INTRODUCTION
Genome sequencing projects have dramatically increased the amount of sequence data available in public databases, providing an opportunity to identify new protein families. The definition of new protein families allows knowledge from studies of one or a few members of a protein family to be applied to many other proteins in the family. The analysis presented here defines an actin-binding module, the actin-depolymerizing factor homology (ADF-H) domain, which is present in each member of a newly identified, extensive protein family. This family consists of three phylogenetically distinct classes: the ADF/cofilins, the twinfilins, and the drebrin/Abp1s. Each class has been used by eukaryotic organisms for more than 1 billion years, since before the divergence of fungi and animals. The ADF-H domain can be considered a functional building block that is arranged differently in each of the three protein classes in the family. In addition to being evolutionarily distinguishable, proteins in each class seem to possess distinct biochemical properties. Thus, ancient duplication of the ADF-H domain allowed the development of functional diversity.
THE ADF-H DOMAIN PROTEIN FAMILY
To regulate the structure and dynamics of the actin cytoskeleton, a plethora of actin-binding proteins has evolved. As the sequence, structural and biochemical data on actin-binding proteins has accumulated, it has become evident that many actin-binding proteins have evolved through the duplication and mutation of DNA sequences encoding a relatively small number of protein motifs or modules (for definitions of motifs and modules, see Henikoff et al., 1997) that interact with monomeric (G) or filamentous (F) actin in a specific manner. Three widely occurring actin-binding motifs and modules that govern the structure and dynamics of the actin cytoskeleton have been described to date: 1) the calponin homology domain, 2) the thymosin-β4 motif, and 3) the gelsolin homology domain (Van Troys et al., 1996, Puius et al., 1998).
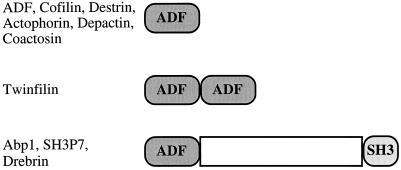
A fourth group of widely occurring actin-binding proteins with sequence and biochemical similarity is the ADF/cofilin group, which facilitates the rapid in vivo disassembly of actin filaments (for a recent review, see Theriot, 1997). Using a comprehensive sequence database search, we have identified 39 proteins that contain a module (an ADF-H domain) with sequence similarity to ADF/cofilins. Examination of the primary structures of these proteins shows that the ADF-H domain protein family can be subdivided into three structurally distinct classes, which we have termed the ADF/cofilins, the twinfilins, and the drebrin/Abp1s (Figure 1).
Figure 1.
Domain structures of the three classes of ADF-H domain proteins. The ADF/cofilin proteins are composed of a single ADF-H domain. Twinfilins are composed of two ADF-H domains arranged in tandem. Members of the drebrin/Abp1 class have an ADF-H domain at the N-terminus of the protein, followed by a variable region and a C-terminal SH3 domain.
ADF/Cofilins
The ADF/cofilins are composed of a single ADF-H domain (Figure 1) and include ADF, cofilin, destrin, actophorin, coactosin, and depactin. ADF/cofilin proteins can interact with both actin monomers and filaments with dissociation constants in the submicromolar range (reviewed by Moon and Drubin, 1995; Theriot, 1997). Furthermore, upon binding to actin filaments, cofilin increases filament twist (McGough et al., 1997) and promotes the turnover of the filaments by increasing the koff rate of the monomers at the minus end of actin filaments (Carlier et al., 1997). Cofilin-induced filament depolymerization provides an essential element of actin dynamics in vivo (Lappalainen and Drubin, 1997), and thus these proteins are required for viability in yeast, worms, and flies (Moon et al., 1993, McKim et al., 1994, Gunsalus et al., 1995).
Twinfilins
To our surprise, sequence database searches revealed proteins that are remarkable in that they are composed of two ADF-H domains (Figure 1). Genes encoding these proteins, which we have named the twinfilins, are found in humans, mice, and yeast (and recently a cDNA clone encoding a probable twinfilin has been obtained from Dictyostelium discoideum [R. Insall, personal communication regarding the University of Tsukuba Dictyostelium cDNA project]). Human twinfilin had been originally identified by screening an embryonic fibroblast expression library with an anti-phosphotyrosine antibody and was named A6. Bacterially expressed A6 was reported to phosphorylate exogenous substrates on tyrosine residues (Beeler et al., 1994). This protein, however, lacks any of the motifs commonly conserved in the catalytic domains of protein kinases, raising doubts about its identification as a protein kinase. Our recent studies have shown that yeast twinfilin localizes to the cortical actin cytoskeleton and that purified twinfilin sequesters actin monomers by forming a tight 1:1 complex with actin monomers (Goode et al., 1998). Thus, twinfilin is indeed a bona fide actin-binding protein. We find no evidence for cross-linking of actin filaments by twinfilin. However, the presence of both ADF-H domains confers tighter actin monomer binding than is observed when only the first domain is expressed (Goode et al., 1998).
