Abstract
We developed a method in preparing size-controllable gold nanoparticles (Au NPs, 2-6 nm) capped with glutathione by varying the pH (between 5.5 and 8.0) of the solution before reduction. This method is based on the formation of polymeric nanoparticle precursors, Au(I)-glutathione polymers, which change size and density depending on the pH. Dynamic light scattering, size exclusion chromatography, and UV-vis spectroscopy results suggest that lower pH values favor larger and denser polymeric precursors and higher pH values favor smaller and less dense precursors. Consequently, the larger precursors led to the formation of larger Au NPs, whereas smaller precursors led to the formation of smaller Au NPs. Using this strategy, Au NPs functionalized with nickel (II) nitriloacetate (Ni-NTA) group were prepared by a mixed-ligand approach. These Ni-NTA functionalized Au NPs exhibited specific binding to 6x-histidine-tagged Adenovirus serotype 12 knob proteins, demonstrating their utility in biomolecular labeling applications.
Introduction
Controlling the size of nanoparticles (NPs) has always been one of the challenges in colloidal science. Changing the size of NPs can result in modulating of their physical and chemical properties. Au NPs have been a good candidate for applications in drug delivery,1 cell imaging,2 and immobilization of proteins for conformational studies,3 among others. Since the discovery of various reducing agents for the gold compounds to form the gold nanoparticles, like sodium citrate, sodium borohydride, phosphorus, alcohols, tannic acid/citrate mixtures, the synthesis and applications of Au NPs of different sizes have flourished.4 More recently, several techniques that focus on the size control via the reduction of tetrachloroauric acid (HAuCl4) were introduced. These include varying the gold to thiol ratio,5 varying the reaction temperature,6 varying the pH by varying the concentration of sodium citrate (reducing agent)7, sonochemical8 and sonoelectrochemical methods,9 seeding methods,10 the use of mixed reverse micelles,11 photochemical methods12 and the use of polymeric ligands such as block copolymers.13
Formation of Au NPs via the reduction of HAuCl4 in the presence of the thiol ligand has been a widely used technique in preparing ligand-stabilized Au NPs.14 Schiffrin and coworkers proposed a two-step mechanism for the formation of Au NP from HAuCl4.15 The first step involves the conversion of Au(III) salt to the Au(I) state (as thiolate salt). The Au(I) thiolate at this point is in a polymeric form. The second step involves the reductive decomposition of the polymeric Au(I) thiolate to form the NP. Although this method has been extensively used, its mechanistic details have not been elucidated.14 One of the most intriguing steps is the formation of polymeric Au(I) thiolate or the NP precursor. Exploration of this step could lead to clues on how to control the size of the resulting NPs, since the properties of the polymeric precursor are dependent on several factors, such as the nature of ligand, ionic strength, solvent conditions, and pH. The variation in the polymeric structure by the nature of the ligand has been well documented in related compounds, such as gold based antiarthritic agents.16-18 For example, Myocrisin assumes a small cyclic structure (tetrameric) due to the repulsive force brought about by the carboxylate groups in adjacent ligands.16
Choosing the right ligand for NP synthesis is key in forming Au NPs with desirable properties. In this study, we chose the naturally occurring peptide ligand, glutathione (GSH). GSH is a ubiquitous tripeptide (γ-Glu-Cys-Gly), which acts primarily as a reducing agent in biological systems. Because of the favorable properties such as the presence of thiol, carboxylic acid and amino groups, water-solubility at relevant biological pH, biological compatibility, and ease of functionalization, GSH is a very attractive ligand in making water-soluble NPs for biological applications. The presence of carboxylic acid and amino groups on GSH ligand complexed with Au(I) has the potential advantage of being a pH-sensitive compound, which can adopt different conformational states and sizes depending on the pH of solution.
The first reported use of GSH as a ligand for Au clusters was by Hainfeld and coworkers19 and further studied by Whetten and coworkers in 1998.20,21 Whetten et al. synthesized the Au NPs via sodium borohydride reduction of the mixture of tetrachloroauric acid and GSH in methanol-water (2:3) and obtained Au NPs with most abundant component having a diameter of 0.9 nm. Tsukuda and coworkers conducted a more extensive electrophoretic separations and high resolution mass spectrometric analyses of the Au NPs, prepared in a similar manner as that of the Whetten's study, to reveal the varied size composition of the NP mixture (i.e. Au10(SG)10, Au15(SG)13, Au18(SG)14…) depending on the reaction temperature (low or room temperature).22,23 In both studies, the size of the NPs formed was between 0.8 nm and 1.1 nm. Although the preparation of the Au NPs was well-established and the composition of the Au NPs was extensively characterized, the issues of size controllability and further functionalization of the Au NPs to suit particular applications, such as biomolecular labeling, were not addressed in these studies. These issues are very important especially for the utility of the Au NPs in applications that require functionality and size control. Herein, we report a technique for which the size of the Au NPs coated with GSH can be controlled by varying the size of the nanoparticle precursor [Au(I)SG] through pH control. The size of the final NPs can be varied from 2 to 6 nm. Furthermore, functionalization of Au NPs coated with GSH is demonstrated by the introduction of nitrilotriacetic acid (NTA) moiety by mixing another ligand (i.e. NTA-Lysine modified ligand) with GSH. To demonstrate the utility of the size-controllable Ni-NTA-functionalized Au NP in the 2-6 nm size regime, a biomolecule, adenovirus serotype 12 (Ad12) knob expressed with 6x-histidine (His) tags, is labeled with 4.4 nm Au NP and analyzed by size exclusion chromatography (SEC) and transmission electron microscopy (TEM).
Ad12 knob protein is a globular knob-like domain located at the ends of trimeric fibers protruding from the adenovirus capsid. It acts as the initiating point for virus infection upon interactions with specific receptors on host cell surfaces.24 The Ad12 knob proteins exist as a trimer with a total molecular weight (MW) of about 60 kDa (Figure 1A). It has C3 symmetry with the His-tagged N-termini clustered near each other (Figure 1B). There are, therefore, three His-tags in close proximity. Our synthesized Ni-NTA functionalized Au NPs show highly specific binding to His tags on Ad12 knob proteins. Labeling of biomolecules with Au NPs is attracting more interests in applications like high-resolution structural analysis,25 medical imaging,26 and molecular sensing.27 These applications require functionalized Au NPs with physical/chemical stability, water solubility, biological compatibility, and site-specific binding.28,29
Figure 1.
