thereby promoting the development of multiorgan autoimmunity.
The molecular mechanism by which Aire induces ectopic transcripts in the thymus remains unclear. Aire contains several domains typical of transcription factors (9, 10). Its SAND domain is shared with Sp100 and a few other transcription factors and is known to be important in the interaction of some of these with DNA, even though the critical DNA-binding residues are absent from Aire. Aire also contains two PHD domains, analogous to those found in transcription and replication factors, which may be responsible for interactions with nucleosomal histones (ref. 11; A. Koh, C.B., and D.M., unpublished data). The presence of these structural elements supports the notion that Aire functions as a transcriptional regulator, but it is unclear whether it acts as a classic transcription factor, binding directly to specific sites in promoter/enhancer elements, or whether it may have more generic effects, modifying transcription on a larger scale (e.g., through long-range chromatin remodeling). The observation that Aire influences the expression of thousands of genes (E.S. Venanzi, R. Melamed, D.M., and C.B., unpublished data) seems more compatible with the second notion, as does the genomic clustering of ectopic expression and Aire-regulated transcription (12, 13).
Within immune system cells, Aire appears to be mostly active in thymic MECs; the low levels found in DCs seem to have no transcriptional (E.S. Venanzi, C.B., and D.M., unpublished data) or functional (4) impact. Aire+ lymph node stromal cells have been reported to express PTAs (14). To address the specificity of Aire's transcriptional impact, we evaluated the consequences of forcing Aire expression in an unrelated epithelial cell: the pancreatic islet β cell. Rather than perform simple transfection into cultured cells, we preferred to drive Aire expression in a normal differentiated epithelium through a transgenic system to recapitulate normal organ development and cell differentiation, thereby providing the best comparison with the natural expression of Aire in thymic epithelium. Should Aire act by specifically transactivating a set of loci, then we would expect to see expression of a largely overlapping set of genes induced in MECs and β cells. However, should Aire operate through more generic transcriptional reprogramming, we would expect its action to be more dependent on the underlying transcriptional program of the cells, and thus have different consequences in the two cellular environments.
With our in vivo system, we found that Aire has a general mechanism of action that can be “transplanted” to tissues other than MECs, but that the cellular environment dictates which genes are most susceptible to Aire's transcriptional promotion.
Results
Generation and Characterization of Rat Insulin Promoter–Aire Transgenic Mice.
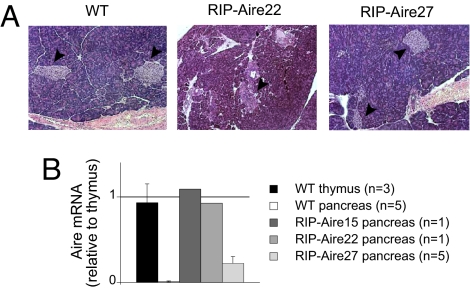
To determine whether Aire's effect could be mirrored in other tissues, we generated mice that expressed Aire at an ectopic site (pancreatic islet β cells). A construct carrying a murine Aire cDNA under the control of the Rat Insulin Promoter (RIP) was microinjected into mouse embryos (of NOD genetic background). Of 30 pups born, 6 had integrated the transgene, judging from polymerase chain reaction (PCR) analysis of tail DNA, and 3 had transmitted the transgene to progeny. Of the three lines derived from these founders, two (RIP-Aire 15 and 22) were afflicted with diabetes at a very early age and were rapidly lost, and so only limited analyses could be performed on these. The third line, RIP-Aire27, did not present with rapid diabetes. Histological analysis was performed to assess islet integrity. The islets of a recently diabetic 4-week-old RIP-Aire22 mouse were considerably smaller than those of the controls, but appeared to be devoid of any immune or inflammatory infiltrate (Fig. 1 A), suggesting that the mechanism mediating early islet loss was not autoimmune, but most likely resulted from direct toxicity of the transgene. However, islets from the young (4-week-old) RIP-Aire27 mice analyzed appeared to be of normal size and distribution (Fig. 1 A), and stained positively for insulin (data not shown). These animals eventually developed the islet infiltration and late diabetes (12–20 weeks) expected on the NOD background, indicating that these β cells have a reasonably normal physiology, including the production of immunogenic autoantigens.
