Abstract
We have developed a novel method that utilizes conformation-switching aptamers for real-time PCR analysis of protein analytes. The aptamers have been designed so that they assume one secondary structure in the absence of a protein analyte, and a different secondary structure in the presence of a protein such as thrombin or PDGF. The protein-bound structure in turn assembles a ligation junction for the addition of a real-time PCR primer. Protein concentrations could be specifically detected into the picomolar range, even in the presence of cell lysates. The method has advantages relative to both immunoPCR (since no signal is produced by background binding) and to the proximity ligation assay (PLA; since only one epitope on a protein surface must be bound, rather than two).
Keywords: Aptamer, real-time PCR, aptamer beacon, conformation-switching aptamer, SELEX, diagnostics
Introduction
ImmunoPCR methods can be used for the sensitive detection of protein analytes [1–4], but they have several distinct disadvantages relative to PCR for the detection of nucleic acids. It is difficult to reproducibly prepare antibody-DNA conjugates; binding and amplification reactions must be separated in time, hampering the development of homogenous or real-time formats; and both specifically and non-specifically bound probes can be amplified, potentially leading to high background signals.
In order to better couple protein detection with nucleic acid amplification, nucleic acid binding species (aptamers) have been adapted to a variety of amplification assays, including exonuclease-protection mediated ligation followed by PCR [5]; and rolling circle amplification [7–8]. Aptamers have also been used for the generation of allosteric ribozymes (aptazymes) that have similarly been coupled to amplification reactions [9–10]. ImmunoPCR methods have also been developed with aptamers, leading to the detection of a thrombin target in the pM range [11].
The most robust method aptamer-mediated amplifications have been obtained using the proximity ligation assay (PLA), in which adjacent binding of two aptamers to a protein or cellular target results in ligation and formation of a unique amplicon which can then be sensitively detected by real-time PCR [12–17]. Fredriksson and co-workers originally showed that aptamer-mediated PLA could be used for the detection of zeptomole amounts of platelet-derived growth factor (PDGF) [12]. Cell surface PLA can also detect as few as 100 B. anthracis and 10 B. subtilis spores, and down to 1 B. cereus spore [15]. While proximity amplification methods are extremely powerful, they can be limited by the need for more than one binding reagent.
We and others have previously reported that aptamers can be engineered so as to transduce the binding energy of ligands into changes in aptamer conformations. The resultant analyte-dependent folding of aptamers has been coupled to optical or electronic signaling [18–26]. Because nucleic acid secondary structure can be readily ‘programmed’, such structure-switching reagents can potentially be generated based on almost any aptamer without prior knowledge of the aptamer’s secondary or tertiary structure [23, 27].
A number of different types of structure-switching aptamers have previously been demonstrated. For example, Nutiu and Li [23] engineered structure-switching aptamers in which ligands stabilized the bound structure and concomitantly displaced an antisense oligonucleotide. Similarly, Bayer and Smolke [27] engineered structure-switching aptamers in which ligands stabilized the bound structure and concomitantly led to formation of an antisense interaction with a mRNA.
Building on these strategies, we now propose to couple conformation-switching aptamers with binding and ligation of an antisense oligonucleotide, ultimately leading to the formation of a novel amplicon for real-time PCR. In this way, we avoid the requirement for two probes currently inherent in the proximity ligation assay and other proximity amplification methods. We have applied our novel real-time detection method to the well-known anti-thrombin and anti-PDGF aptamers in part because of the detailed knowledge that is already available regarding their binding characteristics and structures, but also to more readily compare our results with other biosensor paradigms that have previously been advanced. We find that the conformation-switching aptamers can sensitively and specifically detect thrombin and PDGF both in solution and in cell lysates.
Materials and Methods
Materials
The sequences of the conformation-switching aptamers for the detection of thrombin were as shown in Figures 3, 4, and S1. The substrate for ligation was t.5′P (5′ p-GGTTGGTAGTCTCGAATTGCTCTCT), where 5′ p denotes a 5′ phosphate. Primers for PCR were t.F1 (5′-TGTGGTTGGTGTGGTTGGTT), t.F2 (5′-GGTTGGTTCATGGTCATATTGGT); t.R1(5′-GAGAGCAATTCGAGACTACCAACC) and t.R2 (5′-AGAGAGCAATTCGAGACTACC). All oligonucleotides except MGB probes were purchased from IDT (Coralville, IA).
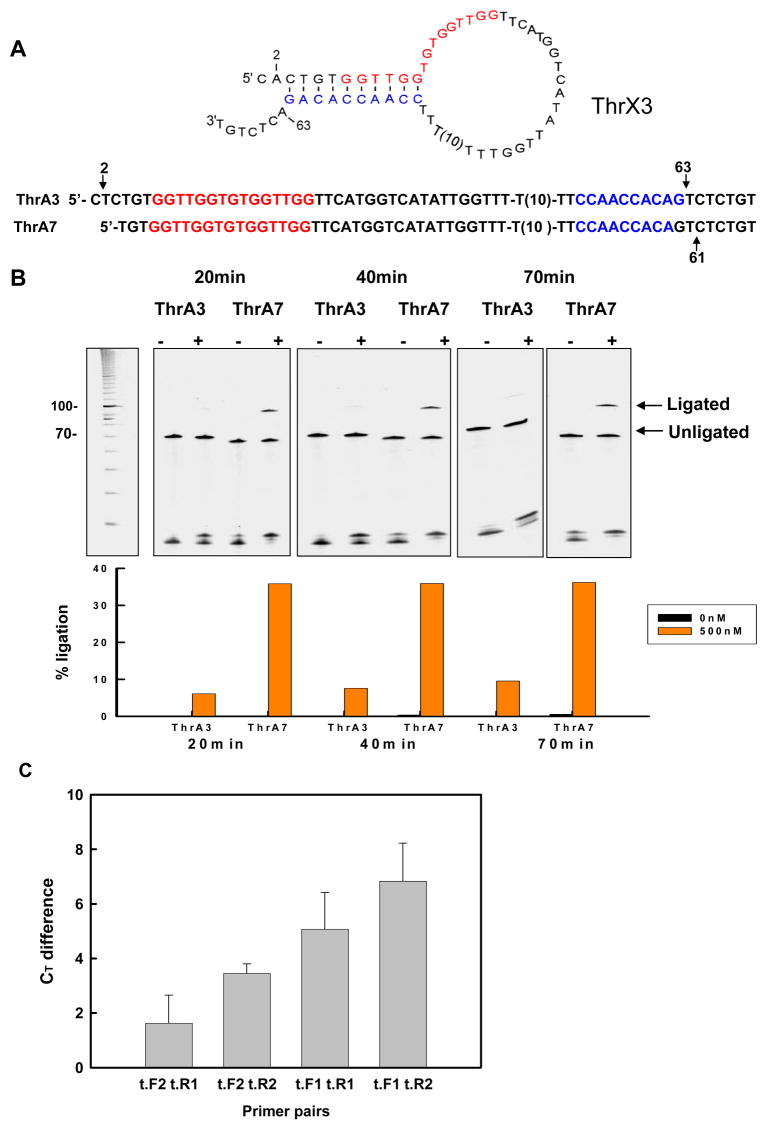
Figure 3.
