Abstract
Salmonella's success at proliferating intracellularly and causing disease depends on the translocation of a major virulence protein, SifA, into the host cell. SifA recruits membranes enriched in lysosome associated membrane protein 1 (LAMP1) and is needed for growth of Salmonella induced filaments (Sifs) and the Salmonella containing vacuole (SCV). It directly binds a host protein called SKIP (SifA and kinesin interacting protein) which is critical for membrane stability and motor dynamics at the SCV. SifA also contains a WxxxE motif, predictive of G protein mimicry in bacterial effectors, but whether and how it mimics the action of a host G protein is not known. We show that SKIP's pleckstrin homology domain, which directly binds SifA, also binds to the late endosomal GTPase Rab9. Knockdown studies suggest that both SKIP and Rab9 function to maintain peripheral LAMP1 distribution in cells. The Rab9:SKIP interaction is GTP-dependent and is inhibited by SifA binding to the SKIP pleckstrin homology domain, suggesting that SifA may be a Rab9 antagonist. SifA:SKIP binding is significantly tighter than Rab9:SKIP binding and may thus allow SifA to bring SKIP to the SCV via SKIP's Rab9-binding site. Rab9 can measurably reverse SifA-dependent LAMP1 recruitment and the perinuclear location of the SCV in cells. Importantly, binding to SKIP requires SifA residues W197 and E201 of the conserved WxxxE signature sequence, leading to the speculation that bacterial G protein mimicry may result in G protein antagonism.
Keywords: GTPase, LAMP1, Rab9, SKIP, lysosome
Gram-negative bacteria that cause a wide variety of diseases use Type III secretion systems to directly deliver virulence determinants into the cytoplasm of their host cells. These secreted proteins can modulate the normal host immune system and/or create a cellular environment that advances pathogen proliferation as well as disease progression. A number of secreted effectors modulate host GTPases (1–3), and a subset of these proteins has been proposed to functionally mimic G proteins themselves, although they do not bind GTP or resemble host G proteins at the level of primary sequence (4).
Salmonella are Gram-negative bacteria which, after they invade the host cell, reside in a membrane bound compartment known as the SCV. The SCV matures in a process whereby it sequentially picks up or loses early and then late endosomal components through vesicular fusion events and trafficks toward a perinuclear position. The maturation of the SCV is arrested at a late endosome-like stage, selectively excluding proteins such as mannose 6-phosphate receptors (MPR) and lysosomal cathepsin proteins (5). The maturation of the SCV also includes movement toward a perinuclear position in the host cell, which appears critical for Salmonella replication (6). Maintenance of SCV membranes and maturation arrest protect Salmonella from the toxic environment of the macrophage and from lysosomes in epithelial cells. Both processes are dependent on the secretion of Salmonella effectors into the host cell via a Type III secretion system. SifA is one such major effector and is required for recruitment of lysosome-associated membrane protein 1 (LAMP1), membrane growth, and maintenance of the SCV.
SifA is known to be a key virulence determinant, as sifA-mutant bacteria show a prominent replication defect in macrophages and are strongly attenuated for virulence in mice (7–10). In addition to its requirement for LAMP1 recruitment and growing and maintaining the SCV membrane, SifA is required for the generation of Sifs which are LAMP1-rich membrane filaments seen to protrude from the SCV, extending along microtubules toward the cell periphery (9, 11). These activities of SifA are mediated by its interaction with a host protein called SKIP (12). SKIP is also important for preservation of the perinuclear localization of SCV. Its pleckstrin homology (PH) domain is known to interact with SifA, resulting in the recruitment of SKIP to the SCV. SKIP also interacts at some level with kinesin, as the N terminus of SKIP pulls down kinesin from lysates, but on SCV membranes SKIP paradoxically acts to exclude kinesin from the SCV, thereby impacting SCV dynamics (12). Boucrot et al. have speculated that SKIP binds to kinesin in a regulatory complex of proteins (12).
The intracellular replication of Salmonella and SCV progression along the endocytic pathway have also been linked to small GTPases called Rabs (reviewed in ref. 13). Rabs have been implicated in the regulation of all steps of endocytic trafficking in uninfected cells, including vesicle formation, intracellular transport (including binding to motor proteins or motor adaptors), vesicle tethering, and vesicle fusion (reviewed in ref. 14). They act through the GTP-dependent recruitment of protein ligands at the appropriate time and place. Rabs and their effectors localize to vacuoles of intracellular pathogens and are important in phagosome trafficking and maintenance (13). The mature SCV interacts with the late endosomal Rabs, Rab7 and Rab9 (13, 15, 16). Rab7 is important for regulating late endosome to lysosomal transport in cells. It appears to recruit LAMP1 to the SCV and has been demonstrated to link the SCV to dynein/dynactin, promoting the early juxtanuclear trafficking of the SCV, via the adaptor protein RILP (Rab7-interacting lysosomal protein) (17). Rab9 has been implicated in MPR trafficking between endosomes and the Golgi in uninfected cells (18), but because MPR does not concentrate in the bacterial vacuole, the function of Rab9 at the SCV is unknown.