Fig. 1.
Characterization of RIP-Aire tg mice. (A) Histological analysis of pancreatic islets. Hematoxylin and eosin staining of 4 week-old pancreatic sections, 10× objective. Arrowheads point to individual islets. The pancreas from the RIP-Aire 22 mouse was fixed in Bouin's solution, resulting in darker staining. (B) RT-PCR analysis of Aire expression in whole pancreas from RIP-Aire mice (whole thymus RNA from a non-tg mouse as a reference). All values are mean ± SD when applicable.
To verify Aire expression in the islets of transgenic (tg) mice, we isolated pancreas RNA from young RIP-Aire animals (age 4 weeks) for quantitative real-time PCR (RT-PCR) analysis. RNA from whole thymus was used for comparison. Aire transcripts were detected in mice from all three lines, at a level comparable to that in the thymus in the two mice afflicted with early diabetes and at ∼20% of that in RIP-Aire27 mice (Fig. 1 B). Because the proportion of Aire+ MEC cells in thymus cells is comparable with that of β cells in the pancreas (∼ 2–3%), these data indicate that β cells in lines 15 and 22 expressed Aire at levels similar to those of normal MECs, and β cells in line 27 expressed Aire at 20% of the level of normal MECs. These two findings suggest that Aire is not well tolerated by pancreatic β cells; nonetheless, RIP-Aire27 mice have detectable Aire expression that is well tolerated in the first weeks of age.
Aire Has a Transcriptional Effect in Islet β Cells.
The effect of Aire expression in β cells was assayed by gene expression profiling using DNA microarrays. Mice of the RIP-Aire27 line, which ostensibly had normal β cell function, were analyzed. Three pairs of 3-week-old wild-type (WT) and RIP-Aire27 littermates were compared (in triplicate, one array per mouse). Between 200 and 400 islets were purified from collagenase digests of each pancreas, total RNA was extracted, and the isolated RNA was amplified and biotinylated for hybridization to Affymetrix M430.v2 arrays that encompass the majority of the mouse genome, allowing a broad examination of transcriptional changes. Raw data were preprocessed for analysis using the RMA algorithm (15) in the GenePattern suite (16). The analyses reported below were performed on the expression values averaged between the three triplicates, primarily within GenePattern (Multiplot and Changes modules). Full datasets are deposited in GenBank under accession no. GSE12073.
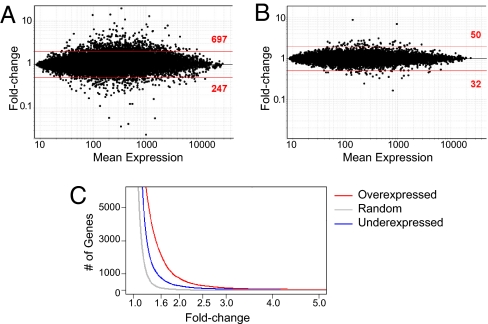
Ectopic expression of Aire had a clear transcriptional effect in β cells. Transcripts were induced or repressed, as seen on the fold-change (FC) versus mean expression plot for RIP-Aire27 relative to nontransgenic (non-tg) islets (Fig. 2 A). At an arbitrary FC threshold of 2, 697 genes were induced and 247 were repressed, a significantly different distribution than that seen in a randomized control dataset (Fig. 2 B); only 50 and 32 probes were induced or repressed at the same cutoff (P < 10−15). These effects covered a wide range of intensities (up to 20-fold; Fig. 2 A), with Aire inducing approximately three times as many genes as it repressed throughout the range of ratios (Fig. 2 C). This predominantly activating effect is similar to what was previously reported for Aire activity in MECs (4, 13), as is the overall size of the effect; a comparison of NOD MECs from Aire knockout (KO) and WT mice on the same M430v2 arrays found that 954 probes were activated and 470 probes were repressed at the same FC threshold of 2 (E.S. Venanzi, R. Melamed, D.M., and C.B., unpublished data).