Effect of antisense sequences on thrombin-dependent ligation. (A) Sequences of the designed constructs. Conventions are as in Figures 1 and S1. (B) Activation of ligation in the presence of thrombin. Ligation assays were carried out as described in Figure S1 except that the ligation time in the presence of 500 nM thrombin was 75min.
Figure 4.
Optimization of designs for real-time PCR. (A) Optimized thrombin-sensing conformation-switching aptamer sequences. ThrA3 and ThrA7 were designed based on ThrX3 and Thr7, respectively, as described in the text. Arrows indicate sequence changes. (B) Activation of ligation in the presence of thrombin. Ligation assays were carried out as described in Figure S1, except that the ligation time in the presence of 500 nM thrombin was varied between 20min and 70min. (C) Amplified ligation signals with different primer sets. Ligations were carried out for 20min with ThrA7 in the presence or absence of thrombin. An aliquot from the ligation reaction in the presence of 500 nM thrombin was used for real-time PCR with various combinations of the primers t.F1, t.F2, t.R1, and t.R2. The Ct difference between amplification in the presence and absence of thrombin (BSA alone) is indicated. Error bars were derived from three determinations.
The MGB probe used for real-time PCR was t.MGB17 (5′-FAM-TTCCAACCACAGTCTCT-MGB-3′) and it was obtained from Applied Biosystems (Foster City, CA). The advantage of the MGB (minor groove binding) probe is that the 3′ adduct can form extremely stable duplexes with single-stranded DNA targets, leading to higher melting temperature and increased specificity compared with unmodified DNA [28, 29]. In addition, the 3′ adduct acts as a quencher, leading to lower background. MGB probes can be as short as 13 nucleotides, compared with 18–22 nt for normal TaqMan probes. MGB probes have been widely used for real-time PCR assays and for quantitation [30–32].
Human α-thrombin, human Factor IXa, and human Factor Xa were purchased form Haematologic Technologies Inc. (Essex Junction, VT).
The sequences of the conformation-switching aptamers for the detection of PDGF were: p.3_13 (5′-AGCCTTTCTCGATCGGATCATTCACAGGCTACGGCACGTAGAGCATCACCATGATCC TGTGTTTCTTCTTCTTTTGATCGGATCATGGTGAT). The substrate for ligation was N.Sub2 (5′p-CCGATCCCTCTTGCTGCCTACGCATGCTGTTACCGTCCGCTACTCATTCC). Primers for PCR were p.F (5′-AGCCTTTCTCGATCGGATCA) and p.R (5′-GGAATGAGTAGCGGACGGTAAC). The TaqMan probe used for real-time PCR was BB-PROBE-12 (5′-FAM-CGATCCCTCTTGCTGCCTACGCA-TAMRA-3′). Again, all oligonucleotides were purchased from IDT (Coralville, IA). PDGF-BB, -AA, and -AB were purchased from R&D Systems (Minneapolis, MN). The PDGF proteins were reconstituted in 4 mM HCl with 0.2% BSA (Invitrogen, Carlsbad, CA), as suggested by the supplier.
Assaying conformation-switching aptamers
In order to test the structure-switching abilities of the designed constructs, ligation was assayed by gel electrophoresis. Pre-ligation mixtures (45 μL) contained 5 μL 10× binding buffer (50mM KCl, 10mM MgCl2, and 20mM Tris.Cl, pH 7.4), 1 μL 10μM designed conformation-switching anti-thrombin aptamer and 39 μL H2O were denatured at 70° C for 3min and cooled to room temperature. The reaction mixture was incubated with or without thrombin for 30 min, and then 1.5 μL 25mM ATP, 1.5 μL 10μM substrate (denatured prior to use) and 1 μL T4 DNA ligase (2U/μL, Epicentre, Madison, WI) were added separately. The final concentration of ATP in the 50 μL ligation mixture was 0.75 mM, conformation-switching aptamer was 0.2 μM, and substrate oligonucleotide was 0.3 μM. Ligation reactions were carried out at room temperature for various times and were terminated by heating to 95° C for 15min. Ligated and unligated species were separated on denaturing (7 M urea) 8% polyacrylamide gels and stained with SybrGold (Molecular Probes, Eugene, OR). The percentage of ligation ( ) in the absence and presence of thrombin was calculated by scanning the stained gel and determining band densities with QuantityOne software (BioRad Laboratories, Hercules, CA). Fold activation was in turn calculated based on the dividing the percentage of ligation in the presence by the percentage of ligation in the absence of thrombin.
Aptamer concentrations for ligation assays were 200nM, higher than that used in real-time PCR assays (0.4nM) in order to be visible by staining.
Detection of thrombin by ligation followed by real-time PCR
Ligation reactions were carried out, and then transferred to real-time PCR. The ligation reactions were similar to those described above except that the final concentration of ThrA7 was 0.4nM and 0.002% BSA was present. The reaction mixture was incubated with thrombin for 30 min, and then ATP, various amounts of substrate oligonucleotide (t.5′P) and 1 μL T4 DNA ligase were added separately. Ligation proceeded at room temperature for 20min, and was terminated by heating at 95° C for 15min. A 2 μL aliquot of the terminated ligation reaction, 15 μL 2xTaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA), 0.5 uL 10μM forward primer, 0.5 μL 10μM reverse primer, 0.5 μL 10μM MGB probe and 11.5 μL H2O were mixed to make a total volume of 30 μL. The final PCR mixture contained 167nM t.F1, 167nM t.R2, and 167nM t.MGB17 in 1x TaqMan Universal PCR Master Mix. Real-time PCR was carried out in a 96-well PCR plate (Applied Biosystems), covered with strip caps (Applied Biosystems). The thermal cycling regime was: initial denaturation for 10min at 95° C, and then cycling for 15s at 95° C and 60s at 60° C, repeated 50 times on a Applied Biosystems HT7900 real-time PCR machine.