Drebrin/Abp1s
Proteins of the drebrin/Abp1 class have a single ADF-H domain at their N-termini, followed by a nonconserved central region and a C-terminal Src homology 3 (SH3) domain (Figure 1). All available biochemical data to date indicate that members of this class bind to F-actin but not G-actin. Yeast Abp1 is linked to the Ras/adenylate cyclase signaling pathway via interactions with Srv2/CAP (Freeman et al., 1996, Lila and Drubin, 1997). A related protein in mouse, found in our database searches, is expressed in all tissues and binds to F-actin (Kessels, unpublished results). This protein was independently discovered by screening for SH3 domain-containing proteins and was named SH3P7 (Sparks et al., 1996). The drebrins are neuron-specific F-actin-binding proteins proposed to provide plasticity to the cytoskeleton and to serve as intracellular regulators of morphogenesis (Ishikawa et al., 1994, Shirao 1995).
EVOLUTION OF THE ADF-H DOMAIN
A protein sequence alignment and subsequent phylogenetic analysis of all ADF-H domains currently present in the public databases group these domains in a manner (Figures 2 and 3A) that agrees with the classification described above, which was based on the domain arrangement of each class of ADF-H domain protein (Figure 1). Based on this analysis, the ADF-H family consists of three phylogenetically distinct classes, the ADF/cofilin class, the drebrin/Abp1 class, and the twinfilin class. Interestingly, members of each class are found in organisms as diverse as mammals and yeast, indicating that each was already present in the common ancestor of all of these organisms. Coactosin from D. discoideum and depactin from Asterias amurensis, which are each composed of a single ADF-H domain, are the only proteins whose phylogenetic classification remains somewhat ambiguous. Statistical analysis of the phylogenetic tree (“bootstrapping”) indicates that coactosin and depactin are, in terms of sequence, more closely related to the drebrin/Abp1 class, whereas their biochemical activities (F- and G-actin binding) appear to be more similar to the members of the ADF/cofilin class (Takagi et al., 1988; de Hostos et al., 1993).
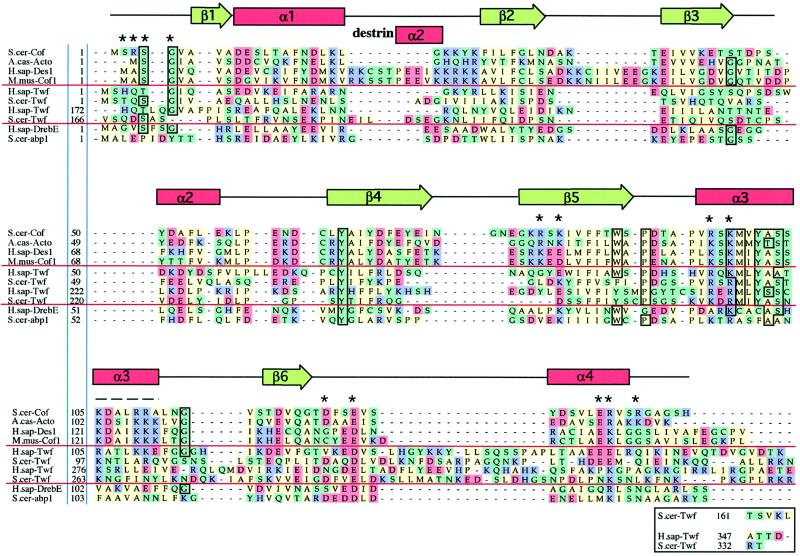
Figure 2.
Sequence alignments of representative examples of ADF-H domains. This alignment was produced essentially according to the methods described for myosin motor domains by Cope et al., (1996). Briefly, the collected sequences were subjected to an initial alignment using the Clustal-W program (Thompson et al., 1994). This initial alignment was refined, thus defining a core ADF-H domain. Acidic (D and E), basic (K, R, and H), uncharged nonpolar (A, I, M, V, L, F, W, and P), and other residues (Y, T, S, G, N, Q, and C) have been colored in red, purple, green, and yellow, respectively. The residues that are >75% conserved throughout the entire ADF-H domain family are boxed. Dashes indicate positions occupied by residues from other ADF/cofilin proteins within the full alignment. The residues shown to be essential for interactions of yeast cofilin with actin (Lappalainen et al., 1997) are indicated by asterisks above the sequences, and the region that has been shown to be important for actin interactions by peptide inhibition studies (Yonezawa et al., 1989) is shown by a dashed line above the sequences. The positions of secondary structure elements based on the yeast cofilin crystal structure (Fedorov et al., 1997) and the nuclear magnetic resonance structure of human destrin (Hatanaka et al., 1996) are shown above the sequences. Protein names, database, and accession numbers for the sequences, respectively, are listed in the legend to Figure 3.