Fig. 2.
Aire induces and represses transcripts in islet β-cells. (A) Scatterplot of Affymetrix M430 2.0 chip probes comparing averaged expression values in islets from RIP-Aire27 tg versus non-tg littermates (x axis, mean expression; y axis, tg vs. non-tg ratio). (B) Same plot as in A, drawn from randomized datasets. (C) Prevalence of islet Aire-activated (red) and repressed (blue) genes over a range of FC variation. Random genes are shown in gray.
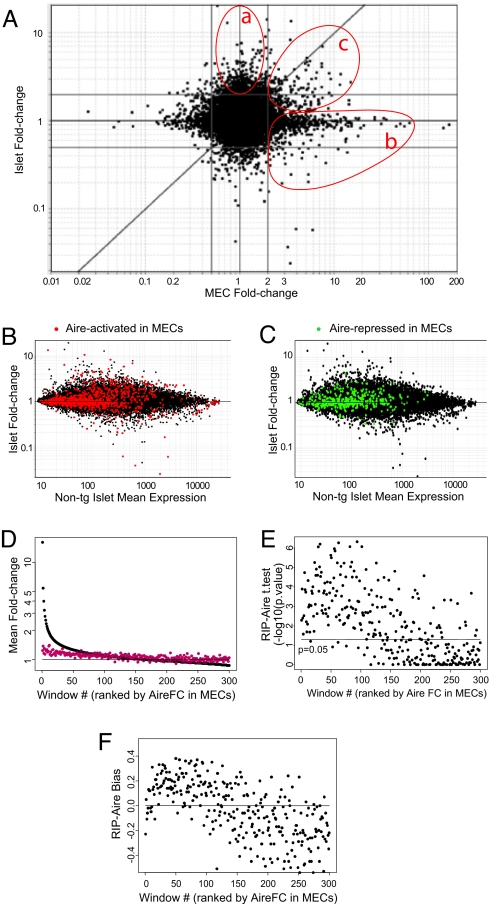
We next explored whether the same transcripts were regulated by Aire in MECs and islets. All comparisons (RIP-Aire27 vs. non-tg islets; Aire WT vs. KO MECs) were performed between tissues from NOD background mice, to eliminate the possibility of strain-specific effects on gene expression profiles. A FC versus FC dot plot illustrates whether a perturbant (here Aire) affects the same genes under two conditions (here MECs vs. islets). In such plots, a distribution of genes along the diagonal x = y line denotes genes similarly affected in the two cell types, whereas changes unique to one or the other condition are reflected by dots lining up along the horizontal or vertical midlines. Fig. 3 A compares the changes induced by Aire in RIP-Aire27 versus non-tg islets with those induced in MECs of WT versus KO mice. The vast majority of Aire's effects were detected in only one of the two cell types (islets or MECs; Fig. 3 A, gate a or b, respectively), as was particularly clear for the transcripts differentially expressed in MECs. The top right quadrant of the plot shows a minor, but clear group of transcripts activated by Aire in both islets and MECs [Fig. 3 A, gate c; genes are listed in supporting information (SI) Table S1]. The differential expression of genes also could be reproduced by RT-PCR analysis, confirming the microarray results (data not shown). These shared changes also were visible on the FC versus mean expression plots, in which the probes highlighted in red (Fig. 3 B) and green (Fig. 3 C) correspond, respectively, to transcripts induced and repressed by Aire in MECs. From these plots, we estimate that only ≈10% of the genes induced in MECs also were induced in β cells. Most were positioned below the x = y reference line, indicating that these common effects were stronger in MECs than in islets, quite possibly due to the lower level of Aire expression in RIP-Aire27 islets than in MECs. Paradoxically, some transcripts induced by Aire in MECs were quite strongly repressed in β cells; however, very few transcripts were corepressed in the two tissues. (Fig. 3 A and C; core pressed genes are listed in Table S2.)