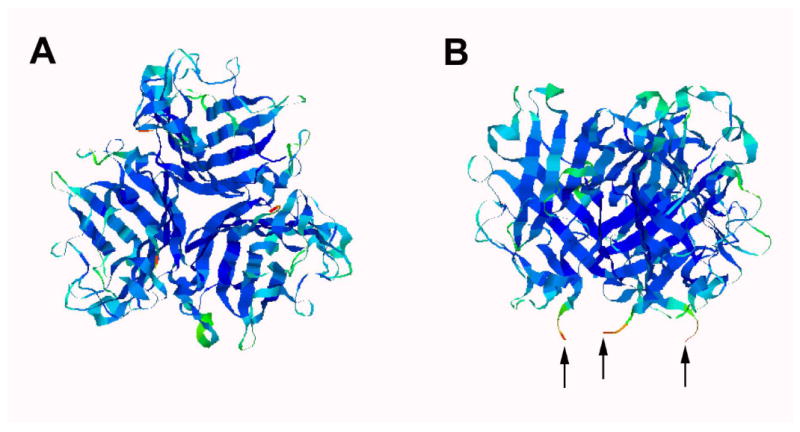
Crystal structure of Ad12 knob protein showing the top view (A) and the side view (B) orientations (Reference 24). The arrows in B indicate the N-termini where His-tags were placed.
Experimental Section
General Remarks
HAuCl4·XH2O was purchased from Strem Chemicals and used as received. Reduced glutathione (GSH), sodium borohydride, N6-carbobenzyloxy-L-lysine, and 4-butyrothiolactone were purchased from Sigma and used as received. UV-Vis spectra were recorded on Agilent 8453 spectrophotometer. TEM specimens were prepared by evaporating sample solutions on plasma-treated Cu mesh grids covered with carbon thin films. Nano-W (Nanoprobes, Inc.) solution was used for negative stain samples. TEM images were taken using a JEM 1200CX (JEOL) operating at 120 kV at low dose (< 10 e/Å2) with a 1k×1k CCD camera (Gatan). The sizes of Au NPs were analyzed with TEM images using ImageJ (NIH). The dynamic light scattering (DLS) data were recorded on a Precision Detectors PD 2000 DLS Plus with CoolBatch 40T. The hydrodynamic radii of polymeric Au(I)SG solutions were analyzed using Precision Deconvolve 32 software.
General Procedure for the Preparation of Au NP coated with GSH
An aqueous solution of tetrachloroauric acid (HAuCl4·XH2O, 1 mL, 0.025 M) was mixed with an aqueous solution of GSH (7.8 mL, 0.019 M). The pH of the resulting mixture was adjusted between 5.3 and 8.0 using 1M NaOH. A freshly prepared NaBH4 solution (2 mg/mL in water, 10-fold molar excess) was added with stirring. The mixture was allowed to react overnight at room temperature. The excess reactants were removed by size exclusion chromatography (GH-25, water) and the gold cluster fraction was collected and concentrated by centrifugal filtration (Molecular weight cutoff or MWCO 50kDa, Millipore Amico Ultra).
Synthesis of Lys-NTA-SH
Lys-NTA-SH ((1S)-N-[5-[(4-Mercaptobutanoyl)amino]-1carboxypentyl]iminodiacetic acid) was synthesized using a modified literature procedure.30-32 Briefly, N6-carbobenzyloxy-L-lysine (Lys(Z)OH) was reacted with bromoacetic acid under basic conditions to form Lys(Z)-NTA (Scheme 1). Lys(Z)-NTA was then deprotected by hydrogenolysis in 210 mL of 20:1 THF-water mixture (instead of the reported 20:1 methanol-water mixture30,31) under 1 atm of H2 for 20h to give Lys-NTA. Reaction of Lys-NTA with 4-butyrothiolactone gave the desired product Lys-NTA-SH in 61% yield (1.0 g).
Scheme 1.
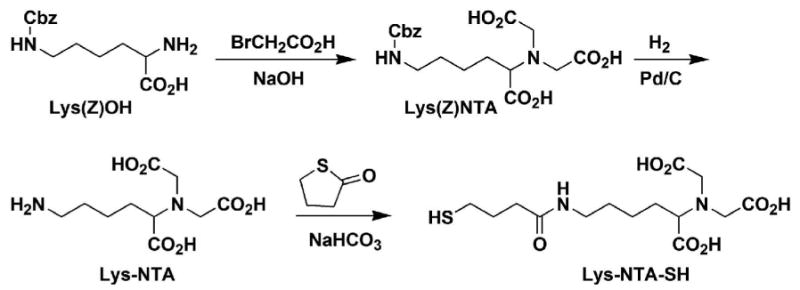
Synthesis of Lys-NTA-SH.
Representative Procedure for the preparation of Au-GSH/Lys-NiNTA
A solution of Lys-NTA-SH (8 mg, 0.022 mmol) in water (1.3 ml) was mixed with a solution of GSH (20 mg, 0.066 mmol) in water (3.4 ml). The mixed solution was added to a solution of hydrogen tetrachloroaurate (HAuCl4, 5 mg, 0.015 mmol) in water (0.5 ml), followed by adjusting the pH to 6.0-6.5 with 1 M NaOH. A solution of sodium borohydride (5.6 mg, 0.15 mmol) in water (2.8 ml) was added with vigorous stirring for 10 min at r.t and the mixture was allowed to react overnight. The Au NPs were purified by gel filtration chromatography (GH-25, water), followed by centrifugal concentration with Centri-prep filter (MWCO 30kDa, Millipore Amico Ultra). The resulting solution was passed through a cation exchange column (CM-Sepharose, Pharmacia) pre-treated with 0.05 M NiCl2 solution. The colored band was collected and used for the next step.
