Abstract
Tagging of proteins in vivo by covalent attachment of a biotin moiety has emerged as a new prospective tool for protein detection and purification. Previously, we established a strategy for expression of in vivo biotinylated proteins in mammalian cells. It is based on coexpression of the protein of interest fused to a short biotin acceptor peptide and biotin ligase BirA cloned in the same vector. We show here that the in vivo biotinylation can be used for immunogold postembedding labeling in immunoelectron microscopy experiments. We show that immunoelectron microscopy with biotinylated nuclear proteins is compatible with a wide range of postembedding methods, facilitating combination of morphological and localization studies in a single experiment. We also show that the method works in both transient transfection and stable cell line expression protocols and can be used for colocalization studies. This manuscript contains online supplemental material at http://www.jhc.org. Please visit this article online to view these materials. (J Histochem Cytochem 56:911–919, 2008)
Keywords: biotin, electron microscopy, postembedding, SUMO2, PML
Epitope tagging is a widely used technology for detection and purification of proteins and their macromolecular complexes from cells (Jarvik and Telmer 1998; Nakatani and Ogryzko 2003). A version of epitope tagging that is based on covalent attachment of biotin to a specific protein of interest and takes advantage of the strong biotin-(strept)avidin has been described (Cull and Schatz 2000). Naturally occurring biotin acceptor domains (BAD domains) fused to the protein of interest have been used to express proteins biotinylated in vivo in a wide range of biological systems including mammalian cells (Parrott and Barry 2000,2001). Several other in vivo biotinylation systems, described more recently (Duffy et al. 1998; Smith et al. 1998; de Boer et al. 2003; Viens et al. 2004), use an artificially optimized short biotin acceptor peptide (BAP) (Schatz 1993; Beckett et al. 1999) that can be targeted by the bacterial enzyme BirA. It has been applied for purification of multiprotein complexes (Duffy et al. 1998) and chromatin immunoprecipitation (Viens et al. 2004; Furuyama and Henikoff 2006; van Werven and Timmers 2006).
Previously, using immunofluorescence, we have shown that the in vivo biotinylation using short biotin acceptor peptides does not affect the intracellular localization of the targeted proteins (Viens et al. 2004). Thus, protein biotinylation in vivo could be used as a tool to study intracellular localization of proteins of interest. Anti-biotin antibodies conjugated to gold particles are commonly used to detect biotinylated nucleic acids probes in ISH experiments at the electron microscopic level (Visa et al. 1993; Arhel et al. 2006). Accordingly, in this report, we studied the possibility of localizing proteins of interest, biotinylated in vivo, by postembedding immunoelectron microscopy, using a high-resolution one-step detection system.
The strategy for expression of in vivo biotinylated proteins, developed in our previous study, is based on an addition of a BAP to the sequence of a protein of interest and transfection of the construct into cells. A bacterial BirA enzyme that covalently attaches biotin to the BAP-fused target protein is expressed from the same vector. Incubation of live cells with biotin [also known as a vitamin H, whose uptake is mediated by a transporter SMVT (Prasad et al. 1998)] allows the BirA enzyme to label with high efficiency the protein of interest in vivo. In this study, we used the CMV.BBHN vector, described previously (Viens et al. 2004), to ask whether our in vivo biotinylation system can be used for postembedding immunoelectron microscopy.
Materials and Methods
Constructs
The vector for in vivo biotinylation, CMV.BBHN, was described in a previous study (Viens et al. 2004). It is a derivative of the pCDNA3 vector (Invitrogen; Carlsbad, CA), containing the cassette with the biotin binding peptide (BAP) MAGLNDIFEAQKIEWHE, reported previously to be an efficient target of the BirA enzyme (Schatz 1993), followed by cloning site, followed by a short internal ribosomal entry site (IRES) that directs initiation of translation of the BirA gene located downstream. This construction ensures that, after insertion of the open reading frame (ORF) of the gene of interest, it will be fused to the BAP and be an efficient target of the BirA enzyme expressed in the close vicinity of the target protein. The CMV.BBHN.H2A and CMV.BBHN.SUMO2 vectors were described in our previous study (Viens et al. 2004). The pBabe-PMLIII-GFP construct for expression of PMLIII-GFP fusion was a generous gift of Dr. S. Minucci, University of Milan, Italy.