Fig. 3.
Aire induces both a known and a novel Aire signature in islets. (A) Scatterplot of a FC induced by Aire in islets (y axis) and MECs (x axis). Islet-specific and MEC-specific Aire-regulated genes are shown on gates a and b, respectively; commonly up-regulated or down-regulated genes are shown on gate c. The gray lines represent a FC of 2 or 0.5. (B) MEC Aire-signature (FC >2) highlighted in red over the FC versus mean expression value plot in islets. (C) MEC Aire-signature (FC <2) highlighted in green over the FC versus mean expression value plot of islets. (D) Mean FC distribution in RIP-Aire (red) and MEC (black) over 300 windows ranked by Aire MEC FC. The horizontal line represents the P = 0.05 significance threshold. (E) Significance of the bias represented in D and F, assessed by t test P value. (F) Bias of the FC distribution over the gene windows defined in D.
To more rigorously estimate the extent of the overlap between Aire-induced genes in MECs and β cells, we performed the scanning-window analysis depicted in Fig. 3 D–F. Transcripts were ranked according to the FC in MECs and analyzed as windows of 100 transcripts along this ranked distribution, evaluating the mean FC and the proportion of transcripts higher in RIP-Aire tg than in non-tg littermates. Genes in these windows with a strong Aire-related FC in MECs exhibited a mean FC >1 in RIP-Aire27 islets (Fig. 3 D); this shift was statistically significant, as determined by one-sample t tests, with P values in the 10−2–10−6 range over the first 100 windows but dropping beyond that (Fig. 3 E). Such a widespread effect of Aire in microarray datasets is compatible with its broad range of activity, reported elsewhere (E.S. Venanzi, R. Melamed, D.M., and C.B., unpublished data). We then calculated the “bias” in each of these windows, that is, the corrected proportion of transcripts with an FC >1. Under the null hypothesis, the random distribution of FCs in the RIP-Aire27 versus non-tg comparison would lead to an equal distribution of FC >1 or <1. In keeping with the distribution of FCs, this analysis detected a strong positive bias over the first 100–120 windows (Fig. 3F); integrating over the first 100 windows shows that 16.8% of transcripts promoted by Aire in MECs also were activated in β cells. Overall, then, Aire has comparably scaled effects in the two types of epithelia, but involving largely different sets of genes.
Aire in β Cells Preferentially Promotes Expression of Tissue-Specific Genes.
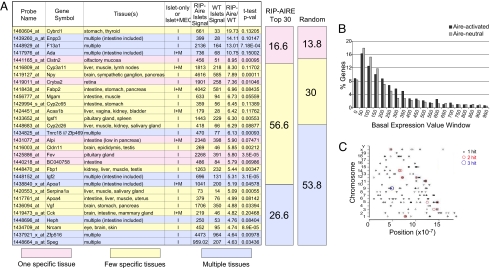
To more closely examine the type of genes activated by Aire in islet β cells of RIP-Aire27 tg mice, we classified the 30 most highly induced genes according to their expression patterns throughout all cells and organs, as defined from the Unigene EST Profile Viewer (National Center for Biotechnology Information) and the SymAtlas database (Genomics Institute at the Novartis Research Foundation; http://symatlas.gnf.org/SymAtlas). Genes were assigned to one of three groups: those with expression restricted to one tissue, those present in a few tissues, or those expressed ubiquitously, as described in Materials and Methods. This partitioning uncovered a pattern biased toward activation of genes with a restricted pattern of expression, with 73.2% of the Aire-induced transcripts corresponding to genes with expression limited to a single or a few specific tissues and 26.6% to ubiquitously expressed genes (Fig. 4 A). The opposite was true for randomly picked genes, with only 43.8% of loci having tissue-specific expression patterns (range, 33%–54% in three independent picks of random genes) and 53.8% being broadly expressed (range, 45%–60%) (Fig. 4 A). This bias was similar to that reported previously for Aire-regulated genes in MEC (4).
