Abstract
Expression of FOXP3, a potent gene-specific transcriptional repressor, in regulatory T cells is required to suppress autoreactive and alloreactive effector T cell function. Recent studies have shown that FOXP3 is an acetylated protein in a large nuclear complex and FOXP3 actively represses transcription by recruiting enzymatic corepressors, including histone modification enzymes. The mechanism by which extracellular stimuli regulate the FOXP3 complex ensemble is currently unknown. Although TGF-β is known to induce murine FOXP3+ Treg cells, TGF-β in combination with IL-6 attenuates the induction of FOXP3 functional activities. Here we show that TCR stimuli and TGF-β signals modulate the disposition of FOXP3 into different subnuclear compartments, leading to enhanced chromatin binding in human CD4+CD25+ regulatory T cells. TGF-β treatment increases the level of acetylated FOXP3 on chromatin and site-specific recruitment of FOXP3 on the human IL-2 promoter. However, the proinflammatory cytokine IL-6 down-regulates FOXP3 binding to chromatin in the presence of TGF-β. Moreover, histone deacetylation inhibitor (HDACi) treatment abrogates the down-regulating effects of IL-6 and TGF-β. These studies indicate that HDACi can enhance regulatory T cell function via promoting FOXP3 binding to chromatin even in a proinflammatory cellular microenvironment. Collectively, our data provide a framework of how different signals affect intranuclear redistribution, posttranslational modifications, and chromatin binding patterns of FOXP3.
Keywords: regulatory T cell, acetylation, TGF-beta signal
FOXP3 mutations in human CD4+ T cells lead to immune dysregulation, polyendocrinopathy, enteropathy, and X-linked autoimmunity (IPEX) syndromes (1–4). Mutations in specific regions of the FOXP3 gene lead to functionally defective proteins unable to inhibit IL-2, IFN-γ, and TNF-α gene expression (1, 2, 5–7). Recent studies from our laboratory have shown that the FOXP3 homoligomer is an integral part of a large ensemble including histone modification enzymes (5, 8). FOXP3 is an acetylated protein, and FOXP3 acetylation is promoted by the histone acetyltransferase TIP60 within the FOXP3 complex (5, 8). FOXP3 exerts a defining role in regulatory T (Treg) cell function and FOXP3 expression levels correlate with the suppressive capability of Treg cells (1, 2, 5, 6). Deacetylase inhibitor treatment promotes FOXP3 acetylation and the generation and function of Treg cells in vivo (9). Treg cells may suppress in both a contact-dependent and -independent manner. Some Treg suppressive functions are modified by the activities of CTLA-4 and by TGF-β, IL-10, and IL-6 signaling pathways (10–16).
However, the mechanism by which the FOXP3 ensemble is regulated by extracellular stimuli is unknown. As a primary antiinflammatory cytokine, TGF-β promotes the differentiation and function of murine Foxp+ Treg cells (17, 18), whereas TGF-β plus IL-6 combined signals promote the induction of RORγ and the differentiation of Th17 cells (19–22). These integrated signals may down-regulate FOXP3 function (19–22). TGF-β-induced FOXP3 may inhibit the differentiation of Th-17 cells by antagonizing the functions of the transcription factor RORγt (23). Although IL-2 is an essential cytokine for the expansion of FOXP3+ Treg cells (24, 25), IL-2 may also antagonize the proinflammatory effects of IL-6 acting in combination with TGF-β (26).
In the present study, we examined how TCR signaling, TGF-β, IL-6, and other exogenous stimuli act on Treg cells to modulate the chromatin binding patterns of FOXP3. TGF-β promotes chromatin binding and promoter occupancy by acetylated FOXP3. Unexpectedly, we observed that IL-6 also enhances the chromatin binding of FOXP3 in the presence of IL-2 signals. This enhanced chromatin binding of FOXP3 is antagonized by histone deacetylation inhibitor (HDACi) treatment. IL-6, together with TGF-β, which is known to provide a set of signals resulting in the generation of proinflammatory Th17 cells, was found to limit but not completely prevent FOXP3 binding to chromatin. The limited binding of FOXP3 to chromatin that occurs under the influence of combined signals of IL-6 and TGF-β can also be reversed by the HDACi sodium butyrate. Our findings suggest that although TGF-β and IL-6 signals affect diverse transcriptional events, these cytokines may also alter FOXP3 function at the posttranslational level by eliciting covalent modifications of FOXP3 and diminishing FOXP3-chromatin binding in human CD4+CD25+ Treg cells.