Stable Cell Line Generation, Transient Transfection, and In Vivo Biotinylation
All cells were grown in DMEM (Gibco; Carlsbad, CA) with 10% FBS (PAN Biotech; Aidenbach, Germany). For transient transfection, a standard calcium phosphate precipitation method was used, and the cells were analyzed 1 or 2 days after transfection. In some experiments, we also used Polyfect (Qiagen; Hilden, Germany). For retroviral transduction, the packaging cell line Phenix-E was transfected with the plasmid DNA of the retroviral vector; 2 days later, the retroviral supernatant was harvested, filter-sterilized, and added to the HeLa or HEK 293 cells. Two days afterward, the transduced cells were sorted using an anti-IL2R antibody coupled to magnetic affinity beads. For biotin labeling, we established in our previous study that overnight incubation with 0.2% biotin yield sufficient biotinylation of the target protein; however, 1-hr biotinylation could be sufficient as well.
Chromatin Preparation and Western Analysis
For chromatin isolation, nuclei were prepared by incubating cells in hypotonic buffer (10 mM Tris HCl, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 0.2 mM PMSF, 15 mM β-mercaptoethanol, 0.1% Triton × 100), and they were resuspended in sucrose buffer (0.34 M sucrose, 10 mM Tris HCl, pH 8, 3 mM MgCl2, 15 mM β-mercaptoethanol, 0.2 mM PMSF) and treated with micrococcal nuclease (Sigma; St. Louis, MO). After stopping the reaction with 10 mM EDTA, the nuclear debris were removed by centrifugation, and the digested chromatin was fractionated on the 10–35% glycerol gradient (with a buffer composition of 20 mM Tris HCl, 0.2 mM EDTA, 300 mM NaCl, and 0.2 mM PMSF) for 5 hr at 40,000 × g using benchtop ultracentrifige (Beckman Optima TL; Fullerton, CA).
Total cell extract for Western analysis was prepared using RIPA buffer (50 mM Tris HCl, pH 8.0, 50 mM NaCl, 1% NP40, 0.5% Na deoxycholate, 0.1% SDS, and 1 mM EDTA). Cells were washed with PBS and incubated with RIPA buffer directly in the 6-well plates for 5 min. The extract was harvested and spun at high speed on a microcentrifuge. SDP-PAGE separation, transfer to a nitrocellulose membrane, blocking, incubation with antibody, and enhanced chemiluminescence detection were performed according to a standard protocol, except that for the detection of biotinylated proteins by the horseradish peroxidase (HRP)-conjugated peroxidase, 500 mM NaCl was added to the washing buffer (PBS + 0.1% Tween). The anti-H2A antibodies were purchased from Abcam (Cambridge, UK) and the streptavidin-HRP conjugate from Sigma.
Immunofluorescence
NIH3T3 cells were grown and transfected with Polyfect on coverslips. The day before fixation, 0.2% biotin was added to the medium. Fixation was accomplished with 4% formaldehyde in PBS for 15 min at 4C. Fixed cells were permeabilized with 0.5% Triton for 15 min, blocked with 1% BSA and 1% FBS, incubated with Cy3-conjugated streptavidin for 1 hr at 37C, and washed with PBS. The cells were observed using a fluorescence microscope, and images were acquired using a CCD camera.
Fixation and Embedding for Electron Microscopy
For Epon embedding, after incubation overnight with 0.2% biotin, cell cultures were fixed for 1 hr at 4C in 1.6% glutaraldehyde (Taab Laboratory Equipment; Reading, UK) in 0.1 M phosphate buffer, pH 7.3. During fixation, cells were scraped from the plastic substratum and centrifuged. Cell pellets were dehydrated in increasing concentrations of ethanol and embedded in Epon. Polymerization was carried out for 48 hr at 64C. Ultrathin sections were collected on formvar-carbon-coated copper grids (200 mesh).
For embedding in Lowicryl K4M (Chemische Werke Lowi; Waldkraiburg, Germany), cell cultures incubated overnight with 0.2% biotin were fixed either in 4% formaldehyde (Merck; Darmstadt, Germany) or in 1.6% glutaraldehyde and dehydrated in increasing concentrations of methanol. Embedding was carried out at low temperature according to Roth (1989). Polymerization was at −30C for 5 days under long-wavelength UV light. Ultrathin sections of Lowicryl-embedded material were collected on formvar-carbon-coated gold grids (200 mesh) and stored until use.