Here we report the discovery of a specific and direct interaction between Rab9 GTPase and the PH domain of SKIP. We additionally show that SKIP and Rab9 are both needed for peripheral LAMP1 distribution, even in the absence of SifA. We demonstrate that SifA can compete with Rab9 for this SKIP binding site both in in vitro binding assays and in host cell lysates. Rab9 antagonizes SifA-induced LAMP1 recruitment and SCV position in cells. We further show that residues W197 and E201 of SifA, conserved in a family of bacterial G protein mimics (4), are critical for the ability of SifA protein to bind the SKIP PH domain. The stronger affinity of SifA:SKIP binding relative to that of Rab9:SKIP binding suggests that competitive displacement by a partial G protein mimic may also be a mechanism for G protein antagonism.
Results
The SKIP Protein PH Domain Specifically Binds to Rab9 in a GTP Dependent Manner and Influences LAMP1-Containing Membrane Dynamics.
SKIP is a recently discovered mammalian protein that appears central to the action of SifA at the SCV (12). Because SKIP is central to the maintenance of the SCV, we investigated whether it interacted with other trafficking proteins known to be recruited to the SCV. Specifically, we investigated late endosomal trafficking protein Rab7 and a second late endosomal Rab, Rab9, reported to be found on Sifs (16). Although the location of Rab7 on Sifs is well established, the presence of Rab9 is unexpected because its major cellular cargo, the MPRs, are not found on the SCV to a significant degree (5). Nonetheless, we have independently confirmed that endogenous Rab9 is present on the SCV and Sifs at later stages of infection (D.M. Catron and K.H., unpublished results). Because SKIP contains a PH domain that binds to SifA, and PH domains have been shown to directly interact with small GTPases (19), we tested whether the SKIP:PH domain could directly bind to host cell Rab7 or Rab9 GTPases.
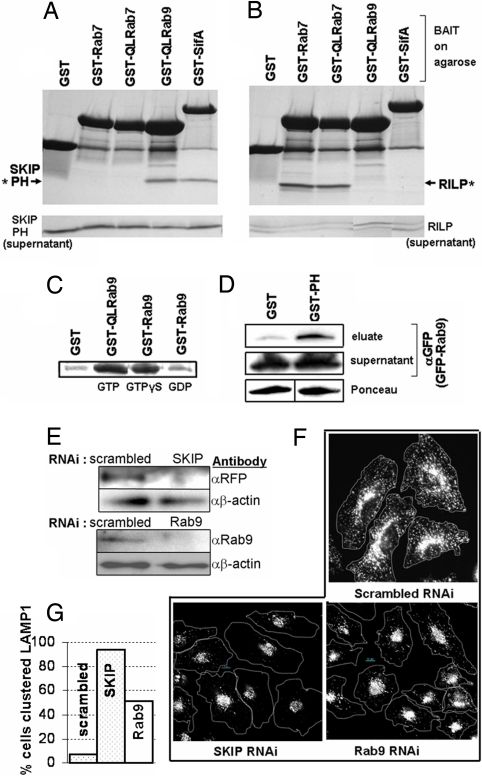
To do this, we bound GST fusions of wild-type or constitutively active (Q67L) Rab7 or constitutively active (Q66L) Rab9 proteins that constantly remain in the GTP-bound active state to agarose beads, loaded them with nonhydrolyzable GTP analogue GTPγS and incubated them with SKIP PH domain protein in pulldown assays. We found that GST-Q66L-Rab9 bound to the SKIP PH domain protein (Fig. 1A), but GST-Q67L-Rab7 did not (Fig. 1A). Control incubations revealed that GST-SifA but not GST alone bound to the SKIP PH domain (Fig. 1A), indicating that SifA binds to SKIP PH directly and confirming that GST-alone had no significant affinity for the SKIP PH domain. Both GST-Q67L-Rab7 and GST-Rab7 proteins were able to bind to a fragment of RILP (amino acid 241 to 320), a known Rab7 effector (Fig. 1B) (20), demonstrating that the Rab7 fusions used in these binding assays were active. Rab7 is the closest Rab9 homologue and is 50% identical to Rab9 (data not shown). Thus its failure to bind to the SKIP PH domain reinforces the idea that the late endosomal Rab9 may specifically bind the SKIP PH domain.
Fig. 1.
Rab9 binds specifically to the PH domain of SKIP, a protein that influences LAMP1 distribution in cells. (A and B) Coomassie stained SDS/PAGE gel shows proteins eluted from washed resin in pulldowns (Upper) or ≈10% of pulldown supernatant (Lower). Resin was loaded with 80 μg of GST, GST-SifA, GST-Q66L-Rab9(GTPγS), GST-Rab7(GTPγS), or GST-Q67L-Rab7(GTP) and incubated with either 53 μg of SKIP PH (amino acids 762–885) (A) or 53 μg of RILP (amino acids 241–320) (B) proteins. Asterisks indicate protein captured and eluted in pulldown. (C) GTP-dependence of binding. SKIP PH from pulldowns is visualized by Coomassie. Resins loaded with 400 μg of GST, GST-Q66L-Rab9, or GST-Rab9 were loaded with the indicated nucleotide and incubated with 400 μg of SKIP PH. (D) SKIP PH can also interact with Rab9 in cell lysates. Resin loaded by using 20 μg of GST or GST-SKIP PH domain was incubated with lysate, created from HeLa cells expressing GFP-Rab9, for pulldown. Approximately 10% of pulldown supernatant is loaded in control blot. (E) Western blots showing knockdown of SKIP or Rab9 protein levels in lysates from cells treated with anti-SKIP, anti-Rab9, or control (scrambled) RNAi. To monitor SKIP knockdown, SKIP RNAi-treated cells were transfected with an RFP-SKIP construct, then lysed and probed by using an anti-RFP antibody. (F) RNAi-treated HeLa cells were fixed, permeabilized, and stained with anti-LAMP1 antibody (white). A dotted line is drawn around cells to indicate the edges. (G) Quantitative analysis of micrographic data, indicating the percentage of cells with clustered LAMP1 in RNAi-treated cells.