Detection of PDGF by ligation followed by real-time PCR
Pre-ligation mixtures contained 5 μL 10x PDGF binding buffer (1.37M NaCl, 101mM Na2HPO4, 18mM KH2PO4 pH 7.4, 27mM KCl, 100mM Tris.Cl, pH 7.4, and 25mM MgCl2) and 1 μL 20nM conformation-switching aptamer p.3_13 and were denatured at 70° C for 3min prior to cooling to room temperature. The reaction mixture was incubated with PDGF-BB for 30min, and then 1.5 μL 25mM ATP (final concentration of 0.75 mM), and 1, 5, 10, or 20 μL 20nM oligonucleotide substrate (N.Sub2, denatured prior to use) and 1 μL T4 DNA ligase (2U/μL, Epicentre, Madison, WI) were added separately to make a final reaction volume of 50μL. The final concentration of BSA in ligation reactions with PDGF was always 0.004%. Ligation was allowed to proceed for 5min, 30min, or 60min at room temperature, and was terminated by heating the reaction to 95° C for 15min. A 2 μL aliquot of terminated ligation reaction was introduced into 1x TaqMan Universal Master Mix (Applied Biosystems) containing 167nM p.F, 167nM p.R and 334nM BB-PROBE-12 and real-time PCR was carried out as detailed for thrombin detection, above.
Ligation in cell lysate
To prepare lysate, approximately 1 × 107 human 293T fibroblast cells (ATCC, Manassas, VA) were collected. Cells were treated with 4mL M-PER mammalian protein extraction reagent (Pierce Biotechnology, Rockford, IL) and shaken at room temperature for 10min. After centrifugation at 10,000rpm for 25min at 4° C, pellets containing dissolved cell membranes and hydrophobic membrane proteins were removed and the supernatant consisting of intracellular proteins and nucleic acids was collected and frozen at −80° C. Total protein in lysate aliquots was quantitated using a bicinchoninic acid total protein assay kit (Pierce Biotechnology, Rockford, IL). The final concentration of lysate proteins in each 50μL ligation reaction mixture was 1 μg/mL. Protein analytes at different concentrations were added to the lysate and ligation and real-time PCR were carried out as above. PDGF samples were always in 0.004% BSA (final concentration in 50μL ligation reaction) and thrombin samples were always in 0.002% BSA (final concentration in 50μL ligation reaction).
Results and Discussion
Adapting conformation-switching aptamers to PCR
Despite the demonstrated advantages of real-time PCR for quantitation, the real-time PCR detection of protein analytes is not routinely practiced. Most schemes for immunoPCR do not directly couple protein-binding to PCR amplification, instead requiring wash steps and other processing prior to amplification. However, conformation-switching aptamers can be used to transduce analyte-binding into optical and other signals [19–21, 24–26]. Therefore, rather than coupling analyte-mediated conformational changes to optical signaling, we instead designed conformation-switching aptamers that could participate in PCR.
A variety of configurations for coupling aptamer conformational changes to PCR can be envisioned, but the simplest configurations would be to have aptamer conformational changes lead to the production of a template for PCR or a primer for PCR. The production of a PCR template would likely yield greater sensitivity of detection, since conformational transduction to produce even a single template should lead to amplification, while multiple primers would have to change conformation in order to support an amplification reaction.
However, it would be difficult to design a PCR template that was only amplified in one conformation, since conformational equilibration even in the absence of analyte would lead to at least some template being available at all times, especially during thermal cycling and in the presence of a DNA polymerase that could read through restrictive conformations. Thus, the use of a single conformation-switching aptamer as a PCR template would of necessity result in high background. Therefore, we decided to require a ligation step to produce competent templates for PCR amplification. In this way, the ligation reaction can be separately optimized to reduce background and no further ligation should occur during thermal cycling. This configuration will also still be useful for the facile detection of analytes, since the ligation reaction can be directly coupled with a real-time PCR without additional wash or other steps.
In our method, antisense sequences (blue, in Figures 1 and 2) added to aptamer (red) termini promote the formation of a non-binding conformation (Figures 1a, 2a), a configuration that is similar to the design of other conformation-switching aptamers [23, 25, 27]. In the presence of analyte, the binding conformation is more favored, and in this conformation a short hairpin stem becomes available for hybridization to an oligonucleotide substrate and subsequent ligation to form a new amplicon (Figures 1b, 2b). Additional sequences added to either end of the aptamer served as primer- and probe-binding sites for real-time PCR. To show the generality of the method, we initially designed two different conformation-switching aptamers, one that bound thrombin and one that bound platelet-derived growth factor (PDGF; Figures 1 and 2, respectively). It should be noted that while one inhibitory conformation is indicated in Figure 1 two different, non-binding secondary structures are possible (similar to conformation-switching aptamers previously designed by Xiao, Piorek, et al., [26]). While both conformation-switching aptamers employ the same basic design, different optimization methods were explored with the two aptamers, and assay results with thrombin and PDGF will be described separately, below.
Figure 1.
Sequence and secondary structure of a designed conformation-switching anti-thrombin aptamer. (A) Inactive conformation in the absence of thrombin. The 3′ end of the aptamer has been extended with an antisense sequence (blue) in order to promote the formation of duplex structure and denaturation of the aptamer. (B) Active conformation in the presence of thrombin. Formation of a quadruplex and binding to thrombin stabilizes a structure in which the extended 3′ end of the aptamer (red) forms a short hairpin structure that can hybridize to a substrate oligonucleotide (green) to form a ligation junction. Following ligation with T4 DNA ligase, the ligated aptamer and substrate can be amplified by PCR using indicated forward (For) and reverse (Rev) primers. The position of the TaqMan MGB probe (Probe) used during real-time PCR amplification is indicated by a dashed red line. The sequence shown is aptamer ThrA7, which is further described in Figure 4a.
Figure 2.
Sequence and secondary structure of a designed conformation-switching anti-PDGF aptamer. (A) Inactive conformation in the absence of PDGF. (B) Active conformation in the presence of PDGF. While the active conformation of the anti-thrombin aptamer is a quadruplex, the active conformation of the anti-PDGF aptamer is a multihelix junction. Features of the design are as in Figure 1.