Antibodies and Immunogold Detection of Tagged Proteins
The following antibodies were used in this study: a goat anti-biotin antibody coupled to 10-nm gold particles (BBInternational; Cardiff, UK), a rabbit polyclonal anti-green fluorescent protein (GFP; Abcam 290), and a goat anti-rabbit antibody coupled to 5-nm gold particles (BBInternational).
For immunodetection of in vivo biotinylated proteins in Epon-embedded cells, grids bearing thin sections were first placed on a drop of 10% H2O2 for 10 min for etching, rinsed in water, and dried. These grids were placed on a drop of 5% BSA for 5 min to suppress nonspecific binding of the antibody and were floated on a drop of 10-nm gold-conjugated anti-biotin antibody (dilution 1/25 in PBS) for 30 min at room temperature. After a 15-min washing in PBS, the grids were rapidly rinsed in a jet of distilled water, air-dried, and counterstained with 4% uranyl acetate. The same labeling protocol was used for the detection of in vivo biotinylated proteins on Lowicryl sections except that the H2O2 treatment was omitted.
For colocalization of biotin-SUMO2 and PML-GFP, glutaraldehyde-fixed Lowicryl-embedded thin sections were first treated with BSA and then reacted with the rabbit anti-GFP antibody (diluted 1/200 in PBS with 1% BSA and 0.1% Triton) for 30 min at room temperature. After a 15-min washing in PBS, grids were incubated for 30 min at room temperature on a mix of 5-nm gold-conjugated anti-rabbit antibody and 10-nm gold-conjugated anti-biotin antibody diluted 1/25 in PBS with 1% BSA and 0.1% Triton. Final washing and staining were as above. The sections were analyzed either with a Zeiss EM902 (Oberkochen, Germany) and photographs were taken on Kodak 4089 films (Rochester, NY) or with a FEI Tecnai Spirit (Hillsboro, OR) and digital images were taken with a SIS MegaviewIII CCD camera (Olympus; Tokyo, Japan).
Quantitative Analysis
For each sample, 15 thin sections were analyzed at a final magnification of ×45,000. The micrographs were taken randomly, but it was ensured that all fields contained nuclear, cytoplasmic, and extracellular (pure resin) areas. Surface areas were determined with Image J, and gold particles were counted by eye. Gold particle density and SD were calculated using Microsoft Excel (Microsoft; Redmond, WA).
Results
Biotinylated Histone H2A Is Incorporated Into Chromatin
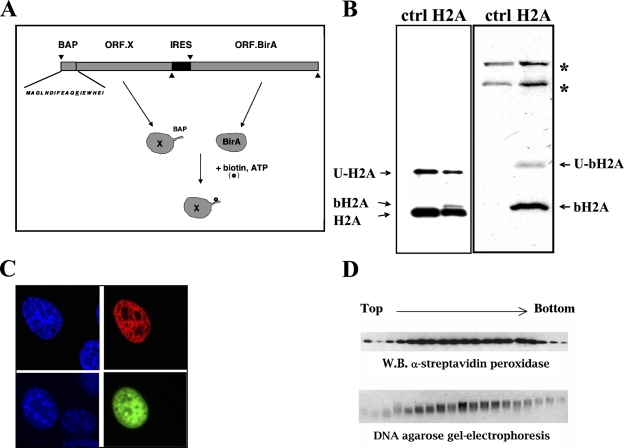
Figure 1A shows the scheme for expression of biotinylated proteins in mammalian cells that was used in this study. The principle is based on coexpression of the target protein fused to a short BAP together with the biotinylating enzyme BirA (Figure 1).
Figure 1.