The binding of mammalian G proteins to their effectors is GTP dependent. We therefore investigated the GTP dependence of the observed interaction between Rab9 and the SKIP PH domain. To test this, we bound GST or GST-Rab9 loaded with either GTPγS or with GDP to resin and incubated this agarose with SKIP PH domain protein in pulldown assays. We also tested whether GST-Rab9(GTPγS) and GST-Q66L-Rab9(GTP) would yield similar results in this assay. We found that recombinant SKIP PH protein was bound similarly by wild-type GST-Rab9 complexed with GTPγS and by constitutively active GST-Q66L-Rab9 complexed with GTP. However, it was not bound by GST-Rab9 complexed with GDP in vitro (Fig. 1C). The GTP-dependent interaction of Rab9 with SKIP PH domain was also seen in cell lysates (Fig. 1D), indicating that it may occur in a complex cellular milieu. Together this data indicates that SKIP may be a specific Rab9 binding protein.
Prior work has demonstrated that SKIP is needed for maintenance of and/or growth of LAMP1-containing SCV and Sif membranes (12). But the function of SKIP in late endosomal membrane dynamics in the absence of infection is largely unknown. To determine the importance of SKIP and Rab9 to the distribution of LAMP1-containing membranes in uninfected cells, we treated cells with SKIP or Rab9 RNAi and immunostained fixed cells. We find that knockdown of SKIP also leads to reorganization of LAMP1 from the periphery to a perinuclear region in cells (Fig. 1 E–G). As previously reported (21), knock down of Rab9 also promotes perinculear clustering of LAMP1 (Fig. 1 E–G). Thus both SKIP and Rab9 appear to be needed to maintain peripheral LAMP1 dynamics in cells.
SifA Competes with Rab9 for Binding to the SKIP PH Domain and SifA Binding Is Dependent on the WxxxE G Protein Mimic Signature Motif.
Because SifA and Rab9 both bind to the PH domain of SKIP, we next determined whether SifA and Rab9 could bind simultaneously to SKIP or if their binding was mutually exclusive (Fig. 2 A–C). For this binding assay GST-Q66L-Rab9 was bound to agarose and incubated with recombinant SKIP PH in the presence or absence of excess SifA protein. Remarkably, we found that the binding of recombinant SKIP PH domain protein to GST-Rab9 was significantly decreased in the presence of excess SifA (Fig. 2B), suggesting that SifA competes the PH domain away from GST-Rab9 (See supporting information (SI) Fig. S1 for model of these results). Excess SifA was also observed to out compete Rab9 for binding to the SKIP PH domain in cell lysates (Fig. 2C). Results for this pulldown from lysate were visualized by Western blotting. Note that the Ponceau stained blot of the pulldown where GST-PH has been incubated with lysate in the presence of excess SifA has two bands in Fig. 2C, as GST-PH has been bound by equal-molar amount of recombinant SifA protein. Because SifA and Rab9 cannot simultaneously bind to the SKIP PH domain, these data suggest that SifA and Rab9 bind to the same site or to overlapping binding sites on SKIP.
Fig. 2.
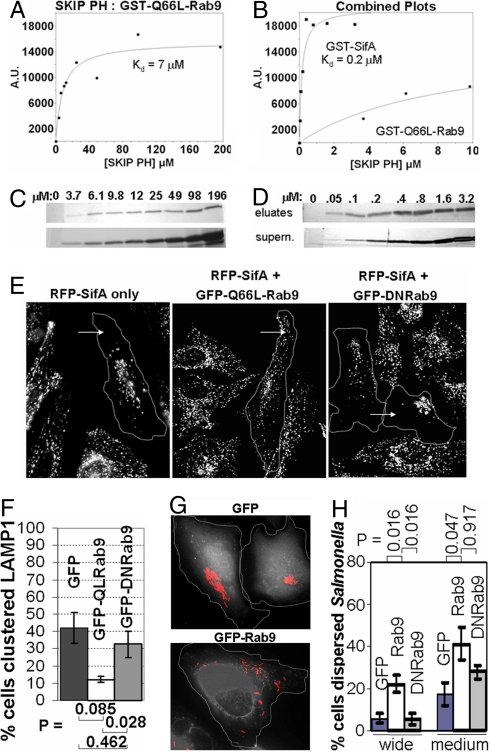
SifA competes with Rab9 for binding to the SKIP PH domain and requires residues of the WxxxE motif for SKIP binding. (A) Individually, SifA and Rab9 can both bind to the SKIP PH domain. (B) Resin loaded with 40 μg of GST-Q66L-Rab9(GTPγS) (Left) or GST (Right) was incubated with 84 μg of SKIP PH ± 350 μg of SifA proteins. Additions of recombinant proteins to incubations are denoted with a plus sign in figures. Proteins were separated by SDS/PAGE and visualized by Coomassie. Approximately 4% of pulldown supernatant is loaded in control blot. (C) SifA/Rab9 competition also occurs in cell lysates. Resin loaded by using 20 μg of GST or GST-SKIP PH was used to probe lysates from HeLa cells expressing GFP-Rab9, in the presence or absence of 100 μg of recombinant SifA protein. Anti-GFP Western blotting was used to visualize GFP-Rab9 in pulldown eluates and supernatants (≈5%). Note that the Ponceau-stained blot of GST-PH incubated with SifA has two bands, as GST-PH has been bound by an equal-molar amount of SifA protein. (D) Residues Trp-197 and Glu-201 of SifA are essential for SKIP PH binding. Resin bound by using 40 μg of GST, GST-SifA, or GST-(W197A,E201A)SifA was incubated with buffer or with SKIP PH for pulldowns. Proteins from eluates (Left) and supernatants (≈10%) (Right) were separated by SDS/PAGE and visualized by Coomassie. Asterisks indicate protein captured and eluted in pulldown.