Fig. 4.
Islet Aire preferentially activates tissue-restricted genes. (A) The 30 most activated genes in transgenic Aire islets, ranked by FC induction and color-coded (blue, multiple tissues; yellow, few tissues; pink, one specific tissue) according to pattern of tissue expression. The proportion of genes belonging to each tissue expression pattern group for the top-30 genes in A or multiple random sets is represented as a column on the right. (B) Distribution of genes activated by islet Aire or neutral to it, according to their basal WT islet Aire expression. The percents of genes over FC activations of 1.7 (Aire-activated, in dark gray) and 0.75–1.3 (Aire-neutral, in light gray) that fall within each incremental 50 basal expression value windows are plotted. (C) Islet Aire-induced genes distributed randomly throughout the genome. The genomic locations of the 200 most activated genes in Aire transgenic islets are shown for all chromosomes. Each single-gene (black), two-gene (red), and three-gene (blue) hit within a 200-kb window is shown as a colored circle.
There was no particular pattern to the tissues in which the Aire-induced genes were normally expressed, including transcripts unique to the intestine, brain, or liver. In particular, this set of the most Aire-responsive genes did not include β cell–specific transcripts; for instance, Aire did not affect the expression of several β cell transcripts, including insulin II (FC = 1.04), islet amyloid polypeptide (FC = 1.03), and glucose-6-phosphatase (FC = 1.04). Similarly, we might have expected genes expressed in the exocrine pancreatic acini to be preferentially affected by Aire in endocrine β cells, which share a developmental origin. This was not the case, however; none of the genes listed in Fig. 4 A were particularly abundant in the exocrine pancreas.
The enrichment of tissue-specific transcripts within those genes most strongly up-regulated by Aire could be a consequence of preferential effects on genes silent in β cells. To address this point, we calculated the proportion of transcripts that fell in discrete ranges of expression in the β cells of normal mice. As illustrated in Fig. 4 B, no overrepresentation of Aire-responsive genes was observed for loci with low (<100) basal islet expression. Thus, Aire is not particularly active on genes silent in β cells.
A previously described feature of Aire-regulated genes is their random distribution throughout all chromosomes in the genome, with local clustering (13). The distribution of the 200 most Aire-activated genes in RIP-Aire27 β cells was widespread along the genome (Fig. 4 C), with no apparent preference for any chromosomes. Several two- and three-gene clusters of Aire-regulated genes were observed (Fig. 4 C). These gene clusters were more common over the distribution of the entire dataset than would be expected by chance (P < .01; Fisher's exact test).
Discussion
We have addressed the specificity of Aire's transcriptional promotion activity by ectopically expressing it in pancreatic islet β cells. Our data are consistent with the concept that Aire uses similar transcriptional regulatory mechanisms in diverse tissues, but with rather different outcomes.
Aire has activities beyond promoting ectopic gene expression; in particular, it induces cell death. Its expression has been associated with apoptosis of MECs, and with reduced survival of cells upon transfection in vitro (7). The phenotype of the two RIP-Aire lines with high transgene expression, with mice developing diabetes very early in life due to β cell loss in the absence of autoimmune infiltrate, is consistent with such a function. Whether cell death results directly from the induction of proapoptotic factors by Aire or indirectly through nonspecific cell stress is unclear at this time. Pancreatic β cells are very sensitive to ER stress, as illustrated by the development of diabetes in Akita mice due to improper protein folding in the ER (17). There is also a long history of highly (and even moderately) expressed transgenes provoking nonimmunologic death preferentially in the pancreas (18). Consequently, it is highly likely that increased Aire-mediated transcription and/or ER overloading in RIP-Aire15 and RIP-Aire 22 islets provoked β cell death.