(A) The principle of expression of biotinylated protein in vivo. Target protein X is fused to a short biotin acceptor peptide (BAP) and is coexpressed with the biotinylating enzyme BirA. BirA uses ATP and biotin (indicated also by a small circle) to biotinylate the target protein X. The principal feature of our design is the placing of the open reading frame of the BirA gene (ORF.BirA) on the same vector downstream of the open reading frame of the target protein X (ORF.X). Their coordinate expression from the same mRNA is ensured by an internal ribosome entrance sequence (IRES). The rationale for this particular design is to provide for a high local concentration of the BirA enzyme in the vicinity of the target protein X, which would ensure efficient biotinylation with minimal amounts of the BirA enzyme. Shown is the sequence of the BAP with the target lysine underlined. The positions of the start codons (Δ) and stop codons (∇) on the ORFs are also indicated. (B) Biotinylated H2A is incorporated into chromatin. Expression of biotinylated H2A (bH2A) in 293T cells detected by Western blot with anti H2A antibodies (left) and horseradish peroxidase (HRP)-streptavidin (right). Ctrl, untransfected cells; H2A, cells transfected with the CMV.BBHN.H2A DNA. Bands corresponding to the endogenous H2A, bH2A, and ubiquitinated H2A (U-H2A) and bH2A (U-bH2A) are shown by arrows. Asterisks indicate endogenously biotinylated proteins (various carboxylases) also present in untransfected cells. (C) Nuclear localization of bH2A as seen by immunofluorescence. Top: HEK 293 cells were transiently transfected with CMV.BBHN.H2A and stained using Cy3-conjugated streptavidin. Bottom: HEK 293 cells were transiently transfected with peGFP.H2A vector and directly visualized under fluoresent microscope. Left panels show DAPI staining and right panels show either Cy3 (top) or GFP (bottom) signals. (D) Co-sedementation in the glycerol gradient. Chromatin was prepared from the transfected cells and partially digested with micrococcal nuclease. It was fractionated on 10–35% glycerol gradient. Twenty fractions were analyzed on the presence of biotinylated histones by Western blotting with HRP-conjugated streptavidin (top). The presence of DNA was also monitored in the same fractions by agarose gel electrophoresis (bottom). A strong correlation in the presence of biotinylated histones (top) and DNA (bottom) in the same fractions strongly indicates that the biotinylated histone is in a complex with DNA.
As a first model of our studies, we used HEK 293 cells, transiently transfected with vector expressing biotinylated H2A histone [CMV.BBHN.H2A vector (Viens et al. 2004)]. Western blot analysis of nuclear extract with HRP-conjugated streptavidin showed a strong band of the expected size appearing in the experimental sample (Figure 1B, right). However, the amount of the biotinylated H2A was small compared with the endogenous H2A, as shown by Western blot with antibodies against H2A (Figure 1B, left, compare the intensities of the bH2A and H2A bands). Immunofluorescent analysis of transfected cells stained with Cy5-conjugated streptavidin showed clear nuclear localization very similar to the localization of the H2A-GFP–fused proteins, analyzed in parallel experiments (Figure 1C). Evidence indicated that the biotinylated H2A (b-H2A) was incorporated into chromatin. First, b-H2A co-sedimented with DNA during glycerol gradient fractionation of chromatin, prepared from the transfected cells and partially digested with micrococcal nuclease (Figure 1D). Second, given that the histones incorporated into nucleosomes resist modest salt treatments, the HEK 293 cells transiently transfected with CMV.BBHN.H2A were permeabilized and washed with buffer containing up to 600 mM NaCl before fixation. These cells were stained with Cy3-conjugated streptavidin to detect the b-H2A. A strong signal was still observed after staining the salt-treated cells with Cy3-conjugated streptavidin (data not shown). Therefore, we concluded that biotinylated histones were specifically incorporated into chromatin.
Protein Biotinylation In Vivo Is Compatible With Immunoelectron Microscopy
To confirm the chromatin incorporation of the in vivo biotinylated H2A, we performed immunoelectron microscopy experiments. We generated a HEK 293 cell line stably expressing b-H2A. These cells and control HEK 293 cells were grown in biotin-containing medium and fixed with either formaldehyde or glutaraldehyde. The cells were scraped and embedded in Lowicryl K4M for immunoelectron microscopy. Ultra-thin sections of embedded material were incubated with anti-biotin antibodies conjugated to 10-nm gold particles, counterstained with uranyl acetate, and observed at 80 kV using a transmission electron microscope.