A family of bacterial G protein mimics have been shown to contain conserved Trp and Glu residues, demonstrated to be essential for activity in proven mimics from this group (4). SifA has been proposed to be a member of this family (4). Thus the corresponding conserved residues in SifA, Trp-197, and Glu-201 were mutated to Ala and the resulting GST-(W197A,E201A)SifA mutant was tested for its ability to interact with recombinant SKIP PH protein (Fig. 2D). For this pulldown assay GST, GST-SifA, or GST-(W197A,E201A)SifA were bound to agarose and incubated with recombinant SKIP PH protein. Wild-type GST-SifA was found to bind to SKIP PH but GST-(W197A,E201A)SifA failed to bind (Fig. 2D), demonstrating the importance of these residues for SifA binding to SKIP. Because expression levels of soluble, full-length fusion protein in E. coli are lower for GST-(W197A,E201A)SifA compared to wild-type GST-SifA, we considered that protein stability may be affected in this mutant (Fig. S2A). However, we were able to obtain sufficient quantities of protein for our assays and have confirmed by dynamic light scattering that the purified mutant and wild-type SifA proteins have similar molecular assemblies in solution and that the protein is not forming large aggregates (Fig. S2B). Our data here demonstrate that conserved residues, important for utilization of downstream effectors by known G protein mimics in this family, are also vital to SifA's interaction with SKIP. However, the combined results of Figs. 1–3 support that SifA functions not as G protein mimic but rather as a G protein antagonist. The data provide the first suggestion in a direct binding assay that the WxxxE motif mutation inactivates bacterial protein function by preventing binding to the host effector.
Fig. 3.
SifA binds more tightly to SKIP PH than Rab9, but SifA-dependent cellular phenotypes can be antagonized by Rab9 overexpression. (A–D) For SKIP PH pulldowns, resin loaded with GST-Q66L-Rab9 (A and C) or with GST-SifA (B and D) was incubated with the indicated concentrations of recombinant SKIP PH protein. SKIP PH protein from pulldown elutions or supernatants (≈10%), was separated by SDS/PAGE and visualized by Coomassie (GST-Q66L-Rab9) or by silver stain (GST-SifA). Eluted SKIP PH protein was quantified by densitometric scanning, and net intensity (A.U.) (y axis) was plotted vs. concentration of SKIP PH added ([SKIP PH]) (x axis). (A and B) Plotted results with data points (filled circles) and fits (solid lines). For comparison, part of the Rab9 binding curve from A was also superimposed onto B. (E) HeLa cells coexpressing either GFP, GFP-Q66L-Rab9, or GFP-DNRab9 were fixed and stained for LAMP1 (white). Transfected cells are outlined with a dotted line. (F) Bar plot showing quantitation of percentage of cells with altered peripheral LAMP1 distribution. (G) HeLa cells expressing either GFP, GFP-Rab9, or GFP-DNRab9 (white) were infected with mCherry Salmonella (red). (H) Bar plot showing percentage of HeLa cells with medium or wide dispersal of Salmonella for cells overexpressing GFP, GFP-Rab9 or GFP-DNRab9 (n = 5 experiments) (see Methods). P values were obtained by using the Wilcoxon–Mann–Whitney test. Error bars are represented as ± SEM.
The SifA:SKIP Interaction Is Higher in Affinity than that of Rab9:SKIP and Excess Rab9 Can Reverse SifA-Induced LAMP1 Recruitment as Well as SCV Position in Cells.
Because the presence of SifA inhibits binding of Rab9 to the SKIP PH domain, we wished to determine relative affinities of SifA for this SKIP domain. To do this, we performed a binding titration to obtain estimated binding constants for the association between recombinant GST-Q66L-Rab9 and the SKIP PH domain as well as between GST-SifA and SKIP PH. We bound GST-Q66L-Rab9, GST-SifA or GST control proteins to agarose, incubated the agarose in the presence of various concentrations of SKIP PH, and used results of the respective pulldowns to set up binding curves. We found that GST-Q66L-Rab9 binds to the SKIP PH domain with an estimated Kd of 7 μM (7.4 ± 3.8 μM) (n = 3). In contrast, GST-SifA binds to SKIP PH much more tightly, with an estimated Kd of 0.2 μM (0.18 ± 0.07 μM Kd) (n = 3) (Figs. 3 A–D). Thus GST-SifA binds to SKIP PH more than an order of magnitude more tightly than GST-Q66L-Rab9 does. This may explain why SifA can efficiently recruit SKIP in the host cell.