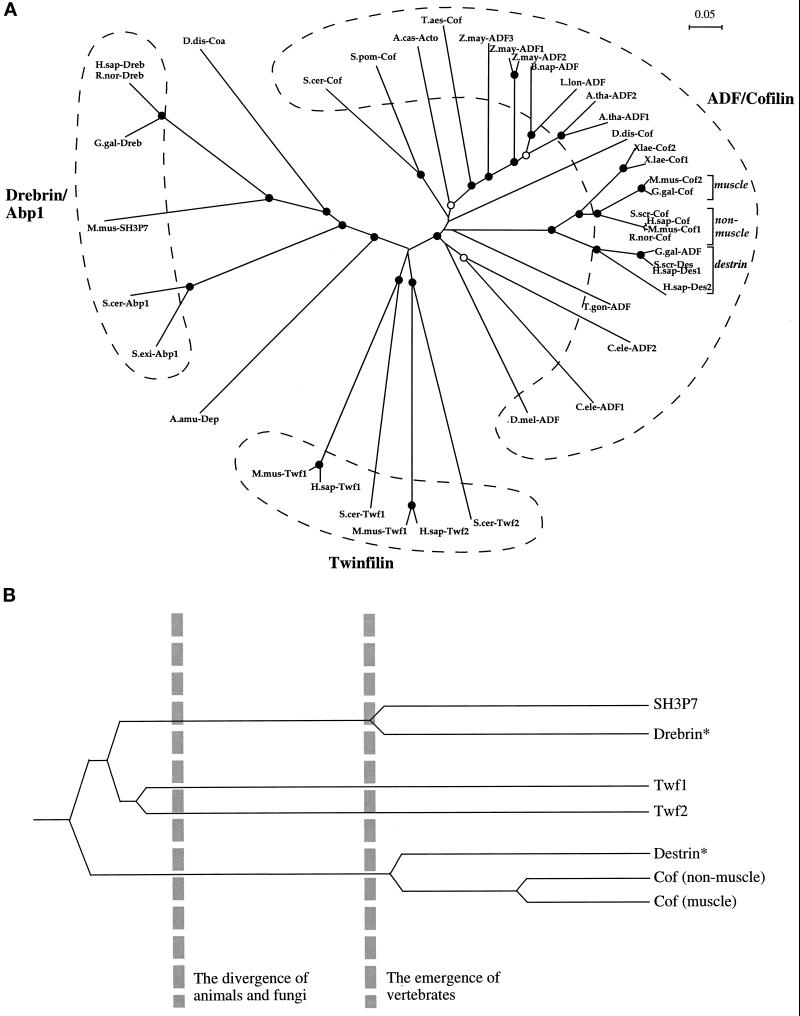
Figure 3.
(A) An unrooted phylogenetic tree of ADF-H domains. This tree was produced by subjecting the alignment depicted in Figure 2 to analysis by the Clustal-W software package (Thompson et al., 1994). An allowance was made for multiple substitutions (Kimura, 1983). Information from intervals in the alignment for which gaps are found in some sequences was included. (Note: the tree architecture is almost identical if gaps are omitted.) The tree was tested (1000 trials) for branching order confidence by bootstrapping (Felsenstein, 1985). Filled circles indicate branch points supported beyond a confidence level of 85%. Empty circles indicate branch points supported beyond the 50% but below the 85% confidence level. Dashed lines indicate the three classes described in this essay. A bar showing 5% divergence is included. Further information on this procedure (as applied to myosin motor domains) can be found on the Worldwide Web at http://www.mrc-lmb.cam.ac.uk/. (B) A simplified, rooted tree depicting the evolution of ADF-H domain proteins in mice. All three families of ADF-H domain proteins were present in the common ancestor of yeast and animals. Distinct members of the ADF/cofilin family in mouse arose after the emergence of vertebrates. The asterisks denote our predictions that destrin- and drebrin-like proteins will be found in mouse, based on the phylogenetic tree shown in A. Protein names, database, and accession numbers for the sequences, respectively, are listed below. Where no database is stated, the accession number refers to GenBank. S. cerevisiae cofilin: Swiss-Prot, Q03048; S. pombe cofilin: DDBJ, D89939; D. discoideum cofilin: Swiss-Prot, P54706; A. castellanii actophorin: Swiss-Prot, P37167; A. thaliana ADF1: U48938; A. thaliana ADF2: U48939; L. longifolium ADF: PIR, S30935; Brassica napus ADF: PIR, S30934; Z. mays ADF1: Swiss-Prot, P46251; Z. mays ADF2: X97725; Z. mays ADF3: X97726; Triticum aestivum cofilin: U58278; Drosophila melanogaster ADF: PIR, A57569; Caenorhabditis elegans ADF1: Swiss-Prot, Q07750; C. elegans ADF2 (Swiss-Prot: Q07749), T. gondii ADF: U62146; H. sapiens destrin 2:(U47924; H. sapiens destrin 1: PIR, A54184; S. scrofa destrin: DDBJ, D90053; Gallus gallus ADF: J02912; R. norvegicus cofilin: Swiss-Prot, P45592; Mus musculus cofilin (nonmuscle isoform): Swiss-Prot, P18760; H. sapiens cofilin: EMBL, X95404; S. scrofa cofilin: M20866; G. gallus cofilin: M55659; M. musculus cofilin (muscle isoform): Swiss-Prot, P45591; X. laevis cofilin 1: U26270; X. laevis cofilin 2: Swiss-Prot, P45593; M. musculus twinfilin (repeat-1): U82324; H. sapiens twinfilin (repeat-1): PIR, A55922; S. cerevisiae twinfilin (repeat-1): SGD, YGR080W; M. musculus twinfilin (repeat-2): U82324; H. sapiens twinfilin (repeat-2): PIR, 55922; S. cerevisiae twinfilin (repeat-2): SGD, YGR080W; H. sapiens drebrin E: Swiss-Prot, Q16643; R. norvegicus drebrin A: Swiss-Prot, Q07266; G. gallus drebrin A, E1 and E2: Swiss-Prot, P18302; M. musculus SH3P7: GenBank, U58884; D. discoideum coactosin: Swiss-Prot, P34121; S. cerevisiae Abp1: EMBL, X51780/Swiss-Prot, P15891; S. exiguus Abp1: Swiss-Prot, P38479; A. amurensis depactin: Swiss-Prot, P20690.