Results
Exogenous Signaling Altered the Intracellular Distribution Patterns of FOXP3.
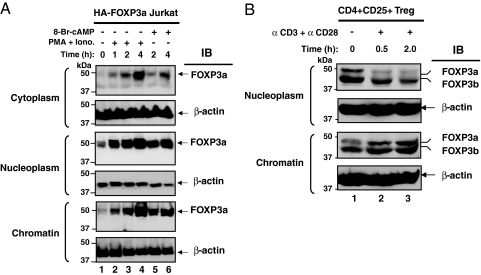
Previous studies identified that FOXP3 is primarily localized in the nuclei of cells, although we and others have noted an additional small cytoplasmic pool of FOXP3 (4, 27). We examined the distribution patterns of FOXP3 in discrete subnuclear compartments after T cell stimulation. A full-length FOXP3 expression construct, pIPHA2FOXP3a, was transiently expressed in Jurkat T cells (5, 8), and cell constituents were separated into nuclear and chromatin fractions based on defined protocols (28, 29). We noted small amounts of full-length FOXP3a in the cytoplasm (Fig. 1A Top) and significantly greater amounts in the nuclear fraction (nucleoplasm) (Fig. 1A Middle) after 1- to 4-h treatments of cells with PMA and ionomycin or with 8-Br-cAMP stimulation. PMA and ionomycin and 8-Br-cAMP trigger potent signaling mechanisms, which we have found to induce significant amounts of FOXP3 within cells.
Fig. 1.
T cell stimulation promotes FOXP3 binding to chromatin. (A) FOXP3 distributes in the cytoplasm (Top), nuclear (Middle), and chromatin (Bottom) fractions after stimulation. Ten million cells transfected with HA-tagged FOXP3a were stimulated with a combination of 50 ng/ml PMA and 1 μM ionomycin or 1 mM 8-bromo-cAMP for the indicated time periods. Cytoplasm, nuclear, and chromatin fractions were prepared from these cells. Equal protein amounts were separated by SDS/PAGE, transferred to nitrocellulose membrane, and immunoblotted with anti-HA-HRP-conjugated antibody, followed by reprobing with anti-β-actin antibody. (B) Stimulation-dependent distribution of FOXP3 in human Treg cells. Ten million human CD4+CD25+ Treg cells were serum starved for 3 h and treated with or without 5 μg of plate-bound anti-CD3 and 2.5 μg of anti-CD28 for the indicated time periods. Equal protein amounts of nuclear and chromatin fractions were separated by SDS/PAGE and immunoblotted with anti-FOXP3 antibody (PCH101, e-bioscience), followed by reprobing with anti-β-actin antibody.
We next examined whether external stimuli can rapidly modify the level of FOXP3 that is bound to chromatin. Chromatin-associated FOXP3 was found to increase over time after PMA-ionomycin treatment and to a lesser extent with 8-Br-cAMP treatment (Fig. 1A Bottom). These results indicate that global T cell stimulation increases the amount of FOXP3 associated with chromatin. Increased amounts of FOXP3 in the chromatin fraction may facilitate its inhibitory role in gene transcription.
To confirm these observations in a physiologically relevant setting, we followed endogenous FOXP3 distribution in human CD4+CD25+ Treg cells. We found that stimulation with anti-CD3 and anti-CD28 antibodies promotes redistribution of FOXP3 from the nuclear fraction (nucleoplasm) (Fig. 1B Upper) to chromatin in a time-dependent manner (Fig. 1B Lower). This data in primary cells supports the conclusion that physiologically relevant T cell receptor signals in human CD4+CD25+ Treg cells can alter the nuclear redistribution of FOXP3 and promote accumulation of FOXP3 on chromatin.
TGF-β Promotes Acetylated FOXP3 Binding to Chromatin.