Optimization of thrombin-sensing conformation-switching aptamers
In addition to adding a ligation junction and sequences for real-time PCR amplification to the aptamers, we found that in order to allow both proteins (the analyte and T4 DNA ligase) to functionally interact with the DNA it was important to include spacer regions. This can be seen by looking at results with a series of conformation-switching anti-thrombin aptamers (Figure S1). In initial designs, several different spacer lengths were used to separate the aptamer and antisense sequences. The spacer sequences were chosen by using the program Mfold (http://www.bioinfo.rpi.edu/applications/mfold/dna/form1.cgi) to evaluate what secondary structures might form. A pyrimidine-rich spacer was found to minimize the formation of alternative secondary structures and to promote the predicted interaction of the aptamer and antisense sequence.
The designs were assayed by looking at thrombin-dependent ligation at 1.5 and 16 hours. The shortest spacer between aptamer and antisense (21 residue spacer; Thr9) gave very little thrombin-dependent ligation. When an additional T10 spacer was added (31 residue spacer; Thr7), though, ligation was more strongly activated by thrombin (11-fold increase at 1.5 hours). When this spacer was extended to 20 residues (41 residue spacer; Thr8), background ligation in the absence of thrombin increased. The longer spacer in Thr8 formed a longer loop structure that apparently destabilized the interactions between the aptamer and the antisense sequence, and thus promoted a ligand-independent, ligation-competent conformation. In all instances, ligation was most strongly activated by thrombin at 1.5 hours, but over time equilibration between the binding and non-binding conformations allowed ligation to occur even in the absence of the analyte, as would be expected for a metastable structure.
In order to further probe the relationship between sequence, conformation-switching, and ligation, a slightly different series of conformation-switching aptamers was synthesized and assayed (Figure 3). In this series, the T10 spacer has been repositioned within the aptamer, nearer the 3′ end. In addition, since it is likely that most optimizations of structure-switching aptamers will center on changing the number and types of base-pairs between the aptamer and the added antisense sequences, we increased the designed base-pairing from 9 bp (Thr7) to 10 bp (ThrX3), 11 bp (ThrX2), and 12 bp (ThrX1). Strong analyte-dependent ligation (36-fold after 75min) could be observed in ThrX3, but not in the other two constructs. The longer antisense sequence could no longer be readily displaced by thrombin-binding. This was especially interesting because the 12 bp stem in ThrX1 was identical to that found in other conformation-switching anti-thrombin aptamer constructs [23, 24]. However, in these previous constructs, base-pairing was with an antisense oligonucleotide in trans, not an antisense sequence in cis, and thus the number of designed base pairs had to be shortened to accommodate the decreased entropy of structural rearrangement. The fact that the position of the spacer and the number of designed base-pairs (9 in Thr7 and 10 in ThrX3) can be changed while structure-switching is retained indicates the robustness of this design approach.
These results indicated that the Thr7 (9 bp hairpin) and ThrX3 (10 bp hairpin) conformation-switching aptamers were both energetically and kinetically poised to switch upon interaction with thrombin. This was important, since analyte responsivity should be highest when the free energy of interaction between the protein and the binding conformation of the aptamer is just slightly larger than the difference between the free energies of folding between the binding and non-binding conformations.
The ThrX3 conformation-switching aptamer was further optimized for real-time PCR by changing the sequences of the probes, primers, and their complementary binding sites. Probe design was guided by two criteria: avoiding secondary structural interference with analyte binding and ligation, and identifying a sequence that could function as a good TaqMan probe. Nucleic acid secondary structures were evaluated using the program Mfold (http://www.bioinfo.rpi.edu/applications/mfold/dna/form1.cgi), while TaqMan functionality (GC content, Tm values) were evaluated using Primer Express Software (Applied Biosystems). Based on an iterative use of these tools, we introduced slight sequence changes into ThrX3 in order to utilize a better probe (and corresponding probe-binding site). This resulted in the production of the conformation-switching anti-thrombin aptamer ThrA3 (Figure 4a) which contained an A to T change in the probe-binding site at position 63 (arrow in Figure 4) and that could therefore utilize a predicted 17 nt real-time PCR probe (5′-TTCCAACCACAGTCTCT; this probe was obtained from Applied Biosystems and was also conjugated to a minor-groove binding compound, as detailed in Materials and Methods). Since position 63 was also involved in the formation of the hairpin stem in the non-binding conformation, position 2 had to be correspondingly changed from A to T, to avoid extending pairing with the antisense sequence.
Since the conformation-switching anti-thrombin aptamer Thr7 had also demonstrated excellent thrombin responsivity, we also attempted to adapt this aptamer to real-time PCR. The T10 spacer was again repositioned to be at the 3′ end of the aptamer (as with ThrX1-ThrX3) and a mutation was again introduced (T61 to C61) to create a probe-binding site (Figure 4a and Figure 1b) that could utilize the same, 17 nt predicted probe sequence described above for ThrX3. The final construct was called aptamer ThrA7.
Both new aptamers, ThrA3 and ThrA7, were assayed for their ability to ligate in the presence of 500nM thrombin (Figure 4b), and were found to be as or more active than the parental sequence. Again, the ability to readily manipulate the sequences of the conformation-switching aptamers without losing thrombin-dependence indicates the robustness of these constructs and of the design process in general.
Earlier ligation times were assayed in order to determine if thrombin-dependent activation could be increased. Both aptamers retained thrombin-dependence. ThrA7 shows greater ligation and thrombin-dependent activation than ThrA3 at all time points. The activation of the conformation-switching aptamers at the shorter time points was much greater than was previously observed at 1.5 hours (compare Figure 4b with Figures 3b and S1b). The conformation-switching aptamer ThrA3 was activated by 82-fold while ThrA7 was activated by 360-fold. These results likely mean that analyte-dependent ligation occurs relatively quickly, while background ligation occurs more slowly. Thus, while larger signals were recovered at longer times, the best signal-to-noise ratios were obtained at short times, strongly arguing for the use of signal amplification methods with these conformation-switching biosensors. In order to facilitate real-time PCR, several different primer sequences were assayed (Figure 4c). Two different forward primers and two different reverse primers were designed, each of which had a predicted Tm near 60° C. Real-time PCRs were carried out with the four possible combinations of forward and reverse primers.