It is important to note that repeats 1 and 2 of twinfilins are distinct from one another and form separate subbranches in the tree (Figure 3). Because each branch contains sequences from such evolutionarily distant organisms as yeast and mammals, it can be concluded that the duplication of the ADF homology domain and the creation of a twinfilin protein was an ancient event that took place (probably only once in evolution) before the divergence of the yeast and animal lineages.
Examination of the phylogenetic trees shown in Figure 3 reveals that all members of ADF/cofilin class have evolved from a single ancestral protein. In multicellular organisms, such as plants and animals, as many as three ADF/cofilin proteins per species have been found. For example, cofilin, destrin 1, and destrin 2 have been identified in Homo sapiens, and ADFs 1–3 have been found in Zea mays. In contrast, there is only a single cofilin in Saccharomyces cerevisiae (and probably in Schizosaccharomyces pombe). These multiple members also arose from single ancestral proteins, distinct examples of which appeared after divergence of the plant and animal kingdoms. In mammals and chicken, the multiple ADF/cofilin proteins fall into at least three distinct subclasses (muscle cofilin, nonmuscle cofilin, and destrin). The gene duplications that gave rise to these three subclasses of vertebrate cofilins took place before divergence of birds and mammals, because the same subclasses exist in both chicken and mammals. Although only up to two of these subclasses have been found to date in a given mammalian species or in chicken, our analysis enables us to predict that at least one member from each of these subclasses will be present in all mammals and all birds. There is an additional subclass of vertebrate ADF/cofilins, made up of two ADF-H proteins, in the toad, Xenopus laevis. It is almost certain that further ADF/cofilins are present in this species as well as members of the drebrin/Abp1 and twinfilin classes.
A simplification of the evolutionary path taken by the ADF-H domain proteins, as determined from the phylogenetic tree, is depicted in Figure 3B. All three classes of these proteins were already present before the divergence of yeast and animals. This implies that each class has performed an important role in eukaryotes for more than 1 billion years. After the emergence of vertebrates, further gene duplication events took place providing additional proteins in the drebrin/Abp1 and the ADF/cofilin classes. These additional proteins presumably helped in the creation of the more complex actin cytoskeletons of multicellular organisms.
STRUCTURE AND FUNCTION OF THE ADF-H DOMAIN
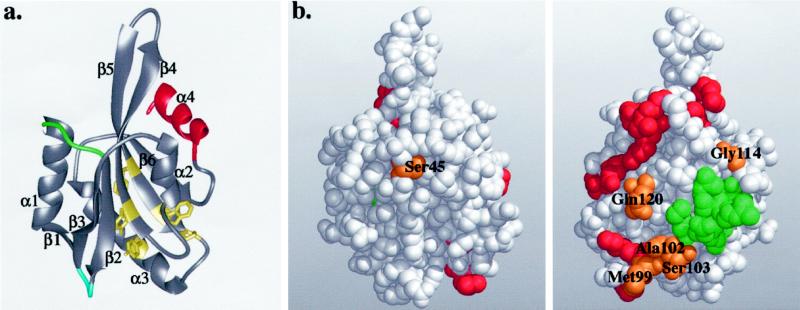
To date, three high-resolution structures of ADF homology domains have been reported. The structure of human destrin was determined by nuclear magnetic resonance (Hatanaka et al., 1996) and was followed by the 2.3-Å resolution x-ray crystal structures of yeast cofilin (Fedorov et al., 1997) and Acanthamoeba actophorin (Leonard et al., 1997). All three structures display a very similar fold, with a central six-stranded mixed β-sheet sandwiched between two pairs of α-helices, one on each face (Figure 4a). The only significant difference between these structures is the presence of an additional α-helix in human destrin between β-strands 1 and 2 (Figures 2 and 4a).