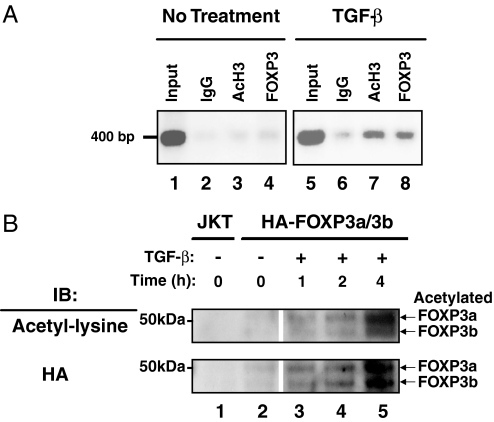
We reported that FOXP3 binds to the IL-2 promoter in in vitro expanded human FOXP3+ T cells (8). Although TGF-β has been implicated in the activation of functional FOXP3-expressing T cells, the molecular process by which this occurs is largely undefined (30). To examine whether TGF-β affects the ability of FOXP3 to bind site-specific chromatin, human CD4+CD25+ Treg cells were incubated in the presence or absence of TGF-β, and FOXP3 binding to the IL-2 promoter was determined by chromatin immunoprecipitation (ChIP) assays. Our data indicate that TGF-β treatment enhances the recruitment of FOXP3 to the IL-2 promoter in human Treg cells (Fig. 2A, compare lanes 4 and 8). TGF-β treatment also increased the acetylation level of histone H3 at the IL-2 promoter (Fig. 2A, compare lanes 3 and 7).
Fig. 2.
TGF-β promotes acetylated FOXP3 binding to chromatin. (A) ChIP was performed by using mIgG, anti-acetyl histone H3 (AcH3), and anti-FOXP3 antibodies with ten million in vitro expanded human CD4+CD25+ Treg cells treated with TGF-β at the indicated time points. The eluted genomic DNA fragment was amplified with hIL-2 promoter-specific primers. (B) Ten million serum-starved human HA-FOXP3a- and -3b-transfected Jurkat T cells were stimulated with or without 1 ng of TGF-β per million cells for indicated time periods. Equal amounts of protein from chromatin fractions were separated by SDS/PAGE, transferred to nitrocellulose, and immunoblotted with anti-acetyl lysine-specific antibody (Ac-K-103, Santa Cruz Biotechnology), followed by reprobing with anti-HA-HRP-conjugated antibody. Acetylated FOXP3a and FOXP3b proteins are indicated.
We also examined whether the binding patterns of FOXP3 to chromatin can be influenced by TGF-β signals. Human CD4+CD25+ Treg cells mainly express two isoforms of FOXP3, namely FOXP3a and FOXP3b (5, 31). We cotransfected Jurkat T cells with HA-tagged FOXP3a and FOXP3b, cultured the transfected cells for 48 hours, and then stimulated the cells with or without TGF-β for the indicated time periods. Equal amounts of total protein from the chromatin fractions were separated and analyzed by Western blotting. We observed a time dependence of TGF-β stimulation that promoted FOXP3 acetylation and chromatin association of FOXP3a and 3b in transfected T cells. As shown in Fig. 2B, significant levels of chromatin-bound and acetylated FOXP3 were apparent between 2 and 4 h after exposure to TGF-β.
TGF-β Treatment Increases the Level of Acetylated FOXP3 Bound to Chromatin.
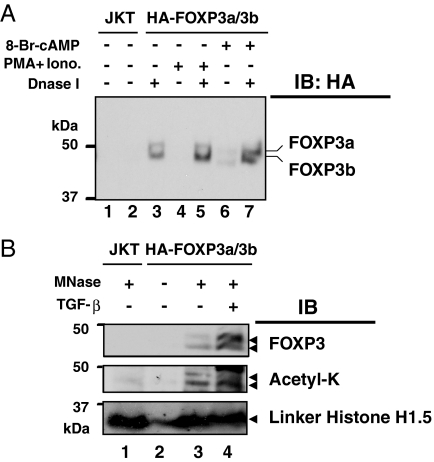
To study the pattern of FOXP3 species bound to chromatin in response to extracellular stimuli, we used DNase I and Micrococcal nuclease (MNase) treatments to digest chromatin completely [supporting information (SI) Fig. S1]. MNase treatment completely digests chromatin in a dose-dependent manner (Fig. S1A) from both control Jurkat T cells and HA-FOXP3a/b-transfected T cells with or without various stimuli (Fig. S1B).
We examined the level of both acetylated and total FOXP3 released from chromatin after DNase I and MNase treatments. DNase I digestion released chromatin-associated FOXP3 (Fig. 3A, compare lanes 2, 4, and 6 with 3, 5, and 7). As expected, PMA plus ionomycin or 8-Br-cAMP stimulation increased the amount of FOXP3 released from the chromatin after DNase I digestion in HA-FOXP3a/b-transfected Jurkat T cells (Fig. 3A, lanes 3, 5, and 7). MNase treatment released FOXP3 from the chromatin-bound fraction (Fig. 3B Top, lane 3), and this treatment released more FOXP3 from the chromatin-bound fraction of TGF-β stimulated cells compared with unstimulated cells (Fig. 3B Top, lanes 3 and 4). Nontransfected Jurkat T cells served as a negative control (Fig. 3B Top, lane 1), and in the absence of MNase, FOXP3 was not released from chromatin (Fig. 3B Top, lane 2). Posttranslational modifications of bound FOXP3 species from the released chromatin bound FOXP3 after MNase treatment were studied and were found to be acetylated (Fig. 3B Middle, lanes 3 and 4).