In order to quantitatively determine the extent of ligation and ligation activation, we relied on delta Ct values. The Ct (cycle threshold) value for a real-time PCR reaction is the number of cycles required to show a signal above background, and is typically determined automatically by the real-time PCR machine. Delta Ct values were calculated by subtracting the Ct value for a reaction with thrombin (cycle threshold should be reached earlier) from the Ct value for the reaction without thrombin (cycle threshold should be reached later). The best pair of primers proved to be t.F1 and t.R2, which generated a Ct difference in the presence of saturating thrombin concentrations (500nM) of 7, which is much higher than that typically observed for the determination of statistically significant changes in mRNA expression by real-time PCR. For example, delta Ct values in the range of −0.9 to 3 were considered significant for quantitating diagnostically important alterations in hemoglobin-alpha mRNA expression levels [33]. Similarly, delta Ct values as low as 0.15 to 0.37 for κ and λ immunoglobulin light chain mRNA expression levels have been considered legitimate for determining B-lymphocyte monoclonality [34]. Hundley, et al. have shown that a delta Ct of less than 2 represents at least a 3.2-fold difference in the expression of myosin binding protein H mRNAs [35].
Thrombin-mediated ligation followed by real-time PCR
The optimized conformation-switching aptamers ThrA3 and ThrA7 and the primer set t.F1 and t.R2 were used in real-time PCR assays for the sensitive detection of thrombin. In these assays, protein-dependent ligation was first carried out, followed by real-time amplification and detection of the ligated products. Thrombin (with BSA as a carrier protein) was incubated with the aptamers for 30 minutes, and T4 DNA ligase and reaction components were then added. Ligation was allowed to proceed for 5-40 minutes and stopped by heat denaturation. A fraction of the ligation reaction was then used to seed a real-time PCR. As a control, a reaction without thrombin (but still containing BSA) was carried out.
Positive delta Ct values were obtained at a variety of oligonucleotide (t.5′P) substrate concentrations irrespective of the length of ligation time, although shorter ligation times tended to provide larger signals (data not shown). Ligation times as short as 5 minutes were attempted to try to further improve the signal-to-noise ratio, as had previously been observed when ligation times were decreased from 1.5 hours to 20 minutes. However, there were larger variances in Ct values at ligation times less than 20 minutes. Because of this, all subsequent assays incorporated a ligation time of 20 minutes.
In order to determine the sensitivity of the method, ligation and real-time PCR assays were performed at a series of thrombin concentrations. Delta Ct values as a function of thrombin concentration are shown in Figure 5a. The variances of the delta Ct values were quite low, the limit of detection (a signal three times the standard deviation of the background) could be detected with as little as 0.8nM thrombin. Moreover, the concentration of thrombin correlated well with the real-time PCR signal between 0.1 and 20nM thrombin.
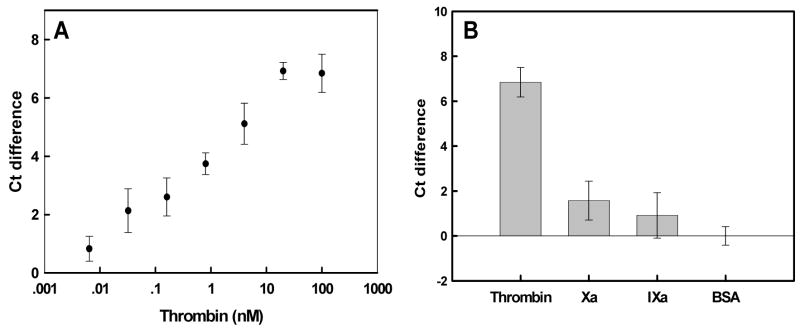
Figure 5.
Real-time PCR amplification of thrombin-dependent ligation reactions. (A) Ct differences as a function of thrombin concentration. Ct differences were calculated relative to BSA alone (0.002% final). (B) Specificity of response. Ligation reactions were carried out in the presence of either 100nM thrombin or similar proteases, such as factor Xa and IXa. Either BSA or no protein controls were also carried out. Error bars were derived from at least three determinations.
The thrombin concentration range in blood plasma is 5-500NIH units/mL, where 1NIH Unit = 0.324+/−0.073ug [36, 37]. Therefore, the lowest physiologically relevant thrombin concentration would be nanomolar to low micromolar concentrations [1, 38], well within the range of our assay.
Our results compare quite favorably with previous aptamer-based thrombin assays. The observed detection limits using the anti-thrombin aptamer and optical biosensors varied from 40nM (capillary electrophoresis with laser-induced fluorescence detection; [39] ) to 1nM (fiber optic biosensor with labeled thrombin; [1]) to 429 ± 63pM (aptamer beacon that yielded changes in FRET; [21]) to 373 ± 30pM (aptamer beacon that dequenched; [22]). For electronic biosensors the values were 2–3 nanomolar [26, 40, 41]. When the anti-thrombin aptamer was adapted to the equivalent of an ELISA the limit of detection was <1nM [42]. The best results have been garnered by other real-time methods: a detection limit of 10 pM in the presence of serum with real-time PCR [11], a detection limit of 30pM by proximity rolling circle amplification [6], and down to several hundred of molecules by exonuclease protection coupled with real-time PCR [5].
The specificity of detection was examined by substituting thrombin with other proteases from the clotting cascade, Factors IXa and Xa (Figure 5b). In the absence of protein or in the presence of carrier BSA, Factor IXa, Factor Xa, or in the absence of protein, the delta Ct signal was very small relative to the value obtained with thrombin. The amplified signal in the absenceof any protein is again higher than that in the presence of BSA.
Taken together, these results suggest that assays with physiological samples should also yield sensitive and specific detection, especially given that our limits of detection seem to be within the same range as thrombin concentrations found in physiological samples, as described above. In order to test this possibility, we carried out assays with thrombin in human 293T fibroblast cell lysates (Figures 6a and 6b). Despite the fact that lysate was in 845-fold excess (by weight), the detection of thrombin in cell lysate was specific and as sensitive as in the absence of lysate. The signals in protein and no protein controls were somewhat higher than those in solution, but the detection limit for thrombin (a signal three times the standard deviation of the background) was 32pM, even lower than that in the absence of lysate (800pM).
Figure 6.
Thrombin detection against a background of cell lysate. (A) Ct differences as a function of added thrombin concentration. Reactions and signal determination were as in Figure 5, except that ligation reactions were carried out in the presence of 1 μg/mL 293T fibroblast cell lysate. Ct differences were calculated relative to BSA in lysate. (B) Specificity of response. BSA (0.002%) in lysate was used as a negative control. Error bars were derived from at least four determinations.