Figure 4.
(a) Ribbon diagram of the yeast cofilin structure. Cofilin has a central mixed β-sheet, which is sandwiched between two pairs of α-helices. The positions of the insertions in mammalian cofilins are in green and blue, and the diverged region in twinfilins is in red. The highly conserved residues that appear to be important for protein stability and correct folding (Tyr64, Phe85, Trp88, Pro90, and Tyr101) are shown in yellow. (b) Space-filling model of yeast cofilin shown in two different orientations (rotated 180° around the y-axis). The residues that are essential for actin interactions in yeast cofilin are highlighted in red. The residues implicated in actin binding by peptide inhibition studies are in green. The highly conserved surface residues (Ser45, Met99, Ala102, Ser103, Gly114, and Gln120) are in orange. All of these residues, except Ser45, are located close to site of cofilin implicated genetically in actin binding, suggesting they also form part of the actin-binding surface. These Figures were produced using Midas Software (University of California San Francisco) running on a Silicon Graphics (Mountain View, CA) Indigo II workstation.
A surprising and remarkable feature of the three-dimensional structures of ADF/cofilin proteins is the similarity of their overall fold to another actin-binding module, the gelsolin homology domain. Members of both actin-binding protein families are built around a central mixed β-sheet and show the same topological connections of secondary structure elements (McLaughlin et al., 1993; Hatanaka et al., 1996).
Because gelsolin consists of multiple segments, and because it appears that different gelsolin segments interact with actin through different interfaces, it is possible that at least one of the gelsolin segments interacts with actin in a manner similar to that of ADF/cofilin proteins. In support of this hypothesis, recent studies have suggested that cofilin and gelsolin segment 2 might interact with actin in a similar manner (Van Troys et al., 1997). In contrast, it appears that the actin-binding surfaces of cofilin and gelsolin segment 1 are significantly different from each other. The gelsolin segment 1–actin interface has been identified from x-ray crystallography studies (McLaughlin et al., 1993). Systematic mutagenesis studies of yeast cofilin suggested that the actin-binding site of cofilin is more extended than the actin-binding site of gelsolin segment 1 and is not as clearly built around the “long” α-helix (α-3 in yeast cofilin) (Lappalainen et al., 1997; Figure 4B). Furthermore, electron microscopy studies on cofilin-decorated actin filaments indicate that the cofilin-binding site on an actin filament is significantly different from the gelsolin segment 1-binding site (McGough et al., 1997). Finally, it should be noted that whereas gelsolin segment 1 is able to interact only with actin monomers (Weeds and Maciver, 1993), members of the ADF/cofilin class of proteins interact tightly with both actin monomers and actin filaments.
Based on the structural homology and the similarity of their biochemical activities (actin binding), it has been proposed that ADF/cofilin proteins and gelsolins have evolved from a common ancestral actin-binding protein (Hatanaka et al., 1996). Primary sequence alignments of ADF/cofilins with gelsolins, however, do not reveal sequence identity between these groups of proteins higher than would be expected between two unrelated proteins. The inclusion of gelsolins in the ADF-H protein family is therefore not justified at this stage, and a meaningful phylogenetic analysis that included gelsolin would be impossible. It must be noted that convergent, as well as divergent, evolution could have given rise to the structural similarities between the proteins. For these reasons, we have in this essay focused only on the three classes of ADF-H domain proteins that clearly evolved from a common ancestral gene.
From the sequence alignment shown in Figure 2 and the structure in Figure 4a, it can be predicted that all ADF-H domains have a similar overall fold. Although, for example, the sequence homology between members of the drebrin/Abp1 and ADF/cofilin classes is low (13–15% identity within and between species), the predicted secondary structure elements identified in the ADF-H domain structural models are well conserved throughout the entire family. Furthermore, predicted loop regions connecting secondary structure elements are less conserved, and insertions are located in the predicted loop regions. Finally, the hydrophobic core elements are well conserved (see below; Figure 4a, residues indicated in yellow).