We next investigated whether we could reverse well established SifA-dependent phenotypes in cells. To do this, we transfected HeLa cells with rfp-sifa in presence or absence of gfp, gfp-rab9, and gfp-dnrab9. As indicated earlier, LAMP1 is a transmembrane glycoprotein that localizes primarily to lysosomes and to late endosomes and is detected in punctuate spots dispersed throughout the cell including its periphery. Further, SifA recruits LAMP1 which becomes clustered into perinuclear regions of the cells (22) (Fig. 3E). This clustering can be significantly reversed by overexpression of GFP-Rab9 (Fig. 3 E–F) but not by GFP or GFP-DNRab9.
We also investigated whether it would be possible to reverse infection phenotypes such as position and stability of the SCV and Sif production, by overexpressing GFP-Rab9 in infected cells. To do this, we transfected HeLa cells with gfp, gfp-rab9, and gfp-dnrab9, infected these cells with Salmonella typhimurium expressing a fluorescent monomeric mCherry reporter, and analyzed infected, transfected cells for proliferation of LAMP1-positive bacteria within the SCV and for Sif production. As indicated earlier, upon Salmonella infection LAMP1 is recruited to SCV and Sif membranes (7, 22–24). Our data indicate that GFP-Rab9 expression had no obvious impact on SCV integrity, based on LAMP1-staining of intracellular bacteria (Fig. S3). There was also little effect on bacterial proliferation in gfp-rab9 and gfp-dnrab9 transfected, infected cells compared to gfp controls (Fig. S4). There was however a measurable effect on the position of the SCV (Fig. 3 G–H) as well as the number of Sif filaments in infected cells (Fig. S5 and Table S1).
To examine the effect of Rab9 overexpression on SCV position, we infected transfected cells and determined the extent to which bacteria were found in a perinuclear foci within them. Prior studies have shown that Salmonella proliferating in SCV are associated in a tight perinuclear region and that this localization depends on both SKIP and SifA, which is likely because of the recruitment of SKIP, by SifA, to the SCV (12). We found that in approximately 20% of cells over expression of GFP-Rab9 resulted in wide dispersion of bacteria through the cell, compared to approximately 6% for GFP controls. Further a moderate dispersion was detected in approximately 40% of cells compared to approximately 17% for GFP controls, suggesting that Rab9 can influence motor dynamics of the SCV.
To examine the effects on Sifs in further detail, we determined the percentage of transfected, infected cells with Sifs on the basis of Sif length. At Sif sizes of greater than or equal to 3, 5 or 10 μm, we detected approximately 30–40% reduction in fraction of infected cells expressing Sifs that could be ascribed to the GTPase activity of Rab9 (P < 0.05; Table S1). Greater Sif reductions were observed at longer Sif lengths (≥15–20 μm) but they were not statistically significant. Smith et al. report that DNRab9 reduced the percentage of infected cells with Sifs compared to cells expressing CFP alone (16). At longer Sif lengths we do see a decrease in Sifs with DNRab9, but a greater decrease is observed with wild-type Rab9 overexpression. It should be noted that Smith et al., did not report the effects of wild-type Rab9 expression on Sif prevalence.
Previous studies and our data reported here have shown that in uninfected cells, Rab9 can control late endosome/lysosomal location in cells (21). We show that Rab9 may influence LAMP1 distribution ascribed to SifA. The SifA effect on SCV localization and Sifs per se during infection is altered by Rab9 overexpression. It is possible that the greater affinity of SifA relative to Rab9 for SKIP, in conjunction with additional bacterial factors, may render reversal of either process inefficient during infection to favor pathogenic proliferation in the SCV.
Discussion
Our data identify an unexpected interaction between two mammalian proteins, SKIP and a small GTPase Rab9, and demonstrate competition for this SKIP interaction by the Salmonella virulence protein SifA. Our data also suggest that the binding of SifA to SKIP (SifA's major host target protein) is a consequence of G protein antagonism. Dixon and coworkers reported that bacterial Type III effector proteins IpgB2, IpgB1 from Shigella and Map from E. coli, when secreted into host cells, function as GTPase mimics in that they use downstream effectors of host GTPases RhoA, Rac1 and Cdc42, without requirement for GTP (4). They further used bioinformatics to predict a diverse and larger family of 24 proteins detected in pathogenic bacteria such as enteropathogenic E. coli, Shigella, Citrobactor and Salmonella. This larger family included SifA (4), an effector known to be essential for intracellular replication of Salmonella in macrophages (8, 22, 25), but there was no knowledge on whether and how it mimicked a G protein.