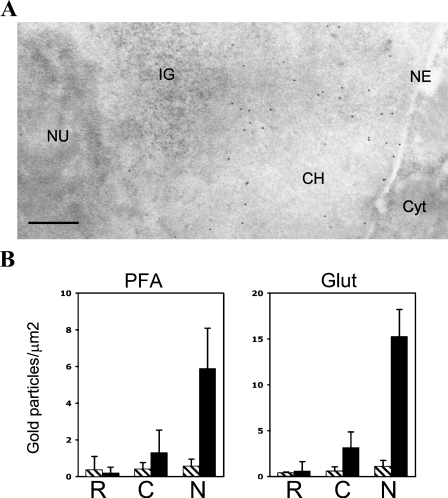
In agreement with the immunofluorescent analysis, we observed a strong labeling of the nuclei in the case of b-H2A–expressing cells and very low staining for the control HEK 293 (Figure 2A and data not shown). Furthermore, in the b-H2A–expressing cells, the labeling was restricted to chromatin-containing areas of the nucleoplasm. Nuclear structures known to be devoid of DNA like the dense-fibrillar and the granular compartments of the nucleolus, the clusters of interchromatin granules (Figure 2A), and nuclear bodies were devoid of gold particles, reinforcing the idea of a specific detection of a chromatin-incorporated biotinylated H2A. Some staining was observed in cytoplasm for both samples, with a somewhat stronger signal in the case of the cells expressing biotinylated H2A. This is most likely because of the presence of endogenous biotinylated proteins in the cytoplasm of mammalian cells (Hollinshead et al. 1997) (see below), whereas the stronger staining of cytoplasm in the experimental sample could be explained by the presence of newly synthesized b-H2A before its import into the nucleus. Finally, almost no signal was observed in the resin, showing low background staining.
Figure 2.
Intranuclear distribution of biotinylated H2A as seen by immunoelectron microscopy. (A) Representative section of a paraformaldehyde-fixed cell embedded in Lowicryl K4M, uranyl acetate staining. Postembedding detection of b-H2A with a gold-conjugated anti-biotin antibody shows numerous gold particles on chromatin-containing areas (CH) of the nucleus. Nuclear regions devoid of DNA like the fibrillo/granular component of the nucleolus (Nu) and a cluster of interchromatin granules (IG) are unlabeled. NE, nuclear envelope; Cyt, cytoplasm. Bar = 0.5 μm. (B) Quantification of labeling by gold particles conjugated with anti-biotin antibodies. The numbers were counted separately for the resin (R), cytoplasm (C), and nucleus (N). Left panel (black and white) inside of every graph shows the results for control cells and right panel (black) for the cells expressing bH2A. Results for both paraformaldehyde (PFA) and glutaraldehyde (GA) fixations are shown.
To support our qualitative observations, we calculated the density of labeling (number of gold particles divided by the observed surface) for control and experimental samples. The results of quantification, presented on the histogram (Figure 2B), are consistent with the absence of difference for the background labeling on the resin and a slight increase in the cytoplasm between the control and experimental samples, whereas a 10-fold increase in the case of nuclear staining of cells expressing biotinylated H2A compared with control cells is evident. Similar results were obtained for lowicryl-embedded cells fixed with glutaraldehyde instead of paraformaldehyde (Figure 2B).
Use of Biotinylated Proteins Is Compatible With Embedding Protocols Preserving Cell Structure
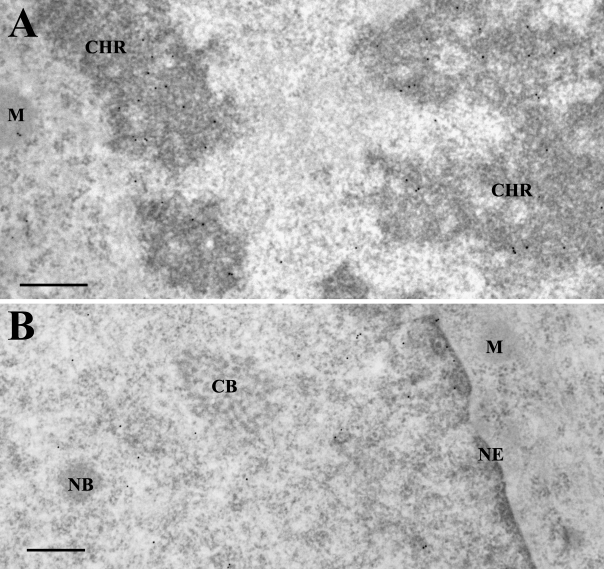
Epoxy resins are preferred for conventional ultrastructural studies because they provide the highest preservation of the cellular structures. More hydrophilic resins, like Lowicryl or LR White, have been developed for immunoelectron microscopy because they favor epitopes/antibodies interactions on thin sections, but with a less optimal ultrastructural preservation. However, a biotin tag is chemically different from usual peptide epitopes, so we asked whether it could tolerate Epon embedding. We used the HEK 293 cell line stably expressing b-H2A and grew it in the biotin-containing medium. The cells were fixed in 1.6% glutaraldehyde, scraped, pelleted, and processed for Epon embedding. After etching with H2O2, ultra-thin sections of Epon-embedded material were incubated with anti-biotin antibodies conjugated to 10-nm gold particles and counterstained with uranyl acetate. As shown in Figure 3, this embedding protocol, which provides a better ultrastructural definition, is also compatible with immunodetection of the b-H2A. We obtained a picture similar to the one previously observed with Lowicryl-embedded cells, with comparable densities of nuclear labelings (16.4 ±5.4 and 15.2 ±2.9 gold particles/μm2 for Epon and Lowicryl embedding of glutaraldehyde-fixed cells).