The sequence alignment also suggests that all ADF-H domains probably interact with actin through a similar interface. The residues essential for the interaction of yeast cofilin with actin (Figures 2, asterisks, and 4b, red) are relatively well conserved in all three classes of ADF-H domain proteins. More specifically, residues Arg96, Lys98, Asp123, and Glu126 (numbering based on positions in yeast cofilin), which are essential for actin monomer and actin filament binding in yeast cofilin (Lappalainen et al., 1997), show extremely high conservation throughout the three protein families. Furthermore, the five N-terminal residues of yeast cofilin, which have been shown to be essential for actin monomer and filament binding, are relatively well conserved in the ADF-H domains (Lappalainen et al., 1997). The most highly conserved residues in this region are Ser4 and Gly5 (numbering based on positions in yeast cofilin), suggesting that these two residues might form an important actin-binding sequence within the N-terminal region of ADF-H domains (the first five residues of cofilin are not shown in Figure 4, because they were found to be disordered in the x-ray crystal structure determination). Interestingly, in vertebrate ADF/cofilin proteins, interactions with actin can be down-regulated by phosphorylation of the aforementioned serine (Agnew et al., 1995). The suggestion that the F-actin interface is conserved, at least between the ADF/cofilin and drebrin/Abp1 classes, is also supported by biochemical data that show that both ADF (Bernstein and Bamburg, 1982) and drebrin (Ishikawa et al., 1994) compete with tropomyosin for actin filament binding.
In addition to the highly conserved residues that have already been shown to be important for the interaction between cofilin and actin, there are also a number of other residues that are well conserved throughout ADF homology domains. These residues fall approximately into two different categories. The first category consists of residues located in the hydrophobic core of cofilin, and these therefore appear to be important for protein stability or correct folding. These residues include Tyr64, Phe85, Trp88, Pro90, and Tyr101 (numbers based on positions in yeast cofilin; Figure 4a, yellow). A critical role of maize ADF residues Tyr82 and Tyr117 (corresponding to residues Tyr64 and Tyr101 in yeast cofilin) in proper protein folding has recently been demonstrated by site-directed mutagenesis (Jiang et al., 1997). The second category of highly conserved residues are those that, because they are exposed on the surface of the protein, may be involved in protein–protein interactions. All of these residues (Met99, Ala102, Ser103, and Gly114) except Ser45 are located close to the actin-binding site as identified by Lappalainen et al., (1997), indicating that these residues might take part in interactions with actin (Figure 4b). Ser45 is located at the opposite side of the molecule. Because this residue is at the end of the β-strand-3 and because this position is in most cases occupied by either serine or glycine, it is possible that a residue lacking a bulky side chain is required at this position for steric reasons. Gln120, which is conserved in all but one of the ADF/cofilin proteins and is replaced by asparagine in drebrins and mSH3P7, but which is not conserved in twinfilins, coactosin, and depactin, is also located close to the actin-binding site of cofilin.
An interesting and instructive sequence (and, by inference, structural) variance among the ADF-H domains is revealed by the alignment. There are two insertions present in the vertebrate ADF/cofilins but not in any other members of the ADF-H domain family. The first insertion is located between α-helix 1 and β-strand-2, and the second insertion is between β-strand-2 and β-strand-3 (Figures 2 and 4a, green and blue loops, respectively). The first insertion forms a short α-helix in human destrin and is always followed by a putative nuclear localization signal (KKRKK) found only in vertebrate ADF/cofilin proteins (Figure 2; note that human destrin-2 is a pseudogene and contains the sequence KKRTK in this region). Nuclear localization of ADF/cofilin has been demonstrated in mammalian cells placed under stress (Ohta et al., 1989). Conceivably, these insertions may play a role in a nuclear function of mammalian cofilins. It is important, however, to note that an identical KKRKK sequence is also found in ActA, a protein that plays central role in the actin-based motility of the intracellular pathogen Listeria monocytogenes. Mutagenesis studies have shown that this lysine-rich sequence in ActA plays an important role in actin filament nucleation (Lasa et al., 1997). It is therefore formally possible that this sequence might have a similar role in mammalian cofilins.
A second structural difference within the ADF-H domain family revealed by our sequence alignment is that β-strand-4 and β-strand-5 within the drebrin/Abp1 class are shortened and/or disrupted by prolines. The cluster of three charged residues preceding Lys81 (based on position in S. cerevisiae) found in all mammalian and avian cofilins is replaced by hydrophobic but flexible amino acids in the drebrin/Abp1 class. Thus, it seems that the rigid handle protruding from the main body of ADF/cofilin is shortened in the entire drebrin/Abp1 family and probably has a different tertiary structure, the functional implications of which need to be explored.
Finally, our sequence alignment shows that the C-terminal regions following β-strand-6 in all twinfilin ADF-H domains are longer and align poorly with other members of ADF-H domain family (Figure 2). It is possible that this region imparts a unique function to these proteins, such as the reported kinase activity (Beeler et al., 1994). Alternatively, the extension that separates the two tandem ADF-H domains in twinfilin might serve as a linker, thereby conferring an appropriate spatial relationship between the two domains.