We propose that SifA antagonizes Rab9 by binding to the SKIP PH domain. PH domains are best known for their binding to phosphoinositides but have been reported to interact with several different types of protein ligands, including a growing list of small G proteins: Arf1, RhoA, Rac1, and Cdc42 (reviewed in 19). Whether the SKIP PH domain:Rab9 interaction occurs in conjunction with phosphoinositides is not known, and the SKIP PH domain is the first shown to be bound to a RabGTPase. Measured affinities for Rab GTPases and their effectors range from nM to low μM Kd (21, 26–28). Further a Kd of approximately 10 μM has been reported for an analogous interaction between Rac GTPases 1, 2, and 3, and the PH domain of phospholipase C β2 protein (PLC-β2) (29) and more recently a crystal structure of the PLC-β2 PH:Rac 1 complex has been determined (30). Our estimated Kd of 7 μM for the Rab9:SKIP PH interaction is within the range of affinities observed for Rab GTPases and their effectors.
Rab9 is best known for its role in recycling MPR protein from late endosomes to Golgi, but recently has also been shown to be important for LAMP1 distribution in cells (21). Like Rab9, SKIP too can influence dynamics of the major late endosomal membrane protein LAMP1. Recruitment of LAMP1, the position of the SCV and formation and/or stability of LAMP1-rich Sifs is mediated by the interaction of SifA and SKIP during infection (12). We show that this can be negatively impacted by Rab9. We therefore hypothesize that both SKIP and Rab9 are needed to sustain proper dynamics of LAMP1. A comprehensive analysis of membrane and luminal cargoes dependent on the Rab9:SKIP PH interaction has yet to be undertaken. SifA and SKIP are known to be important for the generation of LAMP1- and cholesterol-rich (24) Sifs (12) but the details of how this occurs at the molecular level are not known. Rab7, which regulates LAMP1 transport, is pulled down by SifA in cellular lysates (17). This interaction likely occurs via SKIP although not via a direct interaction with the SKIP PH domain. Future studies may reveal whether SifA and/or Salmonella increase the interaction of SKIP with Rab7 and/or its effectors/membrane cargo through additional SKIP domains. SKIP knockdown is also reported to influence Golgi dynamics in cells (12). By binding Rab9 at its PH domain and kinesin and/or putative Golgi effectors at the RUN domain, SKIP may bridge aspects of late/endosomes and Golgi trafficking in cells.
Although prior studies have presented evidence that the bacterial G protein mimic IpgB2 interacts with Rho-kinase to use downstream effector pathways, this is a unique report of the Kd of one such interaction in a direct binding assay. We estimate a 0.2 μM Kd for the interaction between the bacterial G protein mimic SifA and its host target SKIP. Our finding that Rab9 fails to bind SKIP PH domain in the presence of SifA is then well explained by the fact that the estimated Kds for the Rab:SKIP PH interaction is more than a log higher at approximately 7 μM. Thus the Rab9:SKIP PH interaction may be easily disrupted by SifA in cells in an effective strategy of antagonism by Salmonella that cannot be easily reversed during infection. In addition, because the Kd for the interaction of Rab9:TIP47 (Tail interacting protein of 47 kDa) involved in the recycling of the mannose-6-phosphate receptor from the late endosomes to the Golgi is reported to be 0.097 μM (21), transport events in this pathway may not be affected by Rab9:SKIP interactions targeted by SifA.
The larger family of 24 proteins predicted to be G protein mimics in pathogenic bacteria contain a positionally conserved motif of WxxxE (4). Mutational analysis revealed that the tryptophan and glutamic acid residues were essential to G protein mimicry by IgpB1/B2 and Map, suggesting that the whole family functions by a conserved molecular mechanism (4). Our data with SifA suggest that these residues are critical for the binding interaction with the host target protein and leads to speculation that competitive displacement by a partial bacterial G protein mimic may also allow for G protein antagonism. Further the affinity of a bacterial G protein mimic for its host target may be the primary determinant in establishment of a wide range of host-pathogen interactions.
Materials and Methods
Clones.
GFP-hRab7 and GFP-hRab9 constructs were gifts from the Richard E. Pagano lab (Mayo Clinic, Rochester, MN). The cDNA for skip (KIAA 0842) was from Kazusa DNA Research Institute. The sifA ORF was PCR amplified from S. typhimurium strain SL1344. The cDNA for human rilp was from Invitrogen (FL1001). rilp was subcloned amino acid 241 to 320 (20), and the skip PH domain was subcloned amino acid 762 to 885 (12). Details of all subcloning, mutagenesis, and protein expression are provided in SI Methods and Tables S2 and S3.
Recombinant Protein Pulldowns.
GST-fusion protein was bound to Glutathione Agarose Beads (BD Biosciences) by incubating protein and beads overnight at 4°C in binding buffer [150 mM NaCl, 20 mM Tris (pH 7.5), 2 mM DTT, and 0.1% Triton X-100]. Beads were then washed three times with binding buffer to remove unbound protein. GTPase fusions were loaded with nucleotide on resin as described in SI Methods (31). 30 μl of packed resin was used in a typical binding assay with a 200-μl total reaction volume. Agarose complexed with GST-fusion protein was incubated with recombinant tagless test protein (SKIP PH ± SifA, or RILP) in the presence of protease inhibitors (Complete mini EDTA-free, Roche) for an hour at 4°C. After incubation, the agarose was pelleted at low speed, the supernatant was removed, and the agarose was washed to remove unbound test protein. Proteins were eluted by boiling with 20 μl of SDS sample buffer, separated by SDS/PAGE, and visualized by Coomassie or silver stain as appropriate. The amounts of protein used are indicated in figure legends. For the in vitro competition assay (Fig. 2B), recombinant proteins were added to an approximate molar stoichiometry of 1:5.5:11 for GST-Q66L-Rab9:SKIP PH:SifA proteins.