Fig. 3.
Nuclease digestions released acetylated FOXP3 from chromatin. (A) Ten million serum-starved transfected or control cells were stimulated with a combination of 50 ng/ml PMA and 1 μM ionomycin or 1 mM 8-bromo-cAMP alone for the indicated time. Isolated chromatin fractions were digested with 20 units of DNase I. The released chromatin-bound proteins in solution were separated by SDS/PAGE, transferred, and immunobloted with anti-HA-HRP conjugated antibody. (B) MNase-mediated release of acetylated FOXP3 from chromatin. Ten million HA-FOXP3a- and -3b-transfected or untransfected Jurkat T cells were stimulated as indicated. The chromatin fractions were digested with 10 units of MNase (Roche), the released chromatin digested material was tested for FOXP3 by immunoblotting with anti-HA-HRP antibody, and its acetylation was detected by immunoblotting with anti-acetyl lysine-specific mAb-Ac-K-103 (Upstate Biotechnology).
Linker histone H1 can serve as a marker of excised chromatin (29, 32). Linker histone H1 proteins released after MNase digestion were readily identified in the chromatin released from FOXP3-transfected Jurkat T cells (Fig. 3B). In untransfected Jurkat cells, MNase treatment released chromatin that contains linker histone H1 but obviously lacked FOXP3 (Fig. 3B, lane 1). TGF-β treatment increased the levels of total (Fig. 3B Top, lanes 3 and 4) and acetylated (Fig. 3B Middle, lanes 3 and 4) FOXP3 species released from chromatin after MNase treatment, whereas the level of control linker histone H1 remained unchanged (Fig. 3B Lower, lanes 3 and 4). Therefore, these studies unambiguously identify that TGF-β treatment increases acetylated FOXP3 forms directly associated with chromatin.
IL-6 and TGF-β in Combination Negatively Regulate FOXP3 Binding to Chromatin.
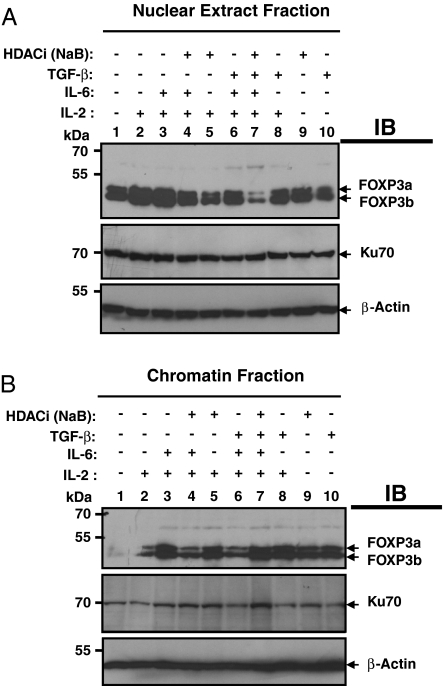
Proinflammatory cytokines such as IL-6 modulate CD4+CD25+ Treg function (33, 34). IL-6 and TGF-β together promote the differentiation of Th-17 cells and suppress the FOXP3 function (19–22). We have identified and subsequently used in our experiments a new CD4+CD25+ T-lymphoid cell line that endogenously expresses both isoforms of human FOXP3. This TCR-expressing, FOXP3+CD4+CD25+, cloned Sezary T cell line (SZ-4), was originally isolated at the University of Pennsylvania by Todd Abrams and colleagues (35, 36). The effect of IL-6 and TGF-β on FOXP3 binding to chromatin was examined. The interaction of FOXP3 with the chromatin fraction in human SZ-4 cells was analyzed after IL-6 stimulation in the presence of high-dose IL-2, and it was found that IL-6 and IL-2 together increased FOXP3 binding to chromatin (Fig. 4B Top, compare lanes 2 and 3). The increased binding to chromatin occurs while the total nuclear pool of FOXP3 protein does not dramatically change (Fig. 4A Top, compare lanes 2 and 3). As expected, we also found that TGF-β alone is sufficient to promote FOXP3 binding to chromatin (Fig. 4B Top, compare lanes 1 and 10). We also studied whether histone deacetylase inhibitors can modify FOXP3 binding to chromatin. NaB treatment alone or in combination with IL-2 promotes FOXP3 binding to chromatin (Fig. 4B Top, compare lanes 1 and 9 with lanes 2 and 5).