CONCLUSIONS
In recent years, with the explosive increase in accessible sequence data, it has become apparent that within the sequences of many proteins can be found motifs and modules that are shared with a number of otherwise unrelated proteins. The availability of sequence data for 39 ADF-H domain proteins has provided us with an opportunity to examine the sequence similarities and the evolutionary relationships within this important family of actin-binding proteins. Further such analysis for other cytoskeleton proteins will provide a better appreciation for which protein motifs and modules were the fundamental cytoskeletal building blocks, and how cytoskeletal complexity evolved from these.
We have shown that the ADF-H domain family is made up of at least three different classes of proteins: the ADF/cofilins, the drebrin/Abp1s, and the twinfilins. Each class was represented in organisms existing before the divergence of the animal, plant, and fungal kingdoms. Multiple examples within each class subsequently arose from gene duplication events after this split and may have fulfilled the needs particular to multicellular lifestyles. Only one member of each of these three classes is present in the unicellular yeast S. cerevisiae, whereas several different members of drebrin/Abp1 and ADF/cofilin proteins are found in more complex organisms.
The sequence alignment of ADF-H domain proteins presented in Figure 2 indicates that it is likely that all ADF-H domains have a similar three-dimensional fold because the extra insertions found in some of these proteins are found only between major secondary structural elements and not within them. Furthermore, the residues that have been shown to be essential for actin interactions in the ADF/cofilin family are also well conserved in the other classes, leading to the prediction that all ADF-H domain proteins interact with actin. This conserved module, however, has provided a framework for three subclasses of actin-binding proteins with differing biochemical properties. Although ADF/cofilins are capable of binding both monomeric and filamentous actin, the drebrin/Abp1s seem to bind to filamentous actin only, and S. cerevisiae twinfilin appears to bind to actin monomers only. Crystal structures of members of the twinfilin and drebrin/Abp1 classes should enhance our understanding how this specificity is achieved.
The actin interactions of the members of the ADF/cofilin family and their role in the regulation of actin filament turnover are well established. It is now important to further elucidate the properties of the other two classes and to elucidate their in vivo functions.
ACKNOWLEDGMENTS
We thank Keith Kozminski for comments on the manuscript. This work was supported by grants from the Human Frontier Science Program (to P.L and M.J.T.V.C), Deutsche Forschungsgemeinschaft (to M.M.K) and the National Institutes of General Medical Sciences (grant GM-42759 to D.G.D).
REFERENCES
- Agnew BJ, Minamide LS, Bamburg JR. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J Biol Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- Beeler JF, LaRochelle WJ, Chedid M, Tronick SR, Aaronson SA. Prokaryotic expression cloning of a novel human tyrosine kinase. Mol Cell Biol. 1994;14:982–988. doi: 10.1128/mcb.14.2.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing-factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- Carlier M-F, Laurent V, Santolini J, Melki R, Didry D, Xia G-X, Chua N-H, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1323. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope MJTV, Whisstock J, Rayment I, Kendrick-Jones J. Conservation within the myosin motor domain—implications for structure and function. Structure. 1996;15:969–987. doi: 10.1016/s0969-2126(96)00103-7. [DOI] [PubMed] [Google Scholar]
- de Hostos EL, Bradtke B, Lottspeich F, Gerisch G. Coactosin, a 17 kDa F-actin binding protein from Dictyostelium discoideum. Cell Motil Cytoskeleton. 1993;26:181–191. doi: 10.1002/cm.970260302. [DOI] [PubMed] [Google Scholar]
- Fedorov AA, Lappalainen P, Fedorov EV, Drubin DG, Almo SC. Structure determination of yeast cofilin. Nat Struct Biol. 1997;4:366–369. doi: 10.1038/nsb0597-366. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies—an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Freeman NL, Lila T, Mintzer KA, Chen Z, Pahk AJ, Ren R, Drubin DG, Field J. A conserved proline rich region of the Saccharomyces cerevisiaecyclase associated protein binds SH3 domains and modulates cytoskeletal localization. Mol Cell Biol. 1996;16:548–556. doi: 10.1128/mcb.16.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode, B.L., Drubin, D.G., and Lappalainen, P. (1998). Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin monomer sequestering protein. J. Cell Biol. (in press). [DOI] [PMC free article] [PubMed]