RNAi and Westerns.
SKIP and scrambled control oligos were decribed by Boucrot et al. (12), and for Rab9 we used: 5′-GGGACAACGGCGACUAUCCUUAUUU-3′. RNAi was performed by using oligofectamine (Invitrogen) according to manufacturer instructions and proceeded for 72 h. For RFP-SKIP transfected cells, cells were treated with RNAi 24 h before RFP-SKIP plasmid transfection and then grown an additional 48 h. Lysates supernatants were probed by using a rabbit anti-RFP antibody (Chemicon AB3216) (1:500). Total cell lysate was probed for Rab9 using a monoclonal mouse anti-Rab9 antibody (SIGMA R5404) (1:200).
Transfection and Infection of HeLa Cells and IFAs.
HeLa cells were plated and transfected with gfp, gfp-rab9, or gfp-dnrab9 (dominant negative, S21N rab9) and/or rfp-sifa. Thirty-six hours after transfection, the cells were infected with S. typhimurium (SL 1344) expressing an mCherry fluorescent protein (32) for 10–12 h. Procedures for HeLa cell infection and for generation of mCherry Salmonella are provided in SI Methods. Ten to twelve hours after infection, cells were washed in PBS and fixed by using 4% paraformaldehyde. Cells were immunostained (SI Methods) and imaged (33) by using mouse anti-hLAMP1 (H4A3-c, Developmental Studies Hybridoma Bank), and a FITC or Cy5-conjugated Anti-Mouse IgG secondary (Jackson Laboratories). To quantitate Salmonella dispersal, in cells containing more than 10 bacteria, a cell was counted as having widely dispersed Salmonella if bacteria were found in all four quadrants of a cell. Medium dispersal includes all cells with bacteria that are less clustered together than typically seen.
Pulldowns from Cell Lysates.
Bait agarose was prepared as above, by using 20 μg of GST or GST-SKIP PH protein and 30 μl of agarose per reaction. HeLa cells transiently expressing GFP-Rab9 were lysed in Triton Lysis Buffer: 1% Triton X-100, 150 mM NaCl, 20 mM Tris (pH 7.5), 1 mM PMSF and Protease inhibitors (Complete mini, Roche) (see SI Methods). Bait agarose was incubated with precleared lysate in the presence of protease inhibitors and 1 mM GTPγS for 2 h at 4°C and pelleted at low speed, the supernatant was removed, and the agarose was washed to remove unbound lysate proteins. Complexes were eluted by boiling with SDS sample buffer, separated by SDS/PAGE, and visualized by Western blot by using rabbit anti-GFP antibody (Molecular Probes) (1:400) and goat anti-Rabbit IgG-HRP conjugate (Bio-Rad) (1:4000). For competition experiments, 100 μg of recombinant SifA protein or an equal volume of buffer was added to the agarose/lysate pulldown mixtures.
Binding Titration and Analysis.
Proteins were complexed to agarose beads as above by using 1–2 μg of GST-SifA or 20 μg of GST-Q66L-Rab9, with 30 μl of beads per reaction. Less than half of the GST-fusion protein added, bound to the beads after an overnight incubation. SKIP PH protein was varied from 0.05 to 6.4 μM for GST-SifA and from 3.6 μM to 196 μM for GST-Q66L-Rab9(GTPγS). Proteins were separated by SDS/PAGE and visualized by silver stain for GST-SifA pulldowns and by Coomassie for GST-Q66L-Rab9 pulldowns. Gels were scanned by using the UVP Bioimaging System (UVP) with LabWorks version 4.0 (Ultra-Violet Products) software. SKIP PH bands in the eluate were quantified by using the Kodak Scientific Imaging System. Data were fit to the equation Kd = (IntensityMax·[PH])/(Kd + [PH]) by using TableCurve 2D version 5.01 software (SYSTAT Software).
Supplementary Material
Acknowledgments.
We thank Julia MacKenzie for assistance with statistical analyses, Dr. Ivan Vorontsov for assistance with dynamic light scattering and Dr. Anjen Chenn and Haldar laboratory members for helpful editorial suggestions. The anti-LAMP1 antibody (H4A3) developed by J. T. August and J. E. K. Hildreth was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. This work was supported by the VA merit award (to K.H.). K.H. acknowledges membership in and support from the Region V Great Lakes Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (National Institute of Allergy and Infectious Diseases Award 1-U54-057153).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801872105/DCSupplemental.