Fig. 4.
IL-6 plus TGF-β down-regulates FOXP3 binding to chromatin that could be reversed by HDACi sodium butyrate. Ten million FOXP3+ SZ-4 T cells were stimulated in the presence or absence of HDACi (10 mM sodium butyrate) along with 20 ng/ml IL-6, 5 ng/ml TGF-β, and 50 U/ml IL-2 as indicated for 4 h in 10% FBS containing RPMI-1640 medium supplied with 10 U/ml IL-2. Cells were washed with cold PBS containing all inhibitors and fractionated into cytoplasmic, nuclear, and chromatin fractions. Equal amounts of proteins were separated by 8% SDS/PAGE, then transferred to nitrocellulose membrane. Both the nuclear fraction (A) and chromatin fraction (B) were immunoblotted successively with antibodies against FOXP3 (221D), Ku-70 (Santa Cruz Biotechnology, sc-17789), and β-actin (Sigma).
We examined whether IL-6 signals could modulate human FOXP3 binding to chromatin in human SZ-4 T cells in the presence of TGF-β. The data showed that the combination of TGF-β and IL-6 treatment, but not either TGF-β nor IL-6 alone limits FOXP3 binding to chromatin even in the presence of high doses of IL-2 (Fig. 4B Top, compare lane 6 with lanes 8 or 3). More interestingly, the decreased binding of FOXP3 to chromatin caused by combinations of TGF-β and IL-6 could be completely reversed by the histone deacetylase inhibitor, NaB (Fig. 4B Top, compare lanes 6 and 7). Increased chromatin-bound FOXP3 may result from redistribution from the nuclear fraction pool because less FOXP3 was detected in the nuclear fraction (nucleoplasm) pool (Fig. 4A Top, compare lanes 6 and 7).
Discussion
We have demonstrated that TGF-β treatment increases the amount of acetylated FOXP3 protein binding to active chromatin sites, whereas the combination of TGF-β and IL-6 signals limit FOXP3 binding to these sites in human T cells. Although portions of the total pool of FOXP3 can exist in diverse nuclear sites such as within the nucleoplasm, active and acetylated FOXP3 is preferentially, but not exclusively, bound to chromatin. Histone deactylase (HDAC) inhibitors affect the chromatin binding pattern of FOXP3. TGF-β and IL-6 cotreatment led to less FOXP3 associated with chromatin, whereas HDAC inhibitors reversed this effect.
The increased FOXP3 level found in the nuclear and/or the chromatin fractions noted in the short time intervals of our studies may occur through enhanced translocation or by stabilization of nuclear pools of FOXP3 protein after posttranslational modifications. Perhaps TGF-beta stimulation limits degradation or acts through some unidentified process that increases the half-life of the FOXP3 RNA species.
Other posttranslational modifications clearly occur on FOXP3. For instance, we have found that chromatin-bound FOXP3 can be phosphorylated on threonine residues (Fig. S2). Phosphorylation is known to induce cellular compartment transitions for other transcription factors (37, 38). Although we have noted acetylation of FOXP3, we are aware and show that histone modifications are also dominantly affected by acetylation. Histone deacetylation is known to repress gene transcription by facilitating the formation of more condensed and compact chromatin structures, which are generally inaccessible to transcription factors and transcriptional activators (39, 40).
Our data showed that FOXP3 proteins in primary CD4+CD25+ Treg cells (5) or in FOXP3 transiently transfected T cells become acetylated after the cells were stimulated with a variety of extracellular signals. TGF-β treatment enhanced both the extent of acetylation and FOXP3 chromatin interactions. This enhanced functional activity of FOXP3 may be relevant to its covalent modifications and is reminiscent of other regulatory proteins. Acetylated functional p53 forms also show increased DNA binding (39, 40). Proinflammatory cytokine signals such as the combination of IL-6 and TGF-β negatively regulate FOXP3 binding to chromatin, a process that can be reversed by HDAC inhibitors. Our work also provides a molecular explanation for how HDAC inhibitors may function. They may directly affect FOXP3 function by promoting its chromatin binding (Fig. 4B Top, lane 9), and/or by antagonizing processes induced by proinflammatory cytokines that limit FOXP3 binding to chromatin (Fig. 4B Top, lane 7).