Labeling of Histidine-tagged Ad12 Knob Proteins
To a solution of His-tagged Ad12 knob proteins (24 μl, 1 mg/ml) at pH 7.5 containing 10 mM phosphate buffered saline (PBS, 150 mM NaCl) was added an aqueous solution of Ni-NTA functionalized Au NPs (40 μl, 1 μM) with an average size of 4.4 nm. The mixed solution was incubated at r.t. for 3 min. The crude mixture was purified by SEC (Superdex 200, Eluant: 10 mM PBS, 140 mM NaCl, pH 7.4) and the labeled fractions were collected and analyzed by TEM.
Dynamic Light Scattering Measurements for the polymeric Au(I)SG
Aqueous solutions of HAuCl4 (10 mg/mL) and GSH (6 mg/ml) were filtered separately through syringe filters (0.22 μm PVDF membrane, Millipore) prior to mixing. To a stirred solution of HAuCl4 (0.5 mL) was added GSH solution (4.5 mL). The resulting mixture was stirred for 1-2 min. The pH of the mixture was adjusted to 4.92 by adding 1M NaOH. The mixture was filtered through a syringe filter (0.45 μm PVDF membrane, Millipore) before measuring the DLS. Mixtures with pH values of 5.08, 5.55, 6.16, 7.25 and 8.11 were also prepared and analyzed using the same procedure as above.
Size Exclusion Chromatographic Analysis of the polymeric Au(I)SG at different pH values
The polymeric Au(I)SG samples at different pH values (4.8, 6.4, and 7.9) were prepared in a similar manner as described above for the DLS samples, but without filtering the solutions. The polymeric solutions (200 μL per injection) were analyzed by SEC-HPLC using Superose 6 column and either 10 mM PBS with 150 mM NaCl (pH 6.4 or 7.9) or 10 mM acetate buffer with 150 mM NaCl (pH 4.8) as the eluting solvent.
Results and Discussion
The Au NPs were synthesized employing a three-step procedure (Scheme 2A). The first step involves the reaction between the GSH and HAuCl4 (6:1 molar ratio), the second step involves the adjustment of pH of the mixture from the first step. Lastly, the Au NPs (1) are formed by addition of NaBH4. This method can be extended to the synthesis of Au NPs with mixed ligands (Scheme 2B). For example, a mixture of GSH and Lys-NTA-SH in the 3:1 molar ratio was used to prepare NTA-functionalized Au NP (2) used in this study for biomolecular labeling.
Scheme 2.
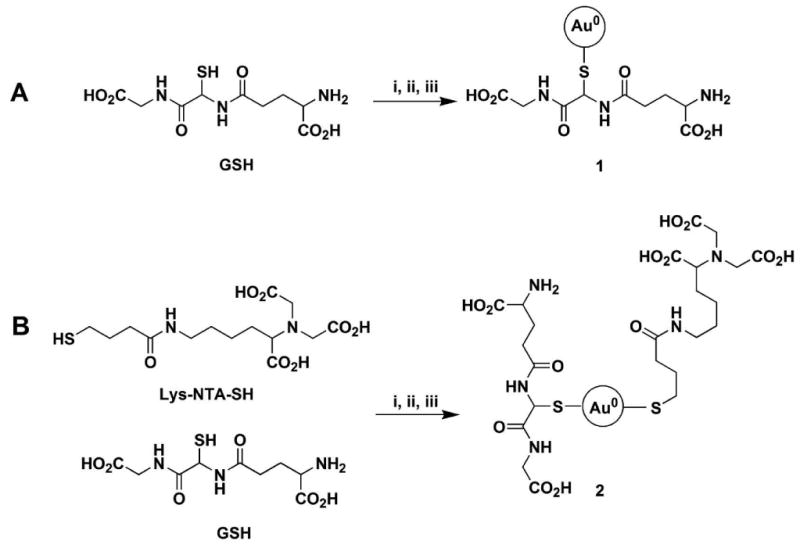
Synthesis of Au NPs. A. Au NP coated with GSH only; B. Au NP coated with 3:1 mixture of GSH and Lys-NTA-SH. Reaction conditions: i. HAuCl4, ii. Adjust pH, iii. NaBH4.
Au NPs coated with GSH (1) were synthesized at various pH values of the pre-borohydride reduced solutions (mixture of HAuCl4 and ligand). The final Au NPs are stable and can be stored at 4°C for three months without aggregation. The change in size of the Au NPs as the pH changes was monitored using UV-vis spectroscopy. There was a blue shift in the conspicuous surface plasmon resonance band from 532 to 507 nm as the pH value was increased from 5.5 to 8.0 (See Figure S1, Supporting Information), indicating that the Au NP size decreases as the pH increases. The exact sizes of 1 were analyzed from the TEM micrographs (Figure 2), and plotted with the pH values as shown in Figure 3. The sizes decreased from 6 to 2 nm with standard deviations of 20-30 % as the pH values increased from 5.3 to 8.0.
Figure 2.
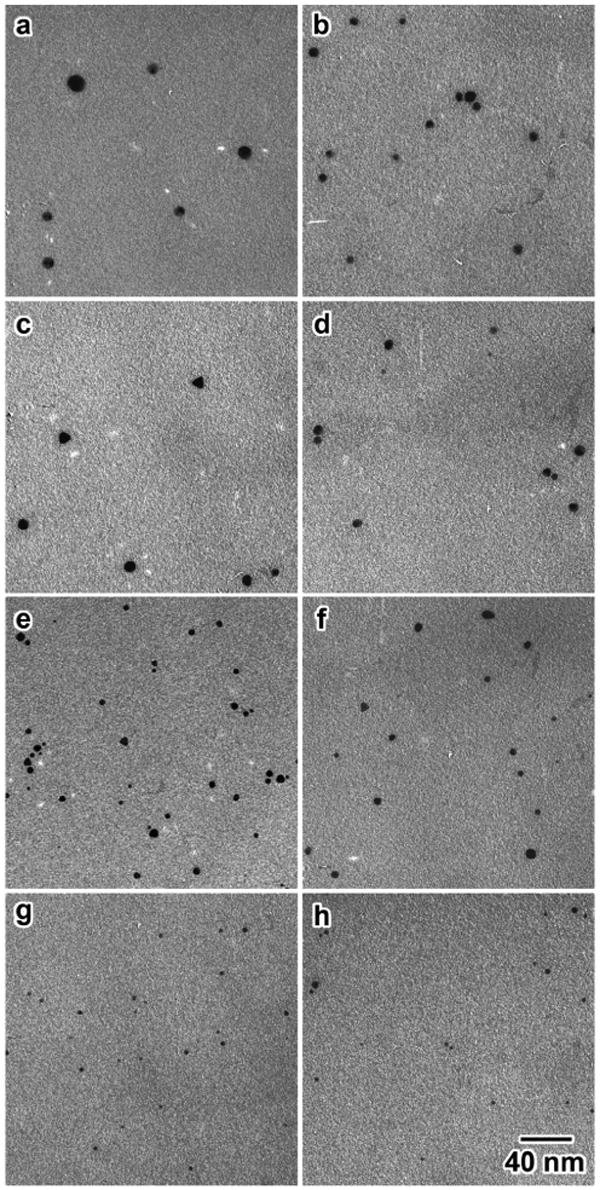
TEM micrographs of the Au NP 1 synthesized at different pH values; in parentheses are the average sizes in nm. (a) 5.5 (6.3), (b) 6.1 (5.4), (c) 6.3 (4.7), (d) 6.5 (6.0), (e) 6.8 (3.6), (f) 7.0 (3.3), (g) 7.5 (2.6), (h) 8 (2.2). All of the micrographs are in the same scale.