References
- 1.Aktories K, Barbieri JT. Bacterial cytotoxins: Targeting eukaryotic switches. Nat Rev Microbiol. 2005;3:397–410. doi: 10.1038/nrmicro1150. [DOI] [PubMed] [Google Scholar]
- 2.Patel JC, Galan JE. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J Cell Biol. 2006;175:453–463. doi: 10.1083/jcb.200605144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murata T, et al. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 4.Alto NM, et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 5.Garcia del-Portillo F, Finlay BB. Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6-phosphate receptors. J Cell Biol. 1995;129:81–97. doi: 10.1083/jcb.129.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salcedo SP, Holden DW. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 2003;22:5003–5014. doi: 10.1093/emboj/cdg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beuzon CR, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein MA, Leung KY, Zwick M, Garcia del-Portillo F, Finlay BB. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol Microbiol. 1996;20:1–164. doi: 10.1111/j.1365-2958.1996.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz-Albert J, et al. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol Microbiol. 2002;44:645–661. doi: 10.1046/j.1365-2958.2002.02912.x. [DOI] [PubMed] [Google Scholar]
- 11.Garcia del-Portillo F, Zwick MB, Leung KY, Finlay BB. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc Natl Acad Sci USA. 1993;90:10544–10548. doi: 10.1073/pnas.90.22.10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boucrot E, Henry T, Borg J-P, Gorvel J-P, Méresse S. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science. 2005;308:1174–1178. doi: 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- 13.Brumell JH, Scidmore MA. Manipulation of Rab GTPase function by intracellular bacterial pathogens. Microbiol Mol Biol Rev. 2007;71:636–652. doi: 10.1128/MMBR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: Achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Méresse S, Steele-Mortimer O, Finlay BB, Gorvel JP. The Rab7 GTPase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. EMBO J. 1999;18:4394–4403. doi: 10.1093/emboj/18.16.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith AC, et al. A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J Cell Biol. 2006;176:263–268. doi: 10.1083/jcb.200611056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison RE, et al. Salmonella impairs RILP recruitment to Rab7 during maturation of invasion vacuoles. Mol Biol Cell. 2004;15:3146–3154. doi: 10.1091/mbc.E04-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lombardi D, et al. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemmon MA. Pleckstrin homology domains: Not just for phosphoinositides. Biochem Soc Trans. 2004;32:707–711. doi: 10.1042/BST0320707. [DOI] [PubMed] [Google Scholar]
- 20.Wu M, Wang T, Loh E, Hong W, Song H. Structural basis for recruitment of RILP by small GTPase Rab7. EMBO J. 2005;24:1491–1501. doi: 10.1038/sj.emboj.7600643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganley IG, Carroll K, Bittova L, Pfeffer S. Rab9 GTPase regulates late endosome size and requires effector interaction for its stability. Mol Biol Cell. 2004;15:5420–5430. doi: 10.1091/mbc.E04-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brumell JH, Rosenberger CM, Gotto GT, Marcus SL, Finlay BB. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell Microbiol. 2001;3:75–84. doi: 10.1046/j.1462-5822.2001.00087.x. [DOI] [PubMed] [Google Scholar]
- 23.Boucrot E, Beuzon CR, Holden DW, Gorvel J-P, Meresse S. Salmonella typhimurium SifA effector protein requires its membrane-anchoring C-terminal hexapeptide for its biological function. J Biol Chem. 2003;278:14196–14202. doi: 10.1074/jbc.M207901200. [DOI] [PubMed] [Google Scholar]
- 24.Nawabi P, Catron D, Haldar K. Esterification of cholesterol by a type III secretion effector during intracellular Salmonella infection. Mol Microbiol. 2008 doi: 10.1111/j.1365-2958.2008.06142.x. in press. [DOI] [PubMed] [Google Scholar]
- 25.Chan K, Kim CC, Falkow S. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect Immun. 2005;73:5438–5449. doi: 10.1128/IAI.73.9.5438-5449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukuda M. Distinct Rab27A binding affinities of Slp2-a and Slac2-a/melanophilin: Hierarchy of Rab27A effectors. Biochem Biophys Res Commun. 2006;343:666–674. doi: 10.1016/j.bbrc.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Junutula JR, et al. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J Biol Chem. 2004;279:33430–33437. doi: 10.1074/jbc.M404633200. [DOI] [PubMed] [Google Scholar]
- 28.Merithew E, Stone C, Eathiraj S, Lambright DG. Determinants of Rab5 interaction with the N terminus of Early Endosome Antigen 1. J Biol Chem. 2003;278:8494–8500. doi: 10.1074/jbc.M211514200. [DOI] [PubMed] [Google Scholar]
- 29.Snyder JT, Singer AU, Wing MR, Harden TK, Sondek J. The pleckstrin homology domain of phospholipase C-β2 as an effector site for Rac. J Biol Chem. 2003;278:21099–21104. doi: 10.1074/jbc.M301418200. [DOI] [PubMed] [Google Scholar]
- 30.Jezyk MR, et al. Crystal structure of Rac1 bound to its effector phospholipase C-β2. Nat Struct Mol Biol. 2006;13:1135–1140. doi: 10.1038/nsmb1175. [DOI] [PubMed] [Google Scholar]
- 31.Hoshino M, et al. Interaction of ras oncogene product p21 with guanine nucleotides. J Biochem (Tokyo) 1987;102:503–511. doi: 10.1093/oxfordjournals.jbchem.a122082. [DOI] [PubMed] [Google Scholar]
- 32.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharjee S, van Ooij C, Balu B, Adams JH, Haldar K. Maurer's clefts of Plasmodium falciparum are secretory organelles that concentrate virulence protein reporters for delivery to the host erythrocyte. Blood. 2007;111:2418–2436. doi: 10.1182/blood-2007-09-115279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.