Antibodies specifically recognizing acetylated FOXP3 will be helpful in defined monitoring of the chromatin bound forms of FOXP3 and its relevance to suppressor cell function in vivo. Understanding how FOXP3+ Treg cells respond to and integrate diverse cytokine signals that occur during inflammation (such as IL-6 plus TGF-β) is clearly needed. Furthermore, it is also critical to elucidate how proinflammatory cytokine signals themselves are tightly regulated, such as by cAMP-elevating immunosuppressive G-protein-coupled receptors, which are now firmly implicated as one of the major regulators of immunosuppression (41, 42). Moreover, the fact that HDACis modulate part of these signaling events at the chromatin level may provide insight into how they function to alter immune processes (9).
Materials and Methods
ChIP.
ChIP assays were carried out with 5–10 million Treg cells with or without stimulation by using EZ-ChIP (cat. 17-371, Upstate Biotechnology) according to the manufacturer's instructions. After sonication on ice, the chromatin solution was centrifuged for 10 min at 6,000 × g. mIgG (Upstate Biotechnology), anti-acetyl histone H3 (Upstate Biotechnology), or anti-FOXP3 (e-Bioscience) were used for immunoprecipitation of the supernatant. Human IL-2 promoter primers were used for amplification of a promoter fragment of 419 bp that was further verified by sequencing (8).
Cell Fractionation.
Ten million cells were stimulated with PMA (50 ng/ml) and 1 μM ionomycin combination or 1 ng/ml TGF-β per million cells, or 1 mM 8-bromo cAMP for indicated time periods in the presence of 500 nM TSA, 10 μM MG-132, 10 mM nicotinamide, 10 mM Na-butyrate, and 1 mM Na3VO4. Cells were collected after centrifuging at 300 × g for 5 min and washed with cold PBS containing all of the above inhibitors. Cells were fractionated according to previously established methods (28, 43) with some modifications. In brief, cells were lysed in 10 mM Hepes buffer (pH 7.6), 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, and 0.1% Nonidet P-40 in the presence of all above inhibitors supplemented with 1 mM PMSF and protease inhibitor mixture (Roche) and were incubated on ice for 10 min. Nuclei were isolated by centrifuging at 800 × g for 5 min at 4°C. The supernatant fraction was further clarified by centrifuging at 10,000 × g for 15 min and was called cytoplasmic extract. Isolated nuclei were washed once with cytosolic buffer and lysed in buffer containing 3M EDTA, 0.2 mM EGTA, 1 mM DTT, and all above mentioned protease, phosphatase, and HDAC inhibitors, incubating on ice for 30 min. After centrifugation at 800 × g for 5 min, the supernatant fraction (nuclear fraction) and the insoluble pellet (chromatin fraction) were collected. The chromatin fraction was washed once with nuclear extraction buffer and was finally dispersed and boiled with Laemmli sample lysis buffer.
DNase I Digestion.
Nuclei were resuspended in 40 μl of DNase I buffer with or without 20 units of DNase I and incubated on ice for 30 min with shaking, followed by the addition of 20 mM EDTA. The supernatant was collected by centrifuging at 800 × g for 5 min. 10% of the DNase-I-digested material was directly lysed by boiling with SDS-sample lysis buffer. The lysates were separated in 8% SDS/PAGE, transferred to nitrocellulose membrane, and immunoblotted with anti-HA-HRP conjugated antibody.
MNase Digestion.
The chromatin fraction was washed two times with 500 μl of MNase digestion buffer containing 10 mM Hepes (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 1 mM PMSF, protease inhibitor mixture (Roche), 500 nM TSA, 5 mM Na-butyrate, 5 mM nicotinamide, and 1 mM Na3VO4, by centrifugation at 800 × g for 5 min. The pellet was dispersed in 100 μl of MNase buffer with or without 10 units of MNase (Roche) and was incubated at room temperature for 30 min with shaking, followed by the addition of 1 mM EGTA. After centrifugation at 800 × g for 5 min, the supernatant fraction was analyzed for nuclease-mediated released protein. Equal protein amounts of material were separated by SDS/PAGE, transferred, and immunoblotted with anti-HA-HRP conjugated antibody. In certain experiments as shown in Fig. S1, after nuclease digestion, genomic DNA was isolated after RNase A and proteinase K treatment by using a QIAamp DNA mini kit for genomic DNA purification (Qiagen). Extracted genomic DNA after MNase digestion was quantitated and was separated on a 1.2% agarose gel.
