Abstract
The childhood cancer neuroblastoma arises in the developing sympathetic nervous system and is a genotypically and phenotypically heterogeneous disease. Prognostic markers of poor survival probability include amplification of the MYCN oncogene and an undifferentiated morphology. Whereas these features discriminate high- from low-risk patients with precision, identification of poor outcome low- and intermediate-risk patients is more challenging. In this study, we analyze two large neuroblastoma microarray datasets using a priori-defined gene expression signatures. We show that differential overexpression of Myc transcriptional targets and low expression of genes involved in sympathetic neuronal differentiation predicts relapse and death from disease. This was evident not only for high-risk patients but was also robust in identifying groups of poor prognosis patients who were otherwise judged to be at low- or intermediate-risk for adverse outcome. These data suggest that pathway-specific gene expression profiling might be useful in the clinic to adjust treatment strategies for children with neuroblastoma.
Keywords: neuroblastoma, pathway analysis, MYCN, Myc, differentiation
Neuroblastoma is the most common solid extracranial childhood malignancy and accounts for ≈15% of all childhood tumor-related deaths (1). The neuroblastoma cells stem from immature or precursor cells of the sympathetic neuronal lineage, and the tumors locate to sites in which sympathetic neuroblasts are found during development, that is, sympathetic ganglia and the adrenal gland (2–4). Phenotypically as well as clinically, neuroblastoma shows considerable heterogeneity. Several critical genetic aberrations have been identified but there are few established molecular markers that associate with outcome, the most important being amplification of the MYCN locus, present in ≈20–30% of all cases, and strongly related to poor clinical outcome (5, 6). Also related to advanced disease and adverse outcome are deletions of chromosomal material on 1p, 11q23 (7) and gain of 17q (8), but despite extensive efforts, no bona fide neuroblastoma-related tumor suppressors or oncogenes have been identified currently at these regions. Another feature related to patient survival is the degree of tumor cell differentiation, which histopathologically ranges from undifferentiated small blue (neuroblastoma) in aggressive tumors, via larger intermediately differentiated (ganglioneuroblastoma), to highly differentiated nondividing cells of ganglioneuromas, which are considered benign tumors (9). Furthermore, a study using a small number of tumors and few marker genes reported a positive correlation between high expression of neuronal differentiation marker genes and favorable outcome (10), an observation also supported by microarray studies (11).
To date, the most functional guideline for neuroblastoma patient treatment and outcome is the risk grouping according to the Children's Oncology Group (COG) (6). This system incorporates tumor stage per the International Neuroblastoma Staging System (INSS), MYCN amplification status, histology according to Shimada (12), and tumor cell ploidy. These guidelines effectively identify high-risk patients, characterized by disseminated disease, frequent MYCN amplification, poorly differentiated tumor cells, and survival probabilities of <40% despite intensive multimodal therapies.
Several microarray studies on global gene expression of neuroblastoma specimens have been published using different technical platforms and analysis methods (11, 13–15). However, the overlap in identified expressed genes with prognostic information between these studies is low. In an interesting recent study, the prognostic impact of previously published gene expression markers for patient outcome was tested by microarray analysis (16). As with the COG risk grade system, it was relatively straight-forward to differentiate high-risk from low-risk tumors; however, the identified markers gave little information regarding outcome of patients with low- and intermediate-stage disease, which is a major clinical issue. In the present study, we have taken an alternative computational approach using a priori biological knowledge and analyzed the data from two clinical neuroblastoma microarray studies (13, 15) from a pathway perspective. The analyses presented here were carried out by using gene expression signatures correlating to two important features of aggressive neuroblastoma: MYCN amplification and stage of differentiation. The results showed that poor outcome neuroblastoma, as well as all MYCN-amplified cases, has elevated signaling through the Myc transcriptional network (Myc, MycN, and MycL target genes) and low expression of lineage marker genes relating to late neuronal differentiation. Interestingly, these gene expression traits were not only present in high-risk tumors, but also in patients with tumors initially diagnosed as low or intermediate risk that ultimately had an adverse outcome.
Results
Strength of Myc Up-regulated Signaling Predicts Clinical Outcome Independently of MYCN Amplification.
One of the most important prognostic factors in neuroblastoma is amplification of the MYCN oncogene. However, the role of the functional MycN protein as a transcription factor in neuroblastoma tumor progression is less clear-cut (17–20). To investigate MycN downstream transcriptional effects on neuroblastoma behavior, we generated a gene expression signature of Myc-dependent transcription based on the entries in the Myc target gene database (21) (www.myccancergene.org) [supporting information (SI) Methods and Table S1]. The transcriptional effects of Myc and MycN appear largely redundant (22–24), and validation of the Myc pathway profile, defined here, with regard to MycN-regulated transcription was established by rank sum calculations in two independent in vitro MYCN overexpression studies, showing increased pathway activity already after eight hours (25, 26) (Fig. S1).