PDGF mediated ligation followed by real-time PCR
As originally described in Figure 2, a conformation-switching anti-PDGF aptamer was generated by appending a 13 nt antisense sequence to the 3′end of the anti-PDGF aptamer via an 18 nt linker region. Again, the aptamer should form a hairpin in the non-binding conformation (Figure 2a), and assume its native binding structure in the presence of PDGF (Figure 2b). In addition, a substrate oligonucleotide (N.Sub2) was designed to form 6 base-pairs at the ligation junction in the binding conformation, and a primer binding site was appended to the 5′ end of the anti-PDGF aptamer to enable real-time PCR amplification. One significant difference from the design of real-time PCR assays with the conformation-switching anti-thrombin aptamer was that the probe binding site for real-time PCR was embedded in the substrate oligonucleotide sequence rather than in the conformation-switching anti-PDGF aptamer sequence. This variation in molecule design should provide significantly greater flexibility in the oligonucleotide probes that can be used and hence in assay development.
As proof of the greater robustness of this design strategy, the initial conformation-switching anti-PDGF aptamers worked well enough to immediately use in real-time PCR without further sequence optimization (Figure 7). Optimization of oligonucleotide substrate (N. Sub2) concentration and ligation time prior to real-time PCR revealed that using 8nM substrate at 60 minutes (rather than 20 minutes, as with the conformation-switching anti-thrombin aptamer) gave the greatest Ct with the least variance (data not shown). Using 8nM substrate concentration and 60min ligation time, real-time PCR was performed using a series of PDGF-BB concentrations. Delta-Ct as a function of PDGF-BB concentration is shown in Figure 7a. The lowest concentration that could be reliably detected was 0.32nM, and the dynamic range of the assay extended to 40nM.
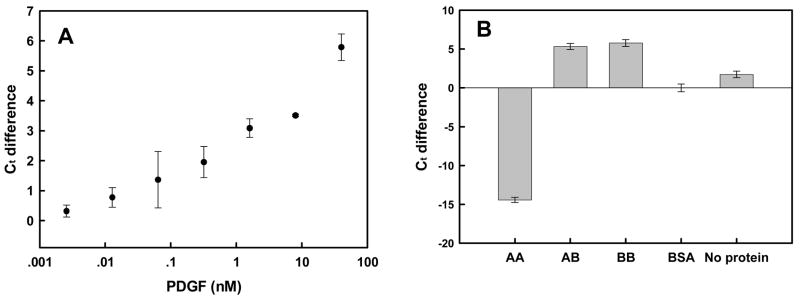
Figure 7.
Real-time PCR amplification of PDGF-dependent ligation reactions. (A) Ct differences as a function of PDGF concentration. Reactions and signal determination were as in Figure 5. Ct differences were calculated relative to BSA alone (0.004% final). (B) Specificity of response. Ligation reactions were carried out in the presence of different PDGF isoforms (40 nM). Either BSA or no protein controls were also carried out. Error bars were derived from at least three determinations.
Again, our results are consistent with the adaptation of this assay to clinical samples. PDGF levels in human serum have been found to be 14.4–24.8ng/mL, or 0.56nM – 1.12nM. The sensitivities observed with our method are within these physiological levels. In addition, serum PDGF levels have been reported to be elevated in many disease states.
These results compare well with most other analytical methods that utilize the same anti-PDGF aptamer. Low nanomolar detection limits have previously been observed with an aptamer beacon developed by Tan’s group [43], with an aptamer-based fluorescence anisotropy assay [44], with a colorimetric determination technique using aptamer-modified gold nanoparticles [45], with a luminescence detection strategy [46], and with an exciton detection strategy [47]. It should be noted, though, that proximity ligation coupled with real time PCR could detect zeptomole amounts of PDGF [12]. However, the proximity ligation assay (PLA) requires two probes, rather than one, to achieve detection, and thus is not as generally applicable as the method we describe.
The specificity of detection was examined by determining delta Ct values with PDGF-AA, AB, BB, or no protein (Figure 7b). In the presence of PDGF-AA, the amplified signal was much lower than the background levels, while the signal in the presence of both PDGF-AB and –BB was significantly higher than background. These results accord with what is known of the anti-PDGF aptamer, as it binds to the B-chain of the protein and has roughly equal affinity for both PDGF-AB and-BB [48]. As we previously observed with the conformation-switching aptamers that recognized thrombin, non-cognate proteins appeared to actually decrease the amplified signal relative to no protein controls, perhaps because protein:DNA interactions inhibited conformational changes that might normally have occurred in solution. Such ‘anti-ligation’ signals have previously been observed in proximity ligation assays [15], and may allow the specific signaling of PDGF-BB relative to PDGF-AA, something that has not been observed in other aptamer-based assays [44].
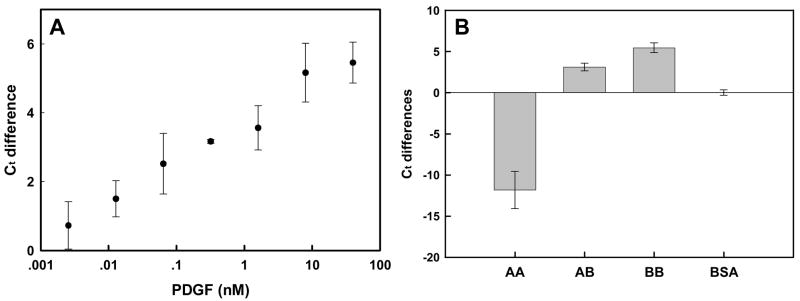
The excellent performance of the conformation-switching anti-PDGF aptamers with respect to sensitivity, specificity, and resilience to interferents suggested that they should also function in the presence of cell lysates. Figures 8a and 8b summarize the dose response and selectivity of PDGF detection in cell lysates. The LOD was 12.8 pM and the delta Ct difference was linearly correlated with the log of the PDGF concentrations within the range of 12.8 pM-8 nM PDGF. These results again demonstrate that our conformation-switching aptamer technology in concert with real-time PCR should allow the sensitive quantitation of PDGF in clinical samples.
Figure 8.