SI Text.
Fig. S1 shows that the chromatin fraction contains DNA which can be digested by MNase treatment. Fig. S2 shows stimulation dependent phosphorylation of FOXP3 in human Treg cells.
Supplementary Material
Acknowledgments.
We thank Dr. Alison H. Banham (John Radcliffe Hospital, Oxford) for providing anti-FOXP3 mAb 221D; Drs. Samik Basu and James L. Riley for providing in vitro expanded CD4+CD25+ Treg cells; and members of the Greene laboratory including Drs. Amy Brown, Kathryn T. Iacono, Gail Massey, Hongtao Zhang, and Zhaocai Zhou for their helpful discussions. A.S. and B.L. are Research Associates in the Department of Pathology and Laboratory Medicine. M.I.G. is the John Eckman Professor of Medical Science at the University of Pennsylvania. This work was supported by Leonard and Madlyn Abramson Family Cancer Research Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806726105/DCSupplemental.
References
- 1.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 2.Sakaguchi S. Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol. 2005;6:345–352. doi: 10.1038/ni1178. [DOI] [PubMed] [Google Scholar]
- 3.Fontenot JD, Rudensky AY. A well adapted regulatory contrivance: Regulatory T cell development and the forkhead family transcription factor Foxp3. Nat Immunol. 2005;6:331–337. doi: 10.1038/ni1179. [DOI] [PubMed] [Google Scholar]
- 4.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 5.Li B, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci USA. 2007;104:4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B, Greene MI. FOXP3 actively represses transcription by recruiting the HAT/HDAC complex. Cell Cycle. 2007;6:1432–1436. [PubMed] [Google Scholar]
- 7.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 8.Li B, et al. FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int Immunol. 2007;19:825–835. doi: 10.1093/intimm/dxm043. [DOI] [PubMed] [Google Scholar]
- 9.Tao R, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 10.Levings MK, et al. Human CD25+CD4+ T suppressor cell clones produce transforming growth factor beta, but not interleukin 10, and are distinct from type 1 T regulatory cells. J Exp Med. 2002;196:1335–1346. doi: 10.1084/jem.20021139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Wahl SM. TGF-beta: The missing link in CD4+CD25+ regulatory T cell-mediated immunosuppression. Cytokine Growth Factor Rev. 2003;14:85–89. doi: 10.1016/s1359-6101(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 12.Read S, et al. Blockade of CTLA-4 on CD4+CD25+ regulatory T cells abrogates their function in vivo. J Immunol. 2006;177:4376–4383. doi: 10.4049/jimmunol.177.7.4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coenen JJ, et al. CTLA-4 engagement and regulatory CD4+CD25+ T cells independently control CD8+-mediated responses under costimulation blockade. J Immunol. 2006;176:5240–5246. doi: 10.4049/jimmunol.176.9.5240. [DOI] [PubMed] [Google Scholar]
- 14.Tang Q, et al. Distinct roles of CTLA-4 and TGF-beta in CD4+CD25+ regulatory T cell function. Eur J Immunol. 2004;34:2996–3005. doi: 10.1002/eji.200425143. [DOI] [PubMed] [Google Scholar]
- 15.Dominitzki S, et al. Cutting Edge: Trans-signaling via the soluble IL-6R abrogates the induction of FoxP3 in naive CD4+CD25 T cells. J Immunol. 2007;179:2041–2045. doi: 10.4049/jimmunol.179.4.2041. [DOI] [PubMed] [Google Scholar]
- 16.Fehervari Z, Sakaguchi S. Control of Foxp3+ CD25+CD4+ regulatory cell activation and function by dendritic cells. Int Immunol. 2004;16:1769–1780. doi: 10.1093/intimm/dxh178. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, et al. A critical function for TGF-beta signaling in the development of natural CD4(+)CD25(+)Foxp3(+) regulatory T cells. Nat Immunol. 2008;9:632–640. doi: 10.1038/ni.1607. [DOI] [PubMed] [Google Scholar]