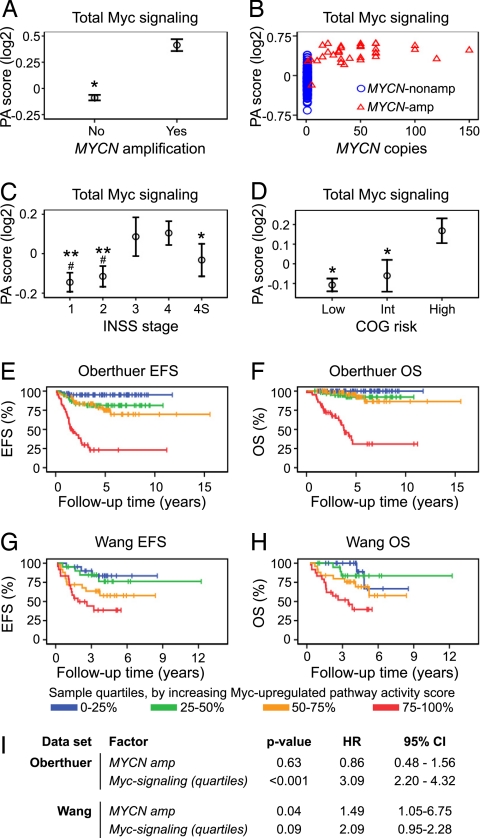
Myc signaling strength, defined as high expression of Myc, MycN, and MycL target genes, was analyzed in the Oberthuer neuroblastoma microarray dataset (13) by using rank-based pathway activity scores for the created Myc target gene signatures; these quantitative measures were then correlated to clinical factors. As expected, Myc-dependent transcriptional targets were significantly overexpressed in the MYCN-amplified tumors (n = 33) compared with nonamplified tumors [n = 217; 1.4-fold difference, P < 0.001, (Fig. 1A)]. This difference was significant both with respect to Myc-up-regulated and Myc-down-regulated signaling; however, the magnitude of difference was larger for Myc-up-regulated compared with Myc-down-regulated signaling (1.7 vs. 1.2 fold, P < 0.001 and P < 0.001, respectively) (Fig. S2). Interestingly, among the tumors harboring MYCN amplification, there was no apparent linear dependence between MYCN gene copy number [as determined by a standardized Protocol for Fluorescence in situ Hybridization (FISH) protocol (13)] and strength in Myc-signaling. Rather, it seemed that Myc-signaling reached a plateau level and additional MYCN copies did not increase the strength of total signaling in the Myc downstream regulatory cascade (see Fig. 1B and Fig. S2). Moreover, in several non-MYCN amplified tumors, the Myc signaling level reached this plateau (Fig. 1B), and high Myc signaling correlated to high INSS tumor stage and high COG risk (Fig. 1 C and D). Again, this trend was stronger for the Myc-up-regulated compared with the Myc-down-regulated gene signature (Fig. S2); consequently, in further analyses of Myc signaling, only the Myc-up-regulated gene signature was considered. To analyze the impact of Myc-induced signaling on patient outcome, we divided the tumor data into quartile groups based on the Myc-up-regulated pathway activity score. Kaplan–Meier analyses of the Oberthuer data using these quartiles showed significant correlation to patient outcome, both when using event-free survival (EFS) (P < 0.001, Fig. 1E), and overall survival (OS) (P < 0.001, Fig. 1F) as the measured endpoint. Interestingly, extremely high Myc signaling (4th quartile; 75–100%) was to a large extent associated with incurable disease (Fig. 1F). Of these tumors (n = 63), as many as 50% were non-MYCN amplified (Table S2), suggesting that tumors with Myc-signaling over a certain level are treatment-resistant. The correlation between high Myc-up-regulated signaling and poor patient prognosis was confirmed when assessing the same calculations for the Wang data [EFS: P < 0.001, (Fig. 1G); OS: P < 0.001, (Fig. 1H), and (Fig. S3)]. Myc-up-regulated pathway activity score was tested as a continuous variable and found to be highly predictive of both patient EFS and OS both for the Oberthuer (EFS: P < 0.001, HR = 12.3, 95% CI 6.7–22.5; OS: P < 0.001, HR = 50.4, 95% CI 19.0–134.0; variable range: −0.95–0.81) and for the Wang data (EFS: P < 0.001, HR = 2,146, 95% CI 41.2–111,750; OS: P < 0.001, HR = 6545, 95% CI 62.1–689,485; variable range: −0.22–0.18**). Furthermore, in multivariate Cox proportional hazards models for EFS and OS, Myc signaling (grouped by quartiles), but not MYCN amplification status was independently informative in the Oberthuer dataset (Fig. 1I, OS: see Table S3). In the Wang data, this was not significant at the 0.05 level (P = 0.09), probably because this patient cohort was selected to have a large proportion of MYCN-amplified cases (Fig. 1I, OS: see Table S3). Taken together, these analyses clearly show that Myc signaling is proportionally correlated to tumor aggressiveness, suggesting that the activity of the Myc gene regulatory pathway carries prognostic information independently of MYCN amplification status. Regarding contributions of individual Myc genes, MYCN expression was high in MYCN-amplified tumors and as shown here correlated to Myc pathway activity score. In non-MYCN amplified cases, the picture seemed more complex. Our data suggest contributions from all Myc-family members and also associated factors such as SKP2 (28) (Fig. S4). This trend was also evident when looking at the highest Myc-signaling tumors only (Fig. S5).