Aire was seen to have a predominantly activating transcriptional effect in β cells, with only one-third of the total number of Aire-regulated genes repressed. This distribution mirrors that elicited by Aire in MECs, and the number of Aire-regulated genes was comparable in MECs and β cells. Also as in MECs, genes activated by Aire in β cells included an overrepresentation of tissue-restricted transcripts. These loci were interspersed throughout the genome, albeit with some degree of local clustering. These characteristics suggest that Aire's mechanism likely is not unique to thymic epithelial cells and does not depend on cofactors present only in those cells, but can be “transplanted” to other tissues, possibly harnessing ubiquitous elements of the transcriptional machinery.
In contrasting to this overall similarity, the actual transcripts that were induced by Aire in β cells were mostly quite different from those up-regulated in MECs. It is theoretically possible that the lower Aire expression in RIP-Aire27 islets compared with MECs could affect the transcriptional profile. Although this could explain why only a small fraction of those genes activated in MECs are similarly induced in islets, the induction of novel targets seems more difficult to reconcile with reduced Aire levels. More directly, gene expression profiles established in conditions in which Aire is less active in MECs, such as Aire+/0 MECs (19, 20) or tetracycline-controlled transgenic expression (M.G., unpublished data), showed the same footprint activity (albeit at a lower level), but no expression of novel genes. It is interesting to note that the pattern of comparable overall impact but different induced genes was also observed in a short-term transfection sytem in cell lines in culture that achieved high Aire expression (J. Abramson, A. Koh, D.M., and C.B., unpublished data), suggesting that factors other than reduced Aire expression play a role in differential Aire targeting. Consequently, we favor the concept that the cellular environment causes these differences.
Overall, our findings indicate that Aire does not behave as a conventional transcription factor that recognizes (alone or in combination with other transcription control elements) particular DNA motifs in enhancer and promoter elements of a well defined operon, a situation that would promote constant effects in different cell types. Rather, the set of genes open to activation by Aire seems to be established as a function of the existing transcriptional program of the cell. This set appears not to be a simple reflection of the baseline transcriptional status, because Aire up-regulates genes that span the whole spectrum of expression levels. A scenario can be imagined in which one (or more) of the various elements that determine the cell's transcriptional program also condition the transcripts or regions of the genome that can be accessible to Aire's action. Tissue-specific supraarchitecture of the chromatin, epigenetic modifications, transcription factor combinations, or regulatory microRNAs all could flag the genomic loci that are susceptible to Aire regulation. Interestingly, the transcription factor STAT3 has similarly been reported to target different genes in different cell contexts (21), whereas FoxN1 interactions with epigenetic signatures are known to result in lineage-specific transcriptional programs (22). The PHD1 domain in Aire has been shown to bind unmethylated histone tails (ref. 11; A. Koh, C.B., and D.M., unpublished data). Because the pattern of histone methylation varies between states, the present results possibly could be explained by Aire's preferential interaction with chromatin regions rich in unmethylated H3. Similarly, the notion that Aire influences transcriptional elongation by releasing the brake on transcription initiation and allowing elongation to proceed (23) would fit with our observations.
In conclusion, our results demonstrate that Aire can induce transcriptional regulation in tissues other than MEC. We envision that this protein harnesses ubiquitous transcriptional regulation machinery while being guided by the cell-specific makeup to produce ectopic gene expression. The future identification of these regulatory elements and how they intersect with Aire will provide further insight into Aire's mechanism of action.
Materials and Methods
Plasmids and Transgenic Mice.