Figure 3.
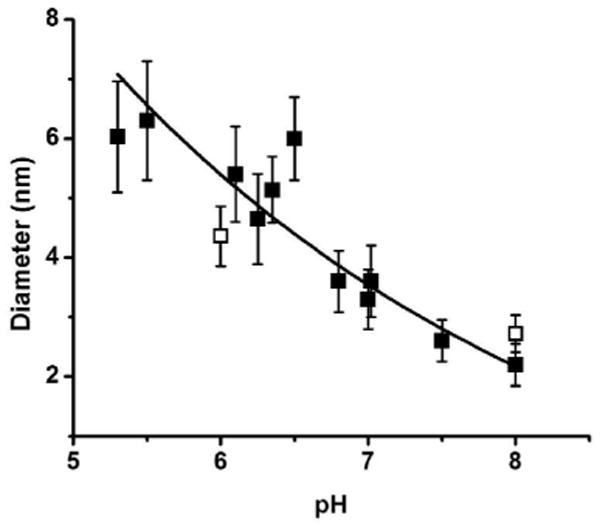
Size dependence of the AuNP 1 with pH as measured by EM. Solid squares correspond to the average sizes of Au NP capped with GSH, while open squares represent the average sizes of Au NP 2. The error bars indicate the standard deviations. About one hundred particles were analyzed for each point.
The same trend is followed when a mixed ligand of 3:1 molar ratio of GSH/Lys-NTA-SH is used, as shown by hollow squares at pH 6 and 8 (Figure 3). Combining GSH with Lys-NTA-SH to form a mixed-ligand Au NP (2) was done in order to functionalize the Au NP and to use as a site-specific labeling agent for electron microscopy applications. A mixture of 3:1 molar ratio of GSH to Lys-NTA-SH proved to be the optimal condition wherein stable clusters with spherical shapes were formed and the size dependence on pH was maintained. This indicates the controllability of the size even in the presence of another ligand by varying the pH of the pre-reduction solution. The effect of the excess GSH ligand was determined by performing control experiments. Before borohydride reduction, the mixture of HAuCl4 and GSH (1:6 molar ratio, pH 8) was filtered using Centricon Ultrafilter (MWCO 5kDa) and washed with water to remove the excess GSH. Dilution of the retentate to the original volume and subsequent reduction with sodium borohydride gave a spectrum similar to that of the “unfiltered” solution (Figure S2, Supporting Information). This indicates that any excess GSH has negligible effect on the size of the resulting Au NP.
The formation of nanoparticle 1 follows the general mechanism proposed previously for the formation of Au NP from Au3+ and thiols.15,20,33 In this mechanism (Scheme 3), Au3+ is reduced by GSH first to Au+1. The Au+1 then forms a polymeric structure ([Au(I)SG]n), where Au+1 ions have a coordination number of 2 and are bridged by the thiolate sulfur atom of GSH.16,34 Addition of sodium borohydride solution decomposes the polymeric structure to form the Au NP.
Scheme 3.
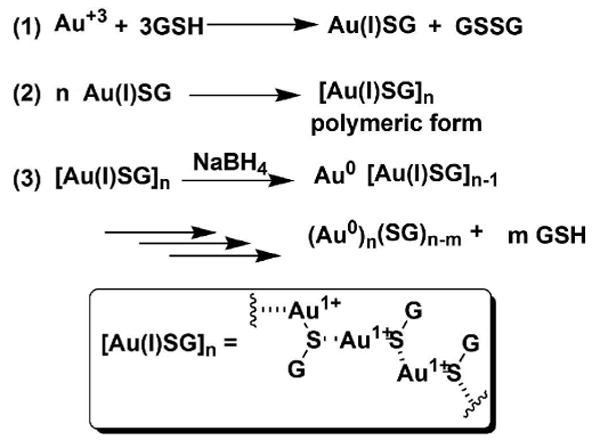
Proposed mechanism for the formation of Au NP from Au3+ and GSH.
The second equation is the key step in this study because the size of the polymer was found to be pH dependent. The polymeric structure of Au(I) thiolates, including Au(I) SG and gold-based anti-arthritic agents, has been established and extensively studied in the past using Mössbauer spectroscopic techniques.34 Although the X-ray structures of some Au(I) thiolates have been solved, that of [Au(I) SG] polymer is yet to be determined. It is likely that the Au(I)SG polymers exist in random coils consistent with these precedent data.