Fig. 1.
Myc-pathway activation correlates to clinical stage, risk assessment, and patient outcome in neuroblastoma. (A and B) Pathway activity (PA) scores for a Myc gene expression signature (up- and down-regulation) in a microarray dataset of 251 neuroblastoma specimens (13) (Oberthuer) correlate to MYCN amplification (*, P < 0.001) (A), but not specifically to MYCN gene copy number (B). (C) High Myc pathway activity score correlates to advanced INSS stage (#, P < 0.001 vs. INSS 3, *, P < 0.01; **, P < 0.001 vs. INSS 4). (D) Myc signaling correlates to high COG risk (*, P < 0.001 vs. COG high risk tumors). (D). Data (A, C and D) are presented as means (open circle) ± two SEM. All P values relate to two-sided Student's t tests. (E and F) Categorization of tumors into groups, based on quartiles of Myc-up-regulated pathway activity scores only, shows that increasing strength of Myc signaling proportionally correlates to adverse patient outcome, when scoring either event-free survival (EFS) (P < 0.001, log-rank test) (E), or overall survival (OS) (P < 0.001, log-rank test) (F). These results were corroborated in an independent dataset of 101 tumors (15) (Wang) (EFS, P < 0.001, and OS, P < 0.001, log-rank tests) (G and H). (I) Cox's proportional hazards models (EFS) show for both datasets that Myc up-regulated signaling is a prognostic factor independently of MYCN amplification status.
Late, Mature, Sympathetic Neuronal Differentiation Predicts Favorable Outcome.
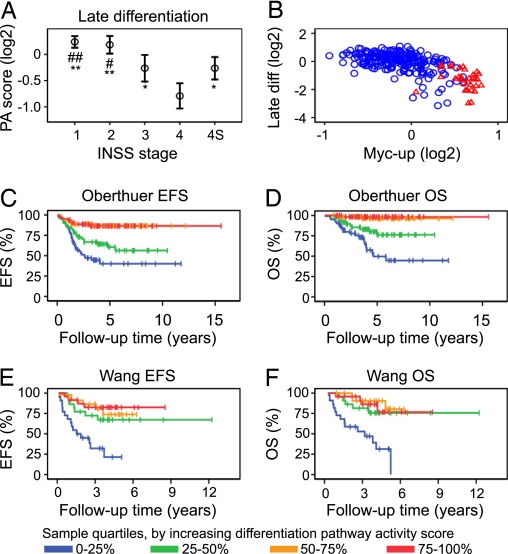
One important histological feature of high stage, aggressive neuroblastoma is lack of neuronal differentiation (12, 27). To investigate the prognostic value of neuroblastoma differentiation status from a pathway perspective, we constructed two gene expression signatures based on our own experience and a review of the literature (2–4, 28–33). One signature represented early, neural crest-associated gene expression and the other late neuronal sympathetic differentiation. By calculating pathway activity scores for these transcriptional signatures, we investigated the correlations of early and late neuronal differentiation-related gene expression to neuroblastoma outcome. Pathway activity scores for the early differentiation gene expression signature (Table S4, n = 7) did not show any correlation to stage, risk, or MYCN amplification in either the Oberthuer or the Wang datasets (Fig. S6); however, a weak negative correlation was seen to the late differentiation signature (Fig. S6). In contrast, pathway activity score calculations for the late differentiation signature (Table S4, n = 9) over the tumors in the Oberthuer data showed that high stage tumors generally had lower expression of these marker genes, as compared to low stage tumors (Fig. 2A). Pathway activity scores based on these genes also showed a negative correlation to risk and a negative correlation to MYCN amplification status (Fig. S7). No correlation between differentiation marker pathway activity score and specific MYCN gene copy number was seen for MYCN-amplified tumors (Fig. S7). Instead, a strong negative correlation to Myc-induced signaling [r = −0.7, P < 0.001, (Fig. 2B)] was seen, where all MYCN-amplified tumors in the Oberthuer dataset (n = 33) showed a below-median pathway activity score for the late differentiation profile. No correlation to Myc-induced down-regulation was seen (data not shown), suggesting that down-regulation of sympathetic marker genes is not under Myc transcriptional control. In the Oberthuer clinical data, low expression of the late neuronal differentiation markers, as determined by grouping tumors based on pathway activity score quartiles, correlated to both decreased event-free survival (P < 0.001, Fig. 2C) and poor overall survival (P < 0.001, Fig. 2D). These findings were corroborated in the Wang data [EFS: P < 0.001, (Fig. 2E), OS: P < 0.001, (Fig. 2F), and (Fig. S8)]. As for the Myc-induced transcriptional profile, analyses of late neuronal differentiation pathway activity scores as a continuous variable was highly predictive of patient EFS and OS both for the Oberthuer (EFS: P < 0.001, HR = 0.50, 95% CI 0.41–0.62; OS: P < 0.001, HR = 0.41, 95% CI 0.32–0.54; variable range: −3.15–1.08) and the Wang (EFS: P = 0.004, HR = 0.20, 95% CI 0.07–0.60; OS: P < 0.001, HR = 0.15, 95% CI 0.04–0.51; variable range: −1.17–0.30) datasets, where low pathway activity score correlated to poor patient outcome. In essence, aggressive neuroblastomas show gene expression relating to an undifferentiated tumor cell phenotype.