PDGF detection against a background of cell lysate. (A) Ct differences as a function of added PDGF concentration. Reactions and signal determination were as in Figure 5, except that ligation reactions were carried out in the presence of 1 μg/mL 293T fibroblast cell lysate. Ct differences were calculated relative to BSA in lysate. (B) Specificity of response. BSA (0.004%) in lysate was used as a negative control. Error bars were derived from at least three determinations.
Conclusions
While conformation-switching aptamers have been adapted to the detection of protein analytes in a variety of formats, there is an inherent difficulty in this approach. In order to have low background and a high signal-to-noise ratio, it is essential that the non-binding conformation of the aptamer be much more stable than the binding conformation. However, the more stable the non-binding conformation of the aptamer is, the more binding energy will be required to stabilize the binding conformation, and the smaller the apparent Kd (and ultimate sensitivity of the analytical method) will be. In order to try to overcome this limitation, we have focused on adaptating conformation-switching aptamers to amplification methods. In this way, we hope to use conformation-switching aptamers with a high signal-to-noise ratio, and to amplify the small signals that occur in the presence of even small amounts of protein analytes. By utilizing different probes for different conformation-switching aptamers it should be straightforward to adapt these assays to a multiplex format, as has recently also been done for the proximity ligation assay [49].
We have previously used a one-piece conformation-switching aptamer and real-time RCA for detection [47]. The method described in the current work, two-piece ligation followed by real-time PCR, has proven to be more sensitive. The calculated LOD for PDGF with one-piece ligation followed by real-time RCA was 0.4nM, while that for two-piece ligation followed by real-time PCR was 12.8pM, about a 30-fold improvement in sensitivity. Just as the proximity ligation assay (PLA) is inherently much more sensitive than immunoPCR because two binding events are required in order to generate a signal, bimolecular formation of a ligation junction is inherently more difficult than unimolecular formation, and hence gives lower background and greater sensitivity.
Supplementary Material
Figure S1. Effect of spacer length on thrombin-dependent ligation. (A) Sequences of thrombin-sensing structure-switching aptamers that have different spacer lengths connecting the antisense sequence (blue) and the aptamer (red). (B) Activation of ligation in the presence of thrombin. Ligation reactions were carried out for 1.5 or 16 hrs. Ligated and unligated species were visualized on 8 % PAGE gels stained with SYBR Gold. The percentage of ligation in the absence and presence of 500 nM thrombin was determined by analysis with QuantityOne software (BioRad Laboratories, Hercules, CA) following scanning of the stained gel.
Acknowledgments
We would like to acknowledge the NIBIB and the Welch Foundation for support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spiridonova VA, Kopylov AM. DNA aptamers as radically new recognition elements for biosensors. Biochem (Mosc) 2002;67:706–709. doi: 10.1023/a:1016110724564. [DOI] [PubMed] [Google Scholar]
- 2.Adler M. Immuno-PCR as a clinical laboratory tool. Adv Clin Chem. 2005;39:239–292. [PubMed] [Google Scholar]
- 3.Niemeyer CM, Adler M, et al. Immuno-PCR: high sensitivity detection of proteins by nucleic acid amplification. Trends Biotechnol. 2005;23:208–216. doi: 10.1016/j.tibtech.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Barletta J. Applications of real-time immuno-polymerase chain reaction (rt-IPCR) for the rapid diagnoses of viral antigens and pathologic proteins. Mol Aspects Med. 2006;27:224–253. doi: 10.1016/j.mam.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Wang XL, Li F, et al. Ultrasensitive detection of protein using an aptamer-based exonuclease protection assay. Anal Chem. 2005;77:2278. doi: 10.1021/ac0494228. [DOI] [PubMed] [Google Scholar]
- 6.Di Giusto DA, Wlassoff WA, et al. Proximity extension of circular DNA aptamers with real-time protein detection. Nucleic Acids Res. 2005;33:e64. doi: 10.1093/nar/gni063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soderberg O, Gullberg M, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Fung CW, et al. Real-Time Rolling Circle Amplification for Protein Detection. Anal Chem. 2007;79:3320–3329. doi: 10.1021/ac062186b. [DOI] [PubMed] [Google Scholar]
- 9.Robertson MP, Ellington AD. In vitro selection of nucleoprotein enzymes. Nat Biotechnol. 2001;19:650–655. doi: 10.1038/90256. [DOI] [PubMed] [Google Scholar]
- 10.Cho EJ, Yang L, et al. Using a deoxyribozyme ligase and rolling circle amplification to detect a non-nucleic acid analyte, ATP. J Am Chem Soc. 2005;127:2022–2023. doi: 10.1021/ja043490u. [DOI] [PubMed] [Google Scholar]
- 11.Fischer NO, Tarasow TM, et al. Protein detction via direct enzymatic amplification of short DNA aptamers. Anal Biochem. 2008;373:121–128. doi: 10.1016/j.ab.2007.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredriksson S, Gullberg M, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 13.Gullberg M, Gustafsdottir SM, et al. Cytokine detection by antibody-based proximity ligation. Proc Natl Acad Sci USA. 2004;101:8420–8424. doi: 10.1073/pnas.0400552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustafsdottir SM, Schallmeiner E, et al. Proximity ligation assays for sensitive and specific protein analyses. Anal Biochem. 2005;345:2–9. doi: 10.1016/j.ab.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Pai S, Ellington AD, et al. Proximity ligation assays with peptide conjugate ‘burrs’ for the sensitive detection of spores. Nucleic Acids Res. 2005;33:e162. doi: 10.1093/nar/gni150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustafsdottir SM, Nordengrahn A, et al. Detection of individual microbial pathogens by proximity ligation. Clin Chem. 2006;52:1152–1160. doi: 10.1373/clinchem.2005.065847. [DOI] [PubMed] [Google Scholar]
- 17.Schallmeiner E, Oksanen E, et al. Sensitive protein detection via triple-binder proximity ligation assays. Nat Methods. 2007;4:135–137. doi: 10.1038/nmeth974. [DOI] [PubMed] [Google Scholar]
- 18.Stojanovic MN, de Prada P, et al. Fluorescent Sensors Based on Aptamer Self-Assembly. J Am Chem Soc. 2000;122:11547–11548. doi: 10.1021/ja0022223. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto R, Baba T, et al. Molecular beacon aptamer fluoresces in the presence of Tat protein of HIV-1. Genes Cells. 2000;5:389–396. doi: 10.1046/j.1365-2443.2000.00331.x. [DOI] [PubMed] [Google Scholar]