To further characterize the structure the precursor Au(I)SG polymers, their relative sizes and spectroscopic behavior were studied at different pH values. The presence of polymeric structures with different sizes at different pH values was followed by DLS. The results of a mixture of HAuCl4 and GSH (1:6 molar ratio) at various pH values (Figure 4A) reveal that the average hydrodynamic radius of the polymeric gold species decreases with increasing pH values. SEC analysis of the polymeric precursors at different pH values confirmed the DLS results (Figure 4B). At low pH (4.8), the mixture is composed of predominantly larger polymeric structures as indicated by the presence of a faster-moving peak with a retention time of 22.9 min, which could be attributed to larger polymers. At moderate pH (6.4), the mixture is possibly composed of a combination of larger and smaller polymers, as evident in the broad peak centered at 23.4 min. Lastly, at high pH (7.9), a slower-moving peak (retention time of 23.7 min) could be attributed to smaller polymers. The excess GSH apparently had no effect on the size of the polymeric precursors. A 1:6 mixture of HAuCl4 and GSH at pH 8 was filtered using Centricon Utrafilter (MWCO 5kDa) and washed with water to remove excess GSH. The hydrodynamic radius of the original solution (24.5± 2.8 nm) and the filtered one gave a similar size (26.5 ± 2.7 nm), indicating that GSH has no effect on the size of the polymers.
Figure 4.
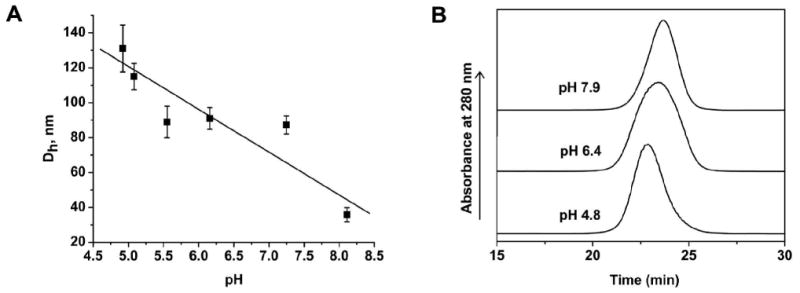
Studies on the dependence of polymeric Au(I)SG precursor size on pH. A. Hydrodynamic radius at different pH values of the polymeric Au(I)SG precursors measured using DLS. Error bars are standard deviation of three trials. B. The size exclusion chromatograms of the polymeric Au(I)SG precursors at pH 4.8, 6.4, and 7.9 with retention times of 22.9, 23.4, and 23.7 min, respectively. The precursors were separated using Superose 6 column with 10 mM phosphate or acetate buffered saline as the elution buffer.
The structural changes in the polymeric precursors can be studied by analyzing the interaction of adjacent Au(I) ions using UV-Vis spectroscopy. A UV-vis spectroscopic investigation of the mixture of tetrachloroauric acid and GSH in 1:6 molar ratio at different pH values showed an absorption at ∼320 nm (shoulder) that diminishes in intensity as the pH increases (Figure 5). This band could be assigned to the dσ*→ pσ transition similar to that observed for diphosphine bridged binuclear Au(I) complexes,35-39 which is an indication of metal-metal interaction. At higher pH values (6-8), the dσ* → pσ absorption band is shifted and considerably smaller but still monotonically changes with pH. At lower pH values (4.72-5.57), the higher intensity of the band suggests the increased presence of metal-metal interactions. The pH range going from 4.7 to 6.0 gave the highest decrease in the intensity of the metal-metal interaction band. In terms of the Au-S-Au angle (φ), the UV-vis data indicates that a smaller angle is favored at lower pH and a larger angle at higher pH (Scheme 4). This also implies that denser polymers are formed at low pH and less dense at high pH.
Figure 5.
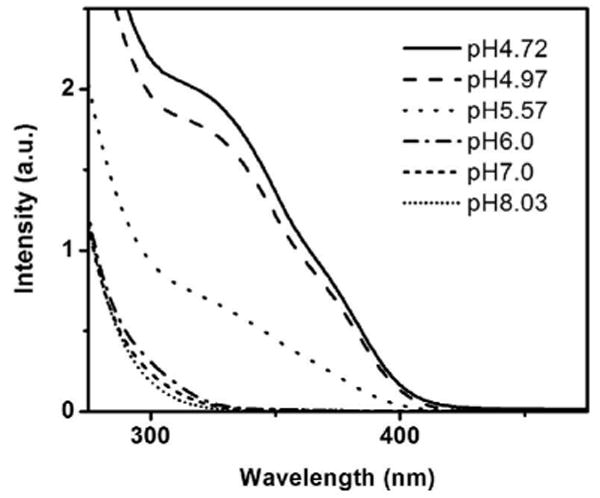
UV-Vis spectra of the polymeric Au(I)SG precursor at different pH values.
Scheme 4.
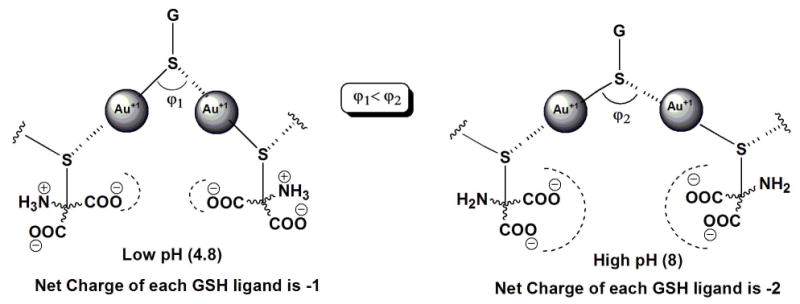
An illustration of the dependence of the Au-S-Au angles and the corresponding intramolecular forces of the NP precursor on pH.
On the basis of the UV-Vis data and results from the DLS and SEC studies, we hypothesize that at lower pH the polymeric precursors have larger and denser structures, while at higher pH the polymeric precursors assume less dense and smaller structures. This could be a consequence of the strength of repulsive electrostatic forces acting between neighboring GSH ligands (Scheme 4). GSH can have four ionization forms (Scheme 5, A-E), since it has four ionizable functional groups (one ammonium group, two carboxylic acid groups, and one thiol group). The first (pK1= 2.12)40 and second (pK2 = 3.53)40 ionizations can be ignored because their pKa values are below the lowest pH value used in this study. Also, the ionization of the thiol group (pK4 = 9.62)40 can be ignored because its pKa is above the highest pH value and coordination with Au(I) converted the thiol into the thiolate form. Thus, the major contributing species in the pH range of interest are species C and D. At low pH (4.8), species C predominates over D and so the net charge on the GSH ligands is about -1. On the other hand, at high pH (8.0), species D predominates over species C and the net charge of the ligand approaches -2. At higher pH values, the monomeric units (Au(I)SG) are negatively charged and so the growth of the polymer is limited by the stronger repulsive interaction; hence, only smaller and denser precursors are formed. At lower pH values, the monomeric units are able to approach a growing polymer due to the less repulsive force and so the precursors formed are larger and less dense.