Fig. 2.
Stage of differentiation correlates to neuroblastoma patient outcome. (A) Pathway activity (PA) scores, calculated over a gene expression signature for late sympathetic neuronal differentiation correlate to low INSS stage in a dataset representing 251 neuroblastoma cases (13) (Oberthuer) (#, P = 0.005; ##, P = 0.001 vs. INSS 3, *, P < 0.01; **, P < 0.001 vs. INSS 4). Presented as means (open circles) ± two SEM. (B) Late differentiation and Myc-up-regulated (Myc-up) signaling pathway activity scores correlate negatively. Blue circles: MYCN-non amplified tumor. Red triangles: MYCN-amplified tumor. (C and D) Categorization of tumors into groups, based on late differentiation pathway activity score quartiles, shows a correlation between decreasing stage of neuronal differentiation and adverse patient outcome scored as either event-free survival (EFS) (P < 0.001, log-rank test) (C) or overall survival (OS) (P < 0.001, log-rank test) (D). (E and F) This result was validated in an independent dataset of 101 tumors (15) (Wang) (EFS, P < 0.001, and OS, P < 0.001, log-rank tests).
Combination of Myc and Differentiation Signatures Identify Patient Subgroups with Differential Outcome.
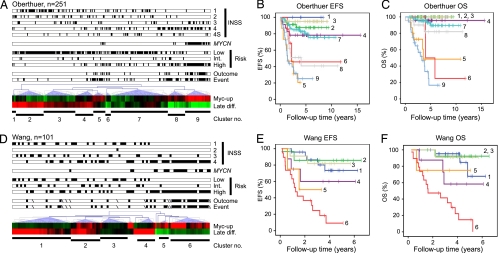
To examine combinatorial effects of Myc-signaling and stage of differentiation in an unsupervised fashion, pathway activity scores were grouped by using hierarchical clustering, similar to methods used when analyzing gene expression. Analysis of these two variables organized the tumors in the Oberthuer dataset into nine clusters, as identified using node correlations (Fig. 3A). The majority of MYCN-amplified tumors grouped into cluster 9, characterized by robust Myc signaling in combination with very low differentiation, and, as expected, this cluster related to poor patient outcome, both when looking at event-free survival and overall survival (Fig. 3 B and C). Similarly, cluster 8 also had low differentiation and high Myc signaling and contained many high stage, high-risk patients. Interestingly, patients in this cluster suffered recurrences and disease progression despite initial therapy (Fig. 3B); however, most benefited from post-recurrence treatment and few succumbed to disease (Fig. 3C). Two other interesting clusters (5 and 6) with poor prognosis were identified (Fig. 3 B and C). Cluster 5 included some MYCN-amplified cases and tumors had high Myc signaling, but median differentiation. Conversely, cluster 6 showed very low differentiation and median Myc signaling. This suggests that the estimation of both Myc signaling and differentiation stage is informative regarding patient outcome independent of each other. The Myc signaling and differentiation clustering method was also informative in the Wang data (Fig. 3D), where most high-risk cases were grouped into clusters 5 and 6. These two clusters featured high Myc signaling in combination with low differentiation (Fig. 3 E and F).
Fig. 3.
Myc-signaling and stage of differentiation define patient subgroups with diverging clinical outcome. Pathway activity (PA) scores for Myc up-regulated signaling (Myc-up) and stage of neuronal differentiation (Late diff.) were used in hierarchical cluster analyses (Euclidian distance, average linkage clustering) of the Oberthuer (A–C) and Wang (D and E) datasets. Identified clusters were used for Kaplan–Meier analyses of event-free survival (EFS) and overall survival (OS) for the Oberthuer (B and C) and Wang (E and F) datasets respectively. Annotations are divided into INSS stage (1, 2, 3, 4, and 4S), MYCN amplification, COG risk (low, intermediate, high), outcome (diseased), and event (recurrence). Each tumor is represented by a filled black box when called into that annotation category. Missing values (Wang) are represented by a diagonal.