The RIP-Aire construct was obtained by substituting the rtTA tetracycline transactivator portion of the RIP-rtTA plasmid (Luis Vence, C.B., and D.M., unpublished data) with mouse Aire cDNA (positions 36–1879 of NM_009646.1). The construct contains the promoter region of the RIP gene (positions 242–946 of J00748), with an intron and polyadenylation site from the rabbit β-globin gene. The fragment was prepared for transgenesis into NOD embryos by AatII and Eco47III digestion, with the 4.1-kb fragment purified on a 0.7% QA-agarose (Qbiogene) containing 2 μg/ml of crystal violet, visualized under natural light, excised from the gel, and purified using a Qiaquick Gel Extraction kit (Qiagen), according to manufacturer's instructions (elution buffer warmed to 50°C). The purified fragment was dialyzed in two exchanges of sterile microinjection buffer (10 mM Tris·HCl [pH 7.6], 0.1 mM EDTA) on a Slide-A-Lyzer MINI (3,500 molecular weight cutoff) dialysis unit (Pierce). The construct was microinjected in the pronucleus of fertilized NOD eggs, and progeny screened with a PCR assay detecting the transgenic Aire construct (primers: 5′GCAACGTGCTGGTTGTTGTG3′ and 5′TGA CTC CAA GTT GCC ATC TG3′).
RNA Isolation and Microarray Hybridization.
RNA was purified from pancreatic islet (purified by collagenase P digestion, Histopaque-1077 density gradient, and hand picking) or total pancreas or thymus following the LiCl/Urea method (24). For microarray probe preparation, the RNA was amplified in two rounds. A MessageAmp aRNA kit (Ambion) was used for the initial first- and second-strand cDNA synthesis, first-round complementary RNA transcription, and subsequent first- and second-round cDNA synthesis. The second round of complementary RNA transcription was performed using a BioArray High-Yield RNA Transcript Labeling Kit (Enzo Diagnostics) to biotinylate the aRNA. The aRNA was purified with a RNAeasy Kit (Qiagen). Biotinylated aRNA was fragmented in 5× fragmentation buffer (200 mM Tris-acetate [pH 8.1], 500 mM KOAc, 150 mM MgOAc) by heating at 94°C for 35 min, and then hybridized to Affymetrix Mouse Genome M430 2.0 chips (Affymetrix).
Microarray Data Analysis.
Microarray data analysis was performed using the GenePattern 2.0 software package obtained from The Broad Institute. The robust multiarray average algorithm RMA (15) was used to normalize the raw probe-level chip data (.CEL files), using the ExpressionFileCreator module from GenePattern. Normalized data were plotted and further analyzed using the Multiplot module from GenePattern (25).
cDNA Synthesis and RT-PCR.
RNA was treated with DNase (Ambion) for 30 min at 37°C to remove contaminating DNA, and then subjected to random-primed cDNA synthesis (SuperScript II; Invitrogen). cDNA was used as a template for amplification by Taqman RT-PCR (Mx3000P; Stratagene). The sequence of primers and probes are available from the authors on request. Data were visualized and analyzed with the MxPro software (Stratagene), using the standard curve method.
Gene Expression Pattern Assignment.
Genes were classified manually according to the expression data reported by the Unigene EST and SymAtlas (http://symatlas.gnf.org/SymAtlas/) databases. Classification into genes with expression restricted to one tissue, present in a few tissues, or expressed ubiquitously was based on both databases concurring on a gene being expressed in a single tissue, in two to five tissues, or in more than five tissues, respectively. A gene was considered expressed in a tissue when ≥ 100 transcripts per million reported in the Unigene database or expression over the SymAtlas-defined baseline was reported for the SymAtlas database. When the two databases differed, the wider pattern of expression was used.
Supplementary Material
Acknowledgments.