Scheme 5.
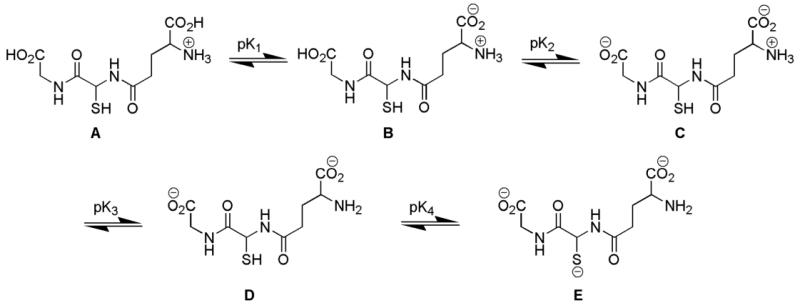
The ionization of GSH. The pK1, pK2, pK3, and pK4 values are 2.12, 3.53, 8.66, and 9.62, respectively (Reference 40).
Higher density of the Au(I)SG polymeric structures at lower pH values is also predicted (Scheme 4). The larger polymeric precursors at low pH lead to the formation of fewer nuclei for borohydride reduction to NPs (Scheme 6). Fewer nuclei which have more Au ions in close proximity result in larger Au NPs. This is consistent with the common principle that fewer nuclei lead to larger particles, since during the growth phase Au(I) is preferentially reduced onto an existing Au(0) surface rather than independently forming a new particle. Conversely, higher pH values favor a lower degree of polymerization of Au(I)SG. Repulsive negative charges increase on the GSH ligand at higher pH values due the deprotonation of the ammonium group (pKa 8.66),40 thus favoring smaller polymeric structures or possibly monomeric Au(I)SG units. This results in the formation of more nuclei, hence leading to smaller NPs (Scheme 6).
Scheme 6.
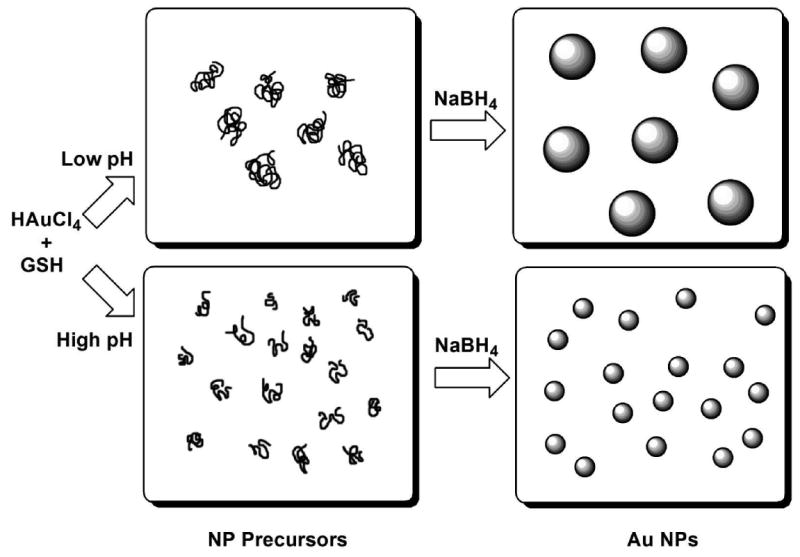
A schematic representation of the proposed mechanism of the formation of size-controllable Au NPs capped with GSH.
This strategy gives an insight into the possibility of tailoring the desired properties such as size, solubility, and functionality of the NPs based on the structure of the ligand and its Au(I) thiolate form. For the size controllability, the structure of the ligand is key. By virtue of its carboxylic acid groups and amino group, GSH responds to the change in pH by changing its conformation due to the repulsive or attractive forces. The size controllability could be potentially extended to the use of ligands that can change conformation upon changing the temperature, or upon exposure to radiation, etc.
Because of the better visibility of larger gold nanoparticles (> 2 nm) in EM, the 4.4 nm Au NPs functionalized with Ni-NTA were used to label His-tagged Ad12 knob protein. The Au NPs used in this investigation were prepared from Au NPs capped with GSH/Lys-NTA-SH mixture (3:1 molar ratio), which was complexed with Ni2+ ion by passing through an ion exchange column pre-treated with a NiCl2 solution.
The binding of the Ni-NTA-functionalized Au NP with a His-tagged protein is governed by the formation of stable coordinate bonds between two imidazoles from two histidines of the His-tagged protein and the Ni2+ center of the Ni-NTA group of the Au NP through a ligand exchange mechanism (Scheme 7). Two water molecules are expelled in the process to maintain the octahedral coordination. Labeling of His-tagged Ad12 knob proteins with 4.4 nm Ni-NTA functionalized Au NPs was confirmed by SEC (see Figure S3 and note, Supporting Information for details of labeling experiment) and TEM analyses. The TEM micrograph of the labeled-protein fraction shows that there is indeed labeling of the Ad12 proteins as evident in the formation of core-shell structures (Figure 6A), which is a consequence of the multiple Ni-NTA functional groups on the surface of each Au NP. The number of knob proteins on the surface of Au NP is limited by steric hindrance. It is also worth mentioning that the binding efficiency of this labeling is very high because all of the Au NPs were encapsulated by the knob proteins.
Scheme 7.