Activation of Myc Signaling and Low Expression of Neuronal Differentiation Genes Predicts Outcome in Low- and Intermediate-Risk Patients.
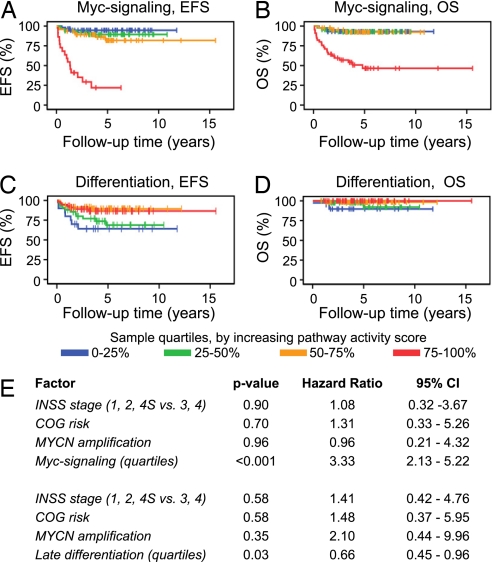
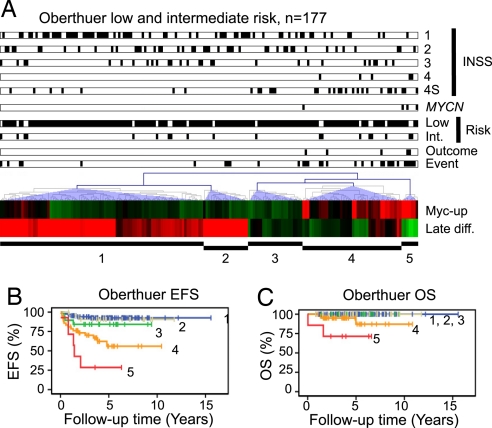
The observation, that the strength of Myc-induced signaling was a prognostic factor independently of MYCN amplification, prompted us to test whether this feature and stage of neuronal differentiation could predict patient outcome for low- and intermediate-risk patients. The Oberthuer dataset contained 177 low and intermediate risk patients, of which four were MYCN amplified. All together these patients suffered 31 unfavorable disease-related events, and five died because of the disease (Table S5). Using the pathway activity score of quartile groups created when analyzing all patients, Kaplan–Meier estimates did indeed show that both strength of Myc signaling and stage of differentiation were proportionally correlated to patient event-free survival (Fig. 4 A and C) and to overall survival (Fig. 4 B and D). Again, analyzing these factors as continuous variables in an univariate Cox regression model using EFS as the endpoint (OS was not evaluated because of the low number of poor outcome patients) showed that patient prognosis was significantly dependent on Myc signaling strength and stage of differentiation (Myc signaling: P < 0.001, HR = 42.2, 95% CI 13.6–131.6; Late differentiation: P < 0.001, HR = 0.46, 95% CI 0.32–0.66). In multivariate Cox's proportional hazard models for EFS using MYCN amplification, INSS stage (high or low), COG risk, and either Myc signaling quartiles or stage of differentiation quartiles as covariates, the latter two came out as the only significant factors (Fig. 4E). Of notable interest, a large proportion of low and intermediate risk patients in the Oberthuer data that suffered unfavorable events (16 of 31) had Myc signaling strength grouped to the fourth quartile; the same quartile as most MYCN-amplified cases (Table S5). Correspondingly, several of these cases were grouped to the first and second differentiation pathway activity score quartiles (7 and 11, respectively). These criteria thus define a low and intermediate risk patient subgroup that share characteristics of aggressive high stage, high-risk tumors (Fig. 5). In the Wang data, only one low risk and two intermediate risk patients suffered relapse, and none of these patients died during the follow-up time. Interestingly, these three patients displayed high Myc signaling or low differentiation (data not shown).
Fig. 4.
Myc-signaling and stage of differentiation correlate to outcome also for low- and intermediate-risk patients. Sample quartiles, based on the entire 251 patient Oberthuer cohort (13), for either Myc up-regulated signaling (distribution: 1st quartile: n = 58; 2nd 50; 3rd 47; 4th 22) (A and B) or late differentiation (distribution: 1st quartile n = 20; 2nd 42; 3rd 59; 4th 56) (C and D) were used for Kaplan–Meier analyses of the 177 low and intermediate risk tumors. (A) P < 0.001, (B) P < 0.001, (C) P = 0.02, (D) P = 0.09, log-rank tests. (E) Cox's proportional hazards models (EFS) by using high/low INSS stage, COG risk, MYCN amplification status, and either Myc-up-regulated or late differentiation quartile groups.
Fig. 5.