Figure 3.
Efficient immunogold localization of biotinylated H2A after Epon embedding. Thin sections of glutaraldehyde-fixed cells embedded in Epon were incubated with 10-nm gold-conjugated anti-biotin antibody to reveal b-H2A. (A) Prophasic cell. Gold particles are concentrated over condensing chromosomes (CHR). (B) Interphasic cell with gold particles scattered over the chromatin-containing regions of the nucleoplasm. Nuclear inclusions [NB, nuclear body; CB, Cajal (or coiled) body] and the cytoplasm, in contrast, are unlabeled. NE, nuclear envelope; M, mitochondrion. Bar = 0.5 μm.
As an example, in a prophasic cell as in Figure 3A, the labeling was concentrated over the condensing chromosomes as expected for a chromatin incorporated b-H2A. Accordingly, in interphasic cells as in Figure 3B, gold particles were confined to chromatin regions, whereas well-preserved nuclear bodies or cytoplasmic areas were essentially unlabeled.
We conclude that the in vivo biotinylation is compatible with embedding methods that preserve morphology better that the methods regularly used for immunoelectron microscopy.
We took advantage of the well-preserved cellular structures to study the biotin distribution in the cytoplasm of untransfected HEK 293 cells, supplemented with biotin (Supplemental Figure 1). If the global density labeling was low at 1.3 ± 0.7 gold particles/μm2, it was unevenly distributed with 5.1 ± 2.5 gold particles/μm2 in mitochondria exceeding by far the labeling of the rest of the cytoplasm at 0.6 ± 0.3 particles/μm2. These endogenously biotinylated mitochondrial proteins have been previously detected by immunoelectron microscopy in kidney cells (Hollinshead et al. 1997) and likely correspond to the upper bands in the Western blot probed with HRP-streptavidin in Figure 1B. They could represent a limitation for detection of specific mitochondrial proteins with our protocol.
Use of In Vivo Biotinylation for Colocalization Experiments in Immunoelectron Microscopy
Our results with b-H2A indicate that in vivo addition of a biotin tag can be used as a new tool for high-resolution localization of proteins. This extension of the repertoire is particularly useful when one is willing to localize, in the absence of a discriminatory antibody, a specific member of a protein family. To show this, we performed colocalization studies. Because the H2A histone is distributed relatively uniformly over the nucleus, we preferred to use proteins with more distinct localization patterns. In our previous immunofluorescent experiments (Viens et al. 2004), biotinylated SUMO2 showed a very specific pattern of localization, reminiscent to the PML body. SUMO-2 and SUMO-3 are 96% identical and therefore hard to discriminate with antibodies and thus could be a good model for our study. Accordingly, we used biotinylated SUMO2 protein as a first target and PML-GFP (isoform III) as a second target. Also, we considered that transient transfection would fit better the colocalization studies, because it is a more practical expression protocol when many different combinations of proteins must be tested.