We acknowledge the expert help of John Stockton (for the manipulated NOD mouse core data) and Vaja Tchipashvili (for the mouse islet core data) of the Juvenile Diabetes Research Foundation Center for Immune Tolerance in Diabetes at Harvard University; Dr. Emily Venanzi for the NOD MEC microarray data; and Rachel Melamed, Scott Davis, Vanessa Tran, Kimie Hattori and Ella Hyatt for help with the computational data analysis and mice. This work was supported by National Institutes of Health Grant R01 DK60027 and Young Chair funds (to D.M. and C.B.), and by the Joslin Diabetes Center's National Institute of Diabetes and Digestive and Kidneys Diseases–funded Diabetes and Endocrinology Research Center.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. GSE12073).
This article contains supporting information online at www.pnas.org/cgi/content/full/0806616105/DCSupplemental.
References
- 1.Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- 2.Bjorses P, et al. Gene defect behind APECED: A new clue to autoimmunity. Hum Mol Genet. 1998;7:1547–1553. doi: 10.1093/hmg/7.10.1547. [DOI] [PubMed] [Google Scholar]
- 3.Ramsey C, et al. Aire-deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum Mol Genet. 2002;11:397–409. doi: 10.1093/hmg/11.4.397. [DOI] [PubMed] [Google Scholar]
- 4.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the Aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 5.Kuroda N, et al. Development of autoimmunity against transcriptionally unrepressed target antigen in the thymus of Aire-deficient mice. J Immunol. 2005;174:1862–1870. doi: 10.4049/jimmunol.174.4.1862. [DOI] [PubMed] [Google Scholar]
- 6.Anderson MS, et al. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23:227–239. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Gray DH, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med. 2007;204:2521–2528. doi: 10.1084/jem.20070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallegos AM, Bevan MJ. Central tolerance to tissue-specific antigens mediated by direct and indirect antigen presentation. J Exp Med. 2004;200:1039–1049. doi: 10.1084/jem.20041457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson TJ, Ramu C, Gemund C, Aasland R. The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor [letter] Trends Biochem Sci. 1998;23:242–244. doi: 10.1016/s0968-0004(98)01231-6. [DOI] [PubMed] [Google Scholar]
- 10.Saltis M, et al. Evolutionarily conserved and divergent regions of the autoimmune regulator (Aire) gene: a comparative analysis. Immunogen. 2008;60:105–114. doi: 10.1007/s00251-007-0268-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Org T, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9:370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derbinski J, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnnidis JB, et al. Chromosomal clustering of genes controlled by the aire transcription factor. Proc Natl Acad Sci U S A. 2005;102:7233–7238. doi: 10.1073/pnas.0502670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JW, et al. Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nat Immunol. 2007;8:181–190. doi: 10.1038/ni1427. [DOI] [PubMed] [Google Scholar]
- 15.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 17.Yoshioka M, Kayo T, Ikeda T, Koizumi A. A novel locus, Mody4, distal to D7Mit189 on chromosome 7 determines early-onset NIDDM in nonobese C57BL/6 (Akita) mutant mice. Diabetes. 1997;46:887–894. doi: 10.2337/diab.46.5.887. [DOI] [PubMed] [Google Scholar]
- 18.Sarvetnick N. Transgenic models of diabetes. Curr Opin Immunol. 1989;2:604–606. doi: 10.1016/0952-7915(90)90018-c. [DOI] [PubMed] [Google Scholar]
- 19.Liston A, et al. Gene dosage-limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J Exp Med. 2004;200:1015–1026. doi: 10.1084/jem.20040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su MA, et al. Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J Clin Invest. 2008;118:1712–1726. doi: 10.1172/JCI34523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarez JV, Frank DA. Genome-wide analysis of STAT target genes: elucidating the mechanism of STAT-mediated oncogenesis. Cancer Biol Ther. 2004;3:1045–1050. doi: 10.4161/cbt.3.11.1172. [DOI] [PubMed] [Google Scholar]
- 22.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oven I, et al. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol. 2007;27:8815–8823. doi: 10.1128/MCB.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auffray C, Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- 25.Hyatt G, et al. Gene expression microarrays: Glimpses of the immunological genome. Nat Immunol. 2006;7:686–691. doi: 10.1038/ni0706-686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.