Myc signaling and stage of differentiation defines patient subgroups with diverging clinical outcome in the cohort of low- and intermediate-risk patients only. Pathway activity scores for Myc-induced signaling (Myc-up) and stage of neuronal differentiation (Late diff.) were used in hierarchical cluster analyses (Euclidian distance, average linkage clustering) of the Oberthuer low to intermediate risk tumors (n = 177) (A). Identified clusters were used for Kaplan–Meier analyses of event-free survival (EFS) and overall survival (OS) for the Oberthuer (B and C). Annotations are divided into INSS stage (1, 2, 3, 4, and 4S), MYCN amplification, COG risk (low, intermediate), outcome (diseased), and event (recurrence). Each tumor is represented by a filled black box when called into that annotation category.
Conclusions
Using high-throughput gene expression analysis of neuroblastoma tumor specimens, a number of studies have defined gene expression signatures capable of dichotomizing favorable vs. poor outcome patients (11, 13, 16, 34, 35). However, these studies have not been able to highlight useful targets for therapeutic intervention and one contributing fact to this dilemma might be that the resulting signature gene lists have little overlap. In this report, we have tested the hypothesis that quantitative analysis of deregulated pathways, rather than deregulated expression of single genes, would be more informative regarding clinical outcome of neuroblastoma patients. Thus, our aim was not to find an optimal gene expression classifier.
Even though amplification of the MYCN oncogene has strong clinical impact (5), high levels of MYCN mRNA or protein in neuroblastoma only seems to correlate to poor outcome in specific patient subsets (17–20). The molecular basis for these counterintuitive results is at present unclear. However, as we show here analysis of overall Myc signaling (increased expression of Myc/MycL/MycN target genes) provides strong prognostic information. Contrary to the binary effect of MYCN amplification on patient outcome, that is, importance of amplification rather than specific gene copy number, Myc signaling shows a more linear correlation to outcome, as confirmed by univariate Cox analysis. Taken together our data clearly demonstrate that activation of the Myc pathway contributes to neuroblastoma aggressiveness.
Morphologic differentiation to ganglioneuromatous histopathology has for decades been recognized as a positive prognostic sign in neuroblastoma (27, 36), but the clinical impact of this knowledge has so far been limited. Here, we demonstrate that poor outcome neuroblastomas with few exceptions show a combined low expression of differentiation marker genes despite differences in genetic background and tumor spread at diagnosis. The reason for the low expression of sympathetic neuronal marker genes in these tumors could in part be that the differentiation stage at tumor initiation is mirrored, but could also be caused by the surrounding microenvironment, for example, hypoxia (30, 37). The negative correlation seen between Myc-induced signaling and stage of differentiation (Fig. 2B) is interesting and suggests a causal link; although, it has been shown that MYCN-overexpressing cells can retain their capacity to differentiate (38). The issue is complicated by the observation that MycN expression during normal sympathetic development is associated with differentiation rather than with proliferation (39).
Recently several studies investigating deregulation of signaling pathways, rather than the importance of expression of individual genes, as central to cancer aggressiveness have been published (40–43). These observations are particularly interesting in light of the development of small inhibitory molecules able to inhibit individual pathways, and it has been shown that pathway analysis can be used to identify appropriate targets for therapeutic intervention (40). In the case of neuroblastoma being an extremely heterogeneous disease, quantitative assessment of pathway involvement hold clinical promise (26, 44). In the present contribution, we demonstrate that a combined analysis of Myc signaling and stage of differentiation can provide independent and complementary information regarding patient outcome (Fig. 3). In particular, these analyses were useful for identifying aggressive neuroblastomas that are initially diagnosed as low or intermediate risk. Analyses of additional pathways would most likely increase the prognostic sensitivity, and might open up for individualized therapeutic intervention regiments.
Materials and Methods
Patient Material.
Two clinical neuroblastoma gene expression datasets were used for the analyses. As described by Oberthuer et al. (251 cases) (13) and Wang et al. (101 cases) (15), the basis of these datasets differ markedly, both with respect to selection of patient cohorts and to the microarray technology used. The patient materials are described further in SI Methods.
Gene Expression Signatures.
Two Myc-related gene expression signatures (up- or down-regulated) were derived from the Myc target gene database (21) based on level of evidence for Myc regulation. The Myc up-regulated signature comprised 83 genes and the Myc down-regulated signature contained 20 genes. On average each of the genes in the Myc signatures had been shown to be Myc regulated in three different cell lines and by three individual studies (SI Methods and Table S1). Based on literature data, two gene expression signatures pertaining to sympathetic neuronal differentiation were constructed; one signature of genes highly expressed in differentiated neuronal sympathetic cells (n = 9), and one signature containing neural crest marker genes (n = 7) (3, 4, 28–30) (Table S4). The gene expression signatures are described further in SI Methods.
Pathway Activity Score Calculations.