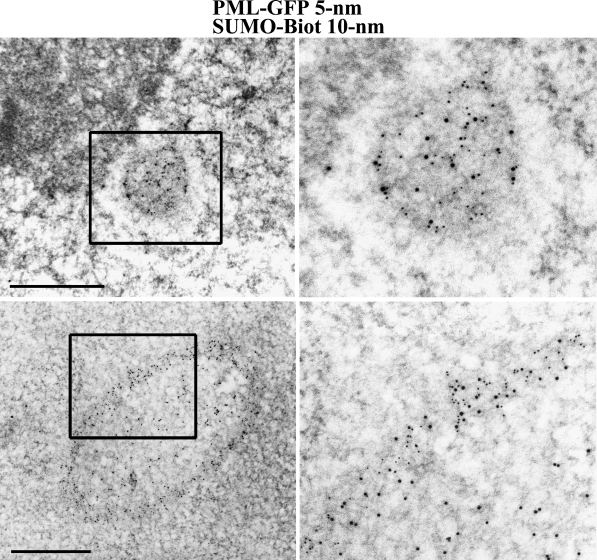
The HEK293 cells were transiently cotransfected with vectors expressing biotinylated SUMO2 and PML-GFP, grown for 2 days in biotin-containing medium, and fixed with 1.6% glutaraldehyde. The cells were scraped and embedded in Lowicryl K4M for optimal detection of the GFP epitopes. Ultra-thin sections of embedded material were incubated with the anti-GFP antibody and subsequently with a secondary anti-rabbit antibody conjugated to 5-nm gold particles and the anti-biotin antibody conjugated to 10-nm gold particles. As seen in Figure 4, we detected two distinct situations, probably linked to the level of expression of the PML-GFP fusion in different cells. In Figure 4A, a canonical PML nuclear body, closely apposed to the nucleolus, is shown. Its size (0.3 μm) and its structure, including a white halo, are, together with its position, strongly suggestive of a preformed PML body on which the biotin-SUMO2 (10-nm gold particles) and PML-GFP (5-nm gold particles) were addressed. This is consistent with our previous immunofluorescent studies and data from the literature indicating that the transfected SUMO2 is localized mostly to the PML bodies (Seeler and Dejean 2003). We also noticed, in a significant proportion of cells, the presence of very large (up to 2 μm) annular nuclear bodies, which are known to be induced by overexpression of some PML isoforms (Condemine et al. 2006). These induced bodies are fibrillar and not surrounded by a white halo. In Figure 4B, we show that these fibrillar bodies are also concentrating the PML-GFP isoform (10-nm particles) and the b-SUMO2 protein (5-nm particles). Interestingly, whereas both exogenous proteins are located in the fibrillar ring of this large PML body, the thin threads of the central network inside the body are decorated only by the 10-nm gold particles of the antibiotin antibody, showing the independent localization of SUMO2 in this compartment. Many proteins found in PML bodies are sumoylated (Seeler and Dejean 2003); therefore, the sumoylation target located in the center of the induced PML body remains to be elucidated. We also noticed that some locations in the fibrillar PML ring were decorated with only one kind (either 10 or 5 nm) of particles. The sumoylation of targets different from PML can partially account for the places labeled exclusively with the 10-nm particles.
Figure 4.
Ultrastructural colocalization of biotin-SUMO2 and a PML-GFP isoform on nuclear bodies. Cells expressing a biotinylated SUMO2 and a PML-III-GFP isoform were fixed in glutaraldehyde and embedded in Lowicryl K4M. Postembedding detection of PML-GFP was indirect with a primary anti-GFP antibody followed by a secondary antibody coupled to 5-nm gold particles. Detection of b-SUMO2 was direct with the anti-biotin antibody coupled to 10-nm gold particles. Top row: Notice colocalization of both tagged proteins on a canonical PML body. Bottom row: A large annular nuclear body is shown. Whereas the two proteins are concentrated in the ring, only the biotinylated SUMO2 is detected within the interior of the body, showing the specificity of the tagging method. Bar = 0.5 μm.
Discussion
The principal result of this study was the demonstration that in vivo biotinylation is a new way to suitably tag a specific protein for immunoelectron microscopy. As with other epitope tagging, in vivo biotinylation provides a general solution for the study of proteins to which antibodies are not available, both for immunofluorescence and immunoelectron microscopy. However, the use of biotin as an alternative to the more traditional epitope tags has several advantages specifically for immunoelectron microscopy. First, it allows for a convenient one-step detection protocol, because of the fact that anti-biotin antibodies conjugated to gold particles are commercially available. Given that the average size of an antibody is 10 nm, the use of one instead of two steps in epitope detection should be of particular interest for the immunoelectron microscopy studies requiring the highest resolution.
Another advantage of biotin is its chemical difference from peptide-based epitope tags. As our results showed, the ability of the anti-biotin antibodies to recognize their ligand is not compromised after Epon embedding, a treatment that preserves ultrastructure but is usually not compatible with immunoelectron microscopy. Therefore, with the use of our system, the immunoelectron microscopy studies can be combined more easily with morphological analysis. As a result, various conditions can be used with this tagging technique, ranging from glutaraldehyde fixation and Epon embedding for highest resolution (Figure 3), glutaraldehyde fixation and Lowicryl-embedding for good preservation, and double labeling (Figure 4), paraformaldehyde fixation, and Lowicryl embedding (Figure 2) for double labeling when antigenicity of second target needs mild fixation conditions. Both stable and transient expression methods can be used in our system. We also envision an additional advantage of our method, not shown in this study: the possibility to study dynamics. After a relatively short incubation in medium with biotin, the cells can be washed and put into a biotin-free medium, and the localization of the biotinylated protein can be followed by EM.