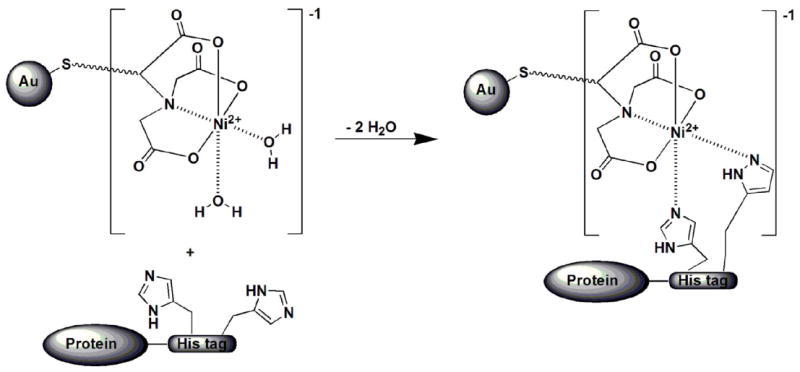
The binding reaction between a His-tagged protein and the Au NP functionalized with NiNTA.
Figure 6.
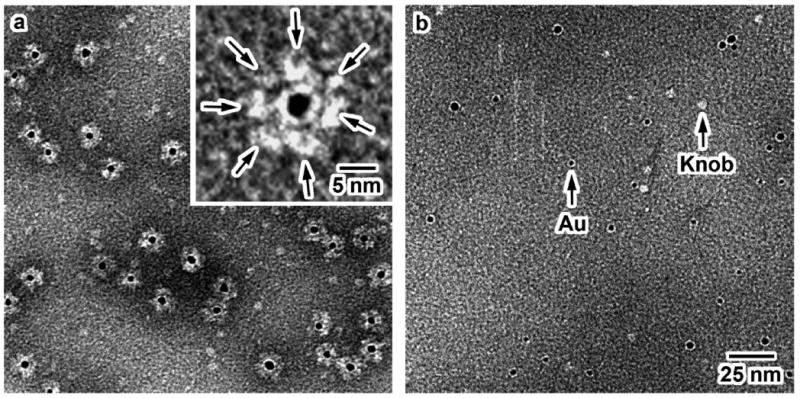
A: TEM micrograph of His-tagged Ad12 knob proteins labeled with Au NP coated with GSH/Lys-Ni-NTA-SH (negatively stained). Inset: Higher magnification image of the core-shell structure formed between the knob proteins and Au NP. Arrows indicate the position of the knob proteins around the black dot (Au NP). B: TEM micrograph of the mixture of non-His-tagged Ad12 knob proteins and Au NPs. Both images are shown in the same scale.
To discount the possibility of nonspecific binding in the labeling experiment, a control experiment was done using Ad12 knob complexes without histidine tags. The absence of core-shell NP-protein structures in the control experiment (Figure 6B) indicates that Ad12 knob complexes without histidine tags do not bind to the Ni-NTA Au NP. This shows that binding of Ni-NTA Au NP to Ad12 knob protein is highly specific for the presence of histidine tag on the protein. Specific binding of NPs to biomolecules has proven to be an indispensable attribute in applications such as electrochemistry41, carbohydrate-protein recognition42, and DNA-based diagnostic applications.43
Conclusion
In conclusion, we have shown that GSH-capped Au NPs of various sizes, in the 2-6 nm size regime, can be prepared by controlling the pH. The size controllability has been attributed to the dependence of the size of the Au NP precursor, Au(I)SG, on pH. The ligand, GSH, plays an important role in the size dependence on pH. Not only does it impart water-solubility to the resulting NPs, but also modulates the size of the NP precursors through the attractive or repulsive forces acting on adjacent ligands at certain pH values. Lower pH values favored the formation of larger and denser polymeric precursors because of the weaker repulsive electrostatic forces, present between adjacent ligands. On the other hand, smaller and less dense polymeric precursors were present at higher pH values due to the stronger repulsive electrostatic forces of the carboxylate groups between adjacent ligands. The larger precursors lead to the formation of larger NPs, whereas smaller precursors lead to the formation of smaller NPs.
The NPs were functionalized by mixing together two types of ligands, GSH and modified lysine bearing the Ni-NTA moiety and a thiol linker. This approach paves the way to preparing Au NPs of various sizes and various functionalities without destroying the desirable properties of solubility and stability. The Ni-NTA functionalized Au NPs were shown to have specific binding to His-tagged Ad12 knob proteins. This labeling technique can have potential biological application in high-resolution structural analysis, medical imaging, drug delivery, molecular sensing, and scaffold-directed assembly where binding specificity and size of NPs are important.
Supplementary Material
The UV-Vis spectra of the Au NPs prepared at different pre-reduction pH values, the UV-Vis spectra of the Au NPs prepared in the presence and absence of excess GSH during reduction, and the chromatographic data of the Au(I)SG polymers at different pH values. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We would like to thank Yanbiao Zhang and Paul I. Freimuth for providing the protein samples. We are grateful to Walter Mangel and Vito Graziano for use of their DLS machine. This work was supported by BNL LDRD Grant 04-055, DOE Grant 06742 and NIH Grants P41EB002181 and R01RR017545.