The activities of the Myc and differentiation pathway gene expression signatures were quantified by defining a rank-based pathway activity score. Genes were ranked according to decreasing expression levels for each microarray separately. The ranks of the genes in a signature were summed up for each tumor. For each gene signature, the rank sums were divided by the average rank sum for the cohort, followed by log2 transformation, thus giving a cohort-comparative score for each tumor. A pathway activity score of zero corresponds to cohort average activity for the pathway, a positive value corresponds to above cohort average activity, and a negative value corresponds to below average activity. For the Myc down-regulated signature ranks were inverted to give a comparable measure of strength of down-regulation. A score for total Myc transcriptional activity was calculated by adding the Myc up- and down-regulated rank sums normalized to number of genes in the transcriptional signature.
Statistical Analyses.
For survival estimates based on dividing patients by quartiles, the entire patient cohort was used, even when subgroups were analyzed. For unsupervised cluster analyses, Myc up-regulated and mature differentiation pathway activity scores were clustered using average linkage and Euclidian distance. All statistical and pathway activity score calculations were performed by using R (www.r-project.org) or SPSS 12.0 (SPSS Inc.). Two-sided Student's t tests were used for between group analyses.
Supplementary Material
Acknowledgments.
We acknowledge The Research Program in Medical Bioinformatics of the Swedish Knowledge Foundation (to E.F.); the Swedish Cancer Society (to S.P. and M.R.); the Children's Cancer Foundation of Sweden (to S.P.); the Swedish Research Council (to S.P.); the Swedish Foundation for Strategic Research through the Lund Strategic Centre for Clinical Cancer Research (CREATE Health) (to S.P. and M.R.); the Royal Physiographic Society (to E.F.); Gunnar Nilssons Cancerstiftelse (to S.P.); the Hans von Kantzow foundation (to S.P.); the Malmö University Hospital research funds (to S.P.); National Institutes of Health (R01-CA87847, U10-CA98543 (Children's Oncology Group) to J.M.M.); the Alex's Lemonade Stand Foundation (to J.M.M.); and the Abramson Family Cancer Research Institute (to J.M.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804455105/DCSupplemental.
High hazard ratios are a consequence of the narrow variable range.
References
- 1.Brodeur GM, Maris JM. In: Principles and Practice of Pediatric Oncology. Pizzo PA, Poplack DG, editors. Philadelphia: JB Lippincott; 2006. pp. 933–970. [Google Scholar]
- 2.De Preter K, et al. Human fetal neuroblast and neuroblastoma transcriptome analysis confirms neuroblast origin and highlights neuroblastoma candidate genes. Genome Biol. 2006;7:R84. doi: 10.1186/gb-2006-7-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoehner JC, et al. A developmental model of neuroblastoma: differentiating stroma-poor tumors' progress along an extra-adrenal chromaffin lineage. Lab Invest. 1996;75:659–675. [PubMed] [Google Scholar]
- 4.Hoehner JC, et al. Developmental gene expression of sympathetic nervous system tumors reflects their histogenesis. Lab Invest. 1998;78:29–45. [PubMed] [Google Scholar]
- 5.Brodeur GM. Neuroblastoma: Biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 6.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 7.Attiyeh EF, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 8.Bown N, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- 9.Shimada H, et al. The International Neuroblastoma Pathology Classification (the Shimada system) Cancer. 1999;86:364–372. [PubMed] [Google Scholar]
- 10.Hedborg F, et al. Biochemical evidence for a mature phenotype in morphologically poorly differentiated neuroblastomas with a favourable outcome. Eur J Cancer. 1995;31A:435–443. doi: 10.1016/0959-8049(95)00025-e. [DOI] [PubMed] [Google Scholar]
- 11.Ohira M, et al. Expression profiling using a tumor-specific cDNA microarray predicts the prognosis of intermediate risk neuroblastomas. Cancer Cell. 2005;7:337–350. doi: 10.1016/j.ccr.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 12.Shimada H, et al. Histopathologic prognostic factors in neuroblastic tumors: Definition of subtypes of ganglioneuroblastoma and an age-linked classification of neuroblastomas. J Natl Cancer Inst. 1984;73:405–416. doi: 10.1093/jnci/73.2.405. [DOI] [PubMed] [Google Scholar]
- 13.Oberthuer A, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- 14.Ohira M, et al. A review of DNA microarray analysis of human neuroblastomas. Cancer Lett. 2005;228:5–11. doi: 10.1016/j.canlet.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 16.Oberthuer A, et al. Classification of neuroblastoma patients by published gene-expression markers reveals a low sensitivity for unfavorable courses of MYCN non-amplified disease. Cancer Lett. 2007;250:250–267. doi: 10.1016/j.canlet.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Asgharzadeh S, et al. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst. 2006;98:1193–1203. doi: 10.1093/jnci/djj330. [DOI] [PubMed] [Google Scholar]