- 20.Hamaguchi N, Ellington AD, et al. Aptamer beacons for the direct detection of proteins. Anal Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- 21.Stojanovic MN, de Prada P, et al. Aptamer-based folding fluorescent sensor for cocaine. J Am Chem Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 22.Li JJ, Fang X, et al. Molecular aptamer beacons for real-time protein recognition. Biochem Biophys Res Commun. 2002;292:31–40. doi: 10.1006/bbrc.2002.6581. [DOI] [PubMed] [Google Scholar]
- 23.Nutiu R, Li Y. Structure-switching signaling aptamers. J Am Chem Soc. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 24.Levy M, Cater SF, et al. Quantum-dot aptamer beacons for the detection of proteins. Chembiochem. 2005;6:2163–2166. doi: 10.1002/cbic.200500218. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Lu Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew Chem Int Ed Engl. 2005;45:90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- 26.Xiao Y, Piorek BD, et al. A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J Am Chem Soc. 2005;127:17990–17991. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- 27.Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 28.Afonina I, Zivarts M, et al. Efficient priming of PCR with short oligonucleotides conjugated to a minor groove binder. Nucleic Acids Res. 1997;25:2657–2660. doi: 10.1093/nar/25.13.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kutyavin IV, Afonina IA, et al. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Decaro N, Elia G, et al. A minor groove binder probe real-time PCR assay for discrimination between type 2-based vaccines and field strains of canine parvovirus. J Virol Methods. 2006;136:65–70. doi: 10.1016/j.jviromet.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoehne M, Schreier E. Detection of Norovirus genogroup I and II by multiplex real-time RT- PCR using a 3′-minor groove binder-DNA probe. BMC Infect Dis. 2006;6:69. doi: 10.1186/1471-2334-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lole KS, Arankalle VA. Quantitation of hepatitis B virus DNA by real-time PCR using internal amplification control and dual TaqMan MGB probes. J Virol Methods. 2006;135:83–90. doi: 10.1016/j.jviromet.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Nussbaumer C, Gharehbaghi-Schnell E, et al. Messenger RNA profiling: a novel method for body fluid identification by real-time PCR. Forensic Sci Int. 2006;157:181–186. doi: 10.1016/j.forsciint.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 34.Stahlberg A, Aman P, et al. Quantitative real-time PCR method for detection of B-lymphocyte monoclonality by comparison of kappa and lambda immunoglobulin light chain expression. Clin Chem. 2003;49:51–59. doi: 10.1373/49.1.51. [DOI] [PubMed] [Google Scholar]
- 35.Hundley AF, Yuan L, et al. Skeletal muscle heavy-chain polypeptide 3 and myosin binding protein H in the pubococcygeus muscle in patients with and without pelvic organ prolapse. Am J Obstet Gynecol. 2006;194:1404–1410. doi: 10.1016/j.ajog.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 36.Aronson DL, Stevan L, et al. Generation of the combined prothrombin activation peptide (F1-2) during the clotting of blood and plasma. J Clin Invest. 1977;60:1410–1418. doi: 10.1172/JCI108902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fenton JW., II Thrombin. Ann NY Acad Sci. 1986;485:5–15. doi: 10.1111/j.1749-6632.1986.tb34563.x. [DOI] [PubMed] [Google Scholar]
- 38.Lee M, Walt DR. A fiber-optic microarray biosensor using aptamers as receptors. Anal Biochem. 2000;282:142–146. doi: 10.1006/abio.2000.4595. [DOI] [PubMed] [Google Scholar]
- 39.German I, Buchanan DD, et al. Aptamers as ligands in affinity probe capillary electrophoresis. Anal Chem. 1998;70:4540–4545. doi: 10.1021/ac980638h. [DOI] [PubMed] [Google Scholar]
- 40.Radi AE, Acero Sanchez JL, et al. Reusable impedimetric aptasensor. Anal Chem. 2005;77:6320–6323. doi: 10.1021/ac0505775. [DOI] [PubMed] [Google Scholar]
- 41.Xiao Y, Lubin AA, et al. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew Chem Int Ed Engl. 2005;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 42.Baldrich E, Restrepo A, et al. Aptasensor Development: Elucidation of Critical Parameters for Optimal Aptamer Performance. Anal Chem. 2004;76:7053–7063. doi: 10.1021/ac049258o. [DOI] [PubMed] [Google Scholar]
- 43.Fang X, Sen A, et al. Synthetic DNA aptamers to detect protein molecular variants in a high-throughput fluorescence quenching assay. Chembiochem. 2003;4:829–834. doi: 10.1002/cbic.200300615. [DOI] [PubMed] [Google Scholar]
- 44.Fang X, Cao Z, et al. Molecular aptamer for real-time oncoprotein platelet-derived growth factor monitoring by fluorescence anisotropy. Anal Chem. 2001;73:5752–5757. doi: 10.1021/ac010703e. [DOI] [PubMed] [Google Scholar]
- 45.Huang CC, Huang YF, et al. Aptamer-modified gold nanoparticles for colorimetric determination of platelet-derived growth factors and their receptors. Anal Chem. 2005;77:5735–5741. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- 46.Jiang Y, Fang X, et al. Signaling aptamer/protein binding by a molecular light switch complex. Anal Chem. 2004;76:5230–5235. doi: 10.1021/ac049565u. [DOI] [PubMed] [Google Scholar]
- 47.Yang CJ, Jockusch S, et al. Light-switching excimer probes for rapid protein monitoring in complex biological fluids. Proc Natl Acad Sci USA. 2005;102:17278–17283. doi: 10.1073/pnas.0508821102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green LS, Jellinek D, et al. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry. 1996;35:14413–14424. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- 49.Fredriksson S, Horecka J, et al. Multiplexed proximity ligation assays to profile putative plasma biomarkers relevant to pancreatic and ovarian cancer. Clin Chem. 2008;54:582–589. doi: 10.1373/clinchem.2007.093195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Effect of spacer length on thrombin-dependent ligation. (A) Sequences of thrombin-sensing structure-switching aptamers that have different spacer lengths connecting the antisense sequence (blue) and the aptamer (red). (B) Activation of ligation in the presence of thrombin. Ligation reactions were carried out for 1.5 or 16 hrs. Ligated and unligated species were visualized on 8 % PAGE gels stained with SYBR Gold. The percentage of ligation in the absence and presence of 500 nM thrombin was determined by analysis with QuantityOne software (BioRad Laboratories, Hercules, CA) following scanning of the stained gel.