- Gunsalus KC, Bonaccorsi S, Williams E, Verni F, Gatti M, Goldberg ML. Mutations in twinstar, a Drosophilagene encoding a cofilin/ADF homologue, results in defects in centrosome migration and cytokinesis. J Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka H, Ogura K, Moriyama K, Ichikawa S, Yahara T, Inagaki F. Tertiary structure of destrin and structural similarity between two actin-regulating protein families. Cell. 1996;85:1047–1055. doi: 10.1016/s0092-8674(00)81305-7. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Greene EA, Pietrokovski S, Bork P, Attwood TK, Hood L. Gene families: the taxonomy of protein paralogs and chimeras. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Hayashi K, Shirao T, Xue Y, Takagi T, Sasaki Y, Kohama K. Drebrin, a development-associated brain protein from rat embryo causes the dissociation of tropomyosin from actin filaments. J Biol Chem. 1994;269:29928–29933. [PubMed] [Google Scholar]
- Jiang C-J, Weeds AG, Khan S, Hussey PJ. F-actin and G-actin binding are uncoupled by mutation of conserved tyrosine residues in maize actin depolymerizing factor (ZmADF) Proc Natl Acad Sci USA. 1997;94:9973–9978. doi: 10.1073/pnas.94.18.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, United Kingdom: Cambridge University Press; 1983. [Google Scholar]
- Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I, Gouin E, Goethals M, Vancompernolle K, David V, Vandekerckhove J, Cossart P. Identification of two regions in the N-terminal domain of ActA involved in the actin comet tail formation by Listeria monocytogenes. EMBO J. 1997;16:1531–1540. doi: 10.1093/emboj/16.7.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard S, Gittis A, Petrella E, Pollard T, Lattman E. Crystal structure of the actin-binding protein actophorin from Acanthamoeba. Nat Struct Biol. 1997;4:369–373. doi: 10.1038/nsb0597-369. [DOI] [PubMed] [Google Scholar]
- Lila T, Drubin DG. Evidence for physical and functional interactions among two Saccharomyces cerevisiaeSH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol Biol Cell. 1997;8:367–385. doi: 10.1091/mbc.8.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Matheson C, Marra MA, Wakarchuk MF, Baillie DL. The Caenorhabditis elegans unc-60gene encodes protein homologous to a family of actin-binding proteins. Mol Gen Genet. 1994;242:346–357. doi: 10.1007/BF00280425. [DOI] [PubMed] [Google Scholar]
- McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Structure of gelsolin segment-1-actin complex and the mechanism of filament severing. Nature. 1993;364:685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- Moon A, Drubin DG. The ADF-cofilin proteins: stimulus responsive modulators of actin dynamics. Mol Biol Cell. 1995;6:1423–1431. doi: 10.1091/mbc.6.11.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AL, Janmey PA, Louie KA, Drubin DG. Cofilin is an essential component of yeast cortical actin cytoskeleton. J Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Nishida E, Sakai H, Miyamoto E. Dephosphorylation of cofilin accompanies heat shock-induced nuclear accumulation of cofilin. J Biol Chem. 1989;264:16143–16148. [PubMed] [Google Scholar]
- Puius YA, Mahoney NM, Almo SC. The modular structure of actin regulatory proteins. Curr Opin Cell Biol. 1998;10:23–34. doi: 10.1016/s0955-0674(98)80083-5. [DOI] [PubMed] [Google Scholar]
- Shirao T. The roles of microfilament-associated proteins, drebrins, in brain morphogenesis. J Biochem. 1995;117:231–236. doi: 10.1093/jb/117.2.231. [DOI] [PubMed] [Google Scholar]
- Sparks AB, Hoffman NG, McConnell SJ, Fowlkes DM, Kay BK. Cloning of ligand targets—systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- Takagi T, Konishi K, Mabuchi I. Amino acid sequence of starfish oocyte depactin. J Biol Chem. 1988;263:3097–3102. [PubMed] [Google Scholar]
- Theriot JA. Accelerating on a treadmill: ADF/cofilin promotes rapid actin filament turnover in the dynamic cytoskeleton. J Cell Biol. 1997;136:1165–1168. doi: 10.1083/jcb.136.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W—improving the sensitivity Of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys MM, Dewitte D, Goethals M, Carlier M-F, Vandekerckhove J, Ampe C. The actin binding site of thymosin β4 mapped by mutational analysis. EMBO J. 1996;15:201–210. [PMC free article] [PubMed] [Google Scholar]
- Van Troys MM, Dewitte D, Verscelde J-L, Goethals M, Vanderkerckhove J, Ampe C. Analogous F-actin binding by cofilin and gelsolin segment-2 substantiates their structural relationship. J Biol Chem. 1997;272:32750–32758. doi: 10.1074/jbc.272.52.32750. [DOI] [PubMed] [Google Scholar]
- Weeds A, Maciver S. F-actin capping proteins. Curr Opin Cell Biol. 1993;5:63–69. doi: 10.1016/s0955-0674(05)80009-2. [DOI] [PubMed] [Google Scholar]
- Yonezawa N, Nishida E, Ohba M, Seki M, Kumagai H, Sakai H. An actin-interacting heptapeptide in the cofilin sequence. Eur J Biochem. 1989;183:235–238. doi: 10.1111/j.1432-1033.1989.tb14918.x. [DOI] [PubMed] [Google Scholar]