References
- 1.Salem AK, Searson PC, Leong KW. Nature Mater. 2003;2:668–671. doi: 10.1038/nmat974. [DOI] [PubMed] [Google Scholar]
- 2.Shukla S, Priscilla A, Banerjee M, Bhonde RR, Ghatak J, Satyam PV, Sastry M. Chem Mater. 2005;17:5000–5005. [Google Scholar]
- 3.Krämer S, Xie H, Gaff J, Williamson JR, Tkachenko AG, Nouri N, Feldheim DA, Feldheim DL. J Am Chem Soc. 2004;126:5388–5395. doi: 10.1021/ja031674n. [DOI] [PubMed] [Google Scholar]
- 4.Handley DA. In: Colloidal Gold: Principles, Methods, and Applications. Hayat MA, editor. Vol. 1. Academic Press, Inc.; San Diego: 1989. pp. 13–30. [Google Scholar]
- 5.Frenkel AI, Nemzer S, Pister I, Soussan L, Harris T. J Chem Phys. 2005;123:184701. doi: 10.1063/1.2126666. [DOI] [PubMed] [Google Scholar]
- 6.Song JH, Kim YJ, Kim JS. Curr Appl Phys. 2006;6:216–218. [Google Scholar]
- 7.Ji X, Song X, Li J, Bai Y, Yang W, Peng X. J Am Chem Soc. 2007 doi: 10.1021/ja074447k. ASAP Article. [DOI] [PubMed] [Google Scholar]
- 8.Mori Y, Kitamoto N, Tsuchiya K. J Chem Eng Jpn. 2005;38:238–288. [Google Scholar]
- 9.Liu YC, Yu CC. J Electroanal Chem. 2005;585:206–213. [Google Scholar]
- 10.Sau TK, Pal A, Jana NR, Wang ZL, Pal T. J Nanoparticle Res. 2001;3:257–261. [Google Scholar]
- 11.Chiang CL. J Colloid Interface Sci. 2001;239:334–341. doi: 10.1006/jcis.2001.7590. [DOI] [PubMed] [Google Scholar]
- 12.Pal A. Mater Lett. 2004;58:529–534. [Google Scholar]
- 13.Sakai T, Alexandridis P. J Phys Chem B. 2005;109:7766–7777. doi: 10.1021/jp046221z. [DOI] [PubMed] [Google Scholar]
- 14.Templeton AC, Wuelfing WP, Murray RW. Acc Chem Res. 2000;33:27–36. doi: 10.1021/ar9602664. [DOI] [PubMed] [Google Scholar]
- 15.Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. J Chem Soc, Chem Commun. 1994:801–802. [Google Scholar]
- 16.Brown K, Parish RV, McAuliffe CA. J Am Chem Soc. 1981;103:4943–4945. [Google Scholar]
- 17.Bachman RE, Bodolosky-Bettis SA, Glennon SC, Sirchio SA. J Am Chem Soc. 2000;122:7146–7147. [Google Scholar]
- 18.Bau R. J Am Chem Soc. 1998;120:9380–9381. [Google Scholar]
- 19.Hainfeld JF, Leone RD, Furuya FR, Powell RD. US Patent: 5,521,289. 1996
- 20.Schaaff TG, Knight G, Shafigullin MN, Borkman RF, Whetten RL. J Phys Chem B. 1998;102:10643–10646. [Google Scholar]
- 21.Schaaff TG, Whetten RL. J Phys Chem B. 2000;104:2630–2641. [Google Scholar]
- 22.Negishi Y, Takasugi Y, Sato S, Yao H, Kimura K, Tsukuda T. J Am Chem Soc. 2004;126:6518–6519. doi: 10.1021/ja0483589. [DOI] [PubMed] [Google Scholar]
- 23.Negishi Y, Nobusada K, Tsukuda T. J Am Chem Soc. 2005;127:5261–5270. doi: 10.1021/ja042218h. [DOI] [PubMed] [Google Scholar]
- 24.Bewley MC, Springer K, Zhang Y, Freimuth P, Flanagan JM. Science. 1999;286:1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- 25.Rice S, Lin AW, Safer D, Hart CL, Naberk N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, PateI E, Cookek R, Taylor EW, Milligan RA, Vale RD. Nature. 1999;402 doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 26.Hainfeld JF, Slatkin DN, Focella TM, Smilowitz HM. Br J Radiol. 2006;79:248–253. doi: 10.1259/bjr/13169882. [DOI] [PubMed] [Google Scholar]
- 27.Aubin ME, Hamad-Schifferli K. Langmuir. 2005;21:12080–12084. doi: 10.1021/la052102e. [DOI] [PubMed] [Google Scholar]
- 28.Hainfeld JF, Powell RD. J Histochem Cytochem. 2000;48:471–480. doi: 10.1177/002215540004800404. [DOI] [PubMed] [Google Scholar]
- 29.Hainfeld JF, Powell RD. Molecular morphology in human tissues: techniques and applications. CRC Press; Boca Raton, FL: 2005. [Google Scholar]
- 30.Du Roure O, Debiemme-Chouvy C, Malthete J, Silberzan P. Langmuir. 2003;19:4138–4143. [Google Scholar]
- 31.Schmitt L, Dietrich C, Tampé R. J Am Chem Soc. 1994;116:8485–8491. [Google Scholar]
- 32.Hochuli E, Dobeli H, Schacher A. J Chromatogr. 1987;411:177–184. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- 33.Schaaff TG, Shafigullin MN, Khoury JT, Vezmar I, Whetten RL, Cullen WG, First PN. J Phys Chem B. 1997;101:7885–7891. [Google Scholar]
- 34.Shaw CFI, Schaeffer NA, Elder RC, Eidsness MK, Trooster JM, Calis GHM. J Am Chem Soc. 1984;106:3511–3521. [Google Scholar]
- 35.Leung KH, Phillips DL, Mao Z, Che CH, Miskowski VM, Chan CK. Inorg Chem. 2002;41:2054–2059. doi: 10.1021/ic011084v. [DOI] [PubMed] [Google Scholar]
- 36.Jaw HRC, Savas MM, Rogers RD, Mason WR. Inorg Chem. 1989;28:1028–1037. [Google Scholar]
- 37.Jaw HRC, Savas MM, Mason WR. Inorg Chem. 1989;28:4366–4369. [Google Scholar]
- 38.King C, Wang JC, Khan MNI, Fackler JP., Jr Inorg Chem. 1989;28:2145–2149. [Google Scholar]
- 39.Che CH, Kwong HL, Poon CK, Yam VWW. J Chem Soc, Dalton Trans. 1990:3215–3219. [Google Scholar]
- 40.Pirie NW, Pinhey KG. J Biol Chem. 1929:321–333. [Google Scholar]
- 41.Kerman K, Chikae M, Yamamura S, Tamiya E. Anal Chim Acta. 2007;588:26–33. doi: 10.1016/j.aca.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Lin PC, Ueng SH, Yu SC, Jan MD, Adak AK, Yu CC, Lin CC. Org Lett. 2007;9:2131–2134. doi: 10.1021/ol070588f. [DOI] [PubMed] [Google Scholar]
- 43.Storhoff JJ, Marla SS, Bao P, Hagenow S, Mehta H, Lucas A, Garimella V, Patno T, Buckingham W, Cork W, Muller UR. Biosens Bioelectron. 2004;19:875–883. doi: 10.1016/j.bios.2003.08.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The UV-Vis spectra of the Au NPs prepared at different pre-reduction pH values, the UV-Vis spectra of the Au NPs prepared in the presence and absence of excess GSH during reduction, and the chromatographic data of the Au(I)SG polymers at different pH values. This material is available free of charge via the Internet at http://pubs.acs.org.