- 18.Li MO, Flavell RA. Contextual regulation of inflammation: A duet by transforming growth factor-beta and interleukin-10. Immunity. 2008;28:468–476. doi: 10.1016/j.immuni.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 20.Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 21.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Kimura A, Naka T, Kishimoto T. IL-6-dependent and -independent pathways in the development of interleukin 17-producing T helper cells. Proc Natl Acad Sci USA. 2007;104:12099–12104. doi: 10.1073/pnas.0705268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou L, et al. TGF-β-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. Cutting Edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J Immunol. 2007;178:4022–4026. doi: 10.4049/jimmunol.178.7.4022. [DOI] [PubMed] [Google Scholar]
- 25.Zheng SG, Wang J, Wang P, Gray JD, Horwitz DA. IL-2 is essential for TGF-beta to convert naive CD4+CD25- cells to CD25+Foxp3+ regulatory T cells and for expansion of these cells. J Immunol. 2007;178:2018–2027. doi: 10.4049/jimmunol.178.4.2018. [DOI] [PubMed] [Google Scholar]
- 26.Horwitz DA, Zheng SG, Wang J, Gray JD. Critical role of IL-2 and TGF-beta in generation, function and stabilization of Foxp3(+)CD4(+) Treg. Eur J Immunol. 2008;38:912–915. doi: 10.1002/eji.200738109. [DOI] [PubMed] [Google Scholar]
- 27.Schubert LA, Jeffery E, Zhang Y, Ramsdell F, Ziegler SF. Scurfin (FOXP3) acts as a repressor of transcription and regulates T cell activation. J Biol Chem. 2001;276:37672–37679. doi: 10.1074/jbc.M104521200. [DOI] [PubMed] [Google Scholar]
- 28.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao K, Kas E, Gonzalez E, Laemmli UK. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selvaraj RK, Geiger TL. A kinetic and dynamic analysis of Foxp3 induced in T cells by TGF-beta. J Immunol. 2007;178:7667–7677. doi: 10.4049/jimmunol.178.12.7667. [DOI] [PubMed] [Google Scholar]
- 31.Allan SE, et al. The role of 2 FOXP3 isoforms in the generation of human CD4+ Tregs. J Clin Invest. 2005;115:3276–3284. doi: 10.1172/JCI24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan J, Xu L, Crawford G, Wang Z, Burgess SM. The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol Cell Biol. 2006;26:155–168. doi: 10.1128/MCB.26.1.155-168.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasare C, Medzhitov R. Toll pathway-dependent blockade of CD4+CD25+ T cell-mediated suppression by dendritic cells. Science. 2003;299:1033–1036. doi: 10.1126/science.1078231. [DOI] [PubMed] [Google Scholar]
- 34.Kretschmer K, et al. Inducing and expanding regulatory T cell populations by foreign antigen. Nat Immunol. 2005;6:1219–1227. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 35.Abrams JT, Balin BJ, Vonderheid EC. Association between Sezary T cell-activating factor, Chlamydia pneumoniae, and cutaneous T cell lymphoma. Ann N Y Acad Sci. 2001;941:69–85. doi: 10.1111/j.1749-6632.2001.tb03712.x. [DOI] [PubMed] [Google Scholar]
- 36.Abrams JT, et al. A clonal CD4-positive T-cell line established from the blood of a patient with Sezary syndrome. J Invest Dermatol. 1991;96:31–37. doi: 10.1111/1523-1747.ep12514693. [DOI] [PubMed] [Google Scholar]
- 37.Chang NS, et al. WOX1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated cell death, and its tyrosine 33-phosphorylated form binds and stabilizes serine 46-phosphorylated p53. J Biol Chem. 2005;280:43100–43108. doi: 10.1074/jbc.M505590200. [DOI] [PubMed] [Google Scholar]
- 38.Utama B, et al. Mechanisms for human cytomegalovirus-induced cytoplasmic p53 sequestration in endothelial cells. J Cell Sci. 2006;119:2457–2467. doi: 10.1242/jcs.02974. [DOI] [PubMed] [Google Scholar]
- 39.Lagger G, et al. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol Cell Biol. 2003;23:2669–2679. doi: 10.1128/MCB.23.8.2669-2679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allison SJ, Milner J. Remodelling chromatin on a global scale: A novel protective function of p53. Carcinogenesis. 2004;25:1551–1557. doi: 10.1093/carcin/bgh212. [DOI] [PubMed] [Google Scholar]
- 41.Sitkovsky MV, Ohta A. The ‘danger’ sensors that STOP the immune response: The A2 adenosine receptors? Trends Immunol. 2005;26:299–304. doi: 10.1016/j.it.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.