- 18.Cohn SL, et al. MYCN expression is not prognostic of adverse outcome in advanced-stage neuroblastoma with nonamplified MYCN. J Clin Oncol. 2000;18:3604–3613. doi: 10.1200/JCO.2000.18.21.3604. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka S, et al. Clinical significance of a highly sensitive analysis for gene dosage and the expression level of MYCN in neuroblastoma. J Pediatr Surg. 2004;39:63–68. doi: 10.1016/j.jpedsurg.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Tang XX, et al. The MYCN enigma: significance of MYCN expression in neuroblastoma. Cancer Res. 2006;66:2826–2833. doi: 10.1158/0008-5472.CAN-05-0854. [DOI] [PubMed] [Google Scholar]
- 21.Zeller KI, et al. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malynn BA, et al. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- 23.Raetz EA, et al. Identification of genes that are regulated transcriptionally by Myc in childhood tumors. Cancer. 2003;98:841–853. doi: 10.1002/cncr.11584. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, et al. Functional homology between N-myc and c-myc in murine plasmacytomagenesis: plasmacytoma development in N-myc transgenic mice. Oncogene. 1992;7:1241–1247. [PubMed] [Google Scholar]
- 25.Koppen A, et al. Dickkopf-1 is down-regulated by MYCN and inhibits neuroblastoma cell proliferation. Cancer Lett. 2007;256:218–228. doi: 10.1016/j.canlet.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 26.Westermann F, et al. High Skp2 expression characterizes high-risk neuroblastomas independent of MYCN status. Clin Cancer Res. 2007;13:4695–4703. doi: 10.1158/1078-0432.CCR-06-2818. [DOI] [PubMed] [Google Scholar]
- 27.Edsjö A, Holmquist L, Pahlman S. Neuroblastoma as an experimental model for neuronal differentiation and hypoxia-induced tumor cell dedifferentiation. Semin Cancer Biol. 2007;17:248–256. doi: 10.1016/j.semcancer.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Gammill LS, Bronner-Fraser M. Neural crest specification: Migrating into genomics. Nat Rev Neurosci. 2003;4:795–805. doi: 10.1038/nrn1219. [DOI] [PubMed] [Google Scholar]
- 29.Gestblom C, et al. In vivo spontaneous neuronal to neuroendocrine lineage conversion in a subset of neuroblastomas. Am J Pathol. 1997;150:107–117. [PMC free article] [PubMed] [Google Scholar]
- 30.Jögi A, et al. Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. Proc Natl Acad Sci USA. 2002;99:7021–7026. doi: 10.1073/pnas.102660199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber K. The sympathoadrenal cell lineage: specification, diversification, and new perspectives. Dev Biol. 2006;298:335–343. doi: 10.1016/j.ydbio.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Krajewski S, Chatten J, Hanada M, Reed JC. Immunohistochemical analysis of the Bcl-2 oncoprotein in human neuroblastomas. Comparisons with tumor cell differentiation and N-Myc protein. Lab Invest. 1995;72:42–54. [PubMed] [Google Scholar]
- 33.Påhlman S, Hedborg F. In: Neuroblastoma. Brodeur G, Tsuchida Y, Sawada T, editors. Amsterdam: Elsevier; 2000. pp. 9–19. [Google Scholar]
- 34.Schramm A, et al. Prediction of clinical outcome and biological characterization of neuroblastoma by expression profiling. Oncogene. 2005;24:7902–7912. doi: 10.1038/sj.onc.1208936. [DOI] [PubMed] [Google Scholar]
- 35.Wei JS, et al. Prediction of clinical outcome using gene expression profiling and artificial neural networks for patients with neuroblastoma. Cancer Res. 2004;64:6883–6891. doi: 10.1158/0008-5472.CAN-04-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cushing H, Wolback S. The transformation of a malignant paravertebral sympathicoblastoma into a benign ganglioneuoma. Am J Pathol. 1927;3:203–216. [PMC free article] [PubMed] [Google Scholar]
- 37.Holmquist-Mengelbier L, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 38.Edsjö A, et al. Neuroblastoma cells with overexpressed MYCN retain their capacity to undergo neuronal differentiation. Lab Invest. 2004;84:406–417. doi: 10.1038/labinvest.3700061. [DOI] [PubMed] [Google Scholar]
- 39.Wakamatsu Y, Watanabe Y, Nakamura H, Kondoh H. Regulation of the neural crest cell fate by N-myc: promotion of ventral migration and neuronal differentiation. Development. 1997;124:1953–1962. doi: 10.1242/dev.124.10.1953. [DOI] [PubMed] [Google Scholar]
- 40.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Ringnér M. Revealing signaling pathway deregulation by using gene expression signatures and regulatory motif analysis. Genome Biol. 2007;8:R77. doi: 10.1186/gb-2007-8-5-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saal LH, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci USA. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watters JW, Roberts CJ. Developing gene expression signatures of pathway deregulation in tumors. Mol Cancer Ther. 2006;5:2444–2449. doi: 10.1158/1535-7163.MCT-06-0340. [DOI] [PubMed] [Google Scholar]
- 44.Liu X, et al. Deregulated Wnt/beta-catenin program in high-risk neuroblastomas without MYCN amplification. Oncogene. 2008;27:1478–1488. doi: 10.1038/sj.onc.1210769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.