Because of the detection of endogenous biotinylated proteins in mitochondria, this organelle is an important limitation of this technique. Nevertheless, our results showed that this approach extends the repertoire of tagging protocols useful for high-resolution localization of specific proteins.
Supplementary Material
Acknowledgments
This work was supported by La Ligue Contre le Cancer and National Center of Biotechnology (Kazakh Republic).
We thank Dr. Saverio Minucci for the generous gift of the PMLIII-GFP fusion expressing vector and anonymous referees for helpful suggestions.
References
- Arhel NJ, Souquere-Besse S, Charneau P (2006) Wild-type and central DNA flap defective HIV-1 lentiviral vector genomes: intracellular visualization at ultrastructural resolution levels. Retrovirology 26:38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D, Kovaleva E, Schatz PJ (1999) A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci 8:921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condemine W, Takahashi Y, Zhu J, Puvion-Dutilleul F, Guegan S, Janin A, de The H (2006) Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res 66:6192–6198 [DOI] [PubMed] [Google Scholar]
- Cull MG, Schatz PJ (2000) Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol 326:430–440 [DOI] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, et al. (2003) Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci USA 100:7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Tsao KL, Waugh DS (1998) Site-specific, enzymatic biotinylation of recombinant proteins in Spodoptera frugiperda cells using biotin acceptor peptides. Anal Biochem 262:122–128 [DOI] [PubMed] [Google Scholar]
- Furuyama T, Henikoff S (2006) Biotin-tag affinity purification of a centromeric nucleosome assembly complex. Cell Cycle 5:1269–1274 [DOI] [PubMed] [Google Scholar]
- Hollinshead M, Sanderson J, Vaux DJ (1997) Anti-biotin antibodies offer superior organelle-specific labeling of mitochondria over avidin or streptavidin. J Histochem Cytochem 45:1053–1058 [DOI] [PubMed] [Google Scholar]
- Jarvik JW, Telmer CA (1998) Epitope tagging. Annu Rev Genet 32:601–618 [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Ogryzko V (2003) Immunoaffinity purification of mammalian protein complexes. Methods Enzymol 370:430–444 [DOI] [PubMed] [Google Scholar]
- Parrott MB, Barry MA (2000) Metabolic biotinylation of recombinant proteins in mammalian cells and in mice. Mol Ther 1:96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott MB, Barry MA (2001) Metabolic biotinylation of secreted and cell surface proteins from mammalian cells. Biochem Biophys Res Commun 281:993–1000 [DOI] [PubMed] [Google Scholar]
- Prasad PD, Wang H, Kekuda R, Fujita T, Fei YJ, Devoe LD, Leibach FH, et al. (1998) Cloning and functional expression of a cDNA encoding a mammalian sodium-dependent vitamin transporter mediating the uptake of pantothenate, biotin, and lipoate. J Biol Chem 273:7501–7506 [DOI] [PubMed] [Google Scholar]
- Roth J (1989) Postembedding labeling on Lowicryl K4M tissue sections: detection and modification of cellular components. In Tartakoff AM, ed. Methods of Cell Biology. New York, Academic Press, 513–551 [DOI] [PubMed]
- Schatz PJ (1993) Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (NY) 11:1138–1143 [DOI] [PubMed] [Google Scholar]
- Seeler JS, Dejean A (2003) Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 4:690–699 [DOI] [PubMed] [Google Scholar]
- Smith PA, Tripp BC, DiBlasio-Smith EA, Lu Z, LaVallie ER, McCoy JM (1998) A plasmid expression system for quantitative in vivo biotinylation of thioredoxin fusion proteins in Escherichia coli. Nucleic Acids Res 26:1414–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, Timmers HT (2006) The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Res 34:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viens A, Mechold U, Lehrmann H, Harel-Bellan A, Ogryzko V (2004) Use of protein biotinylation in vivo for chromatin immunoprecipitation. Anal Biochem 325:68–76 [DOI] [PubMed] [Google Scholar]
- Visa N, Puvion-Dutilleul F, Harper F, Bachellerie JP, Puvion E (1993) Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res 208:19–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.