Abstract
The Alzheimer's Disease Neuroimaging Initiative (ADNI) is a longitudinal multisite observational study of healthy elders, mild cognitive impairment (MCI), and Alzheimer's disease. Magnetic resonance imaging (MRI), (18F)-fluorode-oxyglucose positron emission tomography (FDG PET), urine serum, and cerebrospinal fluid (CSF) biomarkers, as well as clinical/psychometric assessments are acquiredat multiple time points. All data will be cross-linked and made available to the general scientific community. The purpose of this report is to describe the MRI methods employed in ADNI. The ADNI MRI core established specifications thatguided protocol development. A major effort was devoted toevaluating 3D T1-weighted sequences for morphometric analyses. Several options for this sequence were optimized for the relevant manufacturer platforms and then compared in a reduced-scale clinical trial. The protocol selected for the ADNI study includes: back-to-back 3D magnetization prepared rapid gradient echo (MP-RAGE) scans; B1-calibration scans when applicable; and an axial proton density-T2 dual contrast (i.e., echo) fast spin echo/turbo spin echo (FSE/TSE) for pathology detection. ADNI MRI methods seek to maximize scientific utility while minimizing the burden placed on participants. The approach taken in ADNI to standardization across sites and platforms of the MRI protocol, postacquisition corrections, and phantom-based monitoring of all scanners could be used as a model for other multisite trials.
Keywords: MRI, Alzheimer's disease, clinical trials, imaging methods, imaging standardization
Dementia, one of the most feared associates of increasing longevity, represents a pressing public health problem and major research priority. Alzheimer's disease (AD) is the most common form of dementia, affecting many millions around the world. There is currently no cure for AD, but large numbers of novel compounds are currently under development that have the potential to modify the course of the disease and slow its progression. There is a pressing need for imaging biomarkers to improve understanding of the disease and to assess the efficacy of these proposed treatments. Structural magnetic resonance imaging (MRI) has already been shown to be sensitive to presymptomatic disease (1-10) and has the potential to provide such a biomarker. For use in large-scale multicenter studies, however, standardized methods that produce stable results across scanners and over time are needed.
The Alzheimer's Disease Neuroimaging Initiative (ADNI) study is a longitudinal multisite observational study of elderly individuals with normal cognition, mild cognitive impairment (MCI), or AD (11,12). It is jointly funded by the National Institutes of Health (NIH) and industry via the Foundation for the NIH. The study will assess how well information (alone or in combination) obtained from MRI, (18F)-fludeoyglucose positron emission tomography (FDG PET), urine, serum, and cerebrospinal fluid (CSF) biomarkers, as well as clinical and neuropsychometric assessments, can measure disease progression in the three groups of elderly subjects mentioned above. At the 55 participating sites in North America, imaging, clinical, and biologic samples will be collected at multiple time points in 200 elderly cognitively normal, 400 MCI, and 200 AD subjects. All subjects will be scanned with 1.5 T MRI at each time point, and half of these will also be scanned with FDG PET. Subjects not assigned to the PET arm of the study will be eligible for 3 T MRI scanning. The goal is to acquire both 1.5 T and 3 T MRI studies at multiple time points in 25% of the subjects who do not undergo PET scanning [R2C1]. CSF collection at both baseline and 12 months is targeted for 50% of the subjects. Sampling varies by clinical group. Healthy elderly controls will be sampled at 0, 6, 12, 24, and 36 months. Subjects with MCI will be sampled at 0, 6, 12, 18, 24, and 36 months. AD subjects will be sampled at 0, 6, 12, and 24 months.
Major goals of the ADNI study are: to link all of these data at each time point and make this repository available to the general scientific community; to develop technical standards for imaging in longitudinal studies; to determine the optimum methods for acquiring and analyzing images; to validate imaging and biomarker data by correlating these with concurrent psychometric and clinical assessments; and to improve methods for clinical trials in MCI and AD. The ADNI study overall is divided into cores, with each core managing ADNI-related activities within its sphere of expertise: clinical, informatics, biostatistics, biomarkers, and imaging. The purpose of this report is to describe the MRI methods and decision-making process underlying the selection of the MRI protocol employed in the ADNI study.
MATERIALS AND METHODS
The MRI portion of the ADNI study was divided into three phases: development, preparation, and execution of the study itself. In this report we outline activities of the first two phases. Members of the MRI core established a basic set of requirements that guided the protocol development process. The overarching principle was to maximize scientific value while minimizing patient burden. Specific guidelines were:
The MRI data acquired by ADNI must be consistent across sites and over time. That is, similar image qualities (contrast-to-noise, spatial resolution, resistance to artifact, reliability, speed, etc.) must be achieved across sites and platforms over time at each field strength.
Based on responses to an initial questionnaire, virtually all participating clinical enrollment sites had access to at least one MRI scanner from GE Healthcare, Philips Medical Systems, or Siemens Medical Solutions. Consequently scanners from only these three vendors were supported. A variety, but not all, of the MRI platforms from each vendor were supported. Specifically, some older platforms (e.g., Siemens Vision, or GE Healthcare systems running software earlier than 9.1) were not supported.
Modification to an existing product pulse sequence on a particular vendor platform was encouraged, but only if it was both practical and would substantially benefit the study.
The study emphasis was on brain morphometry; hence the most important image set was the T1-weighted 3D volumetric acquisition. Isotropic voxels were desired to avoid a directional bias, but not required. The target voxel size was approximately 1 mm3, with a maximum of 1.5 mm in any one direction.
The whole brain must be covered without image wrap. Given the large number and variety of participating sites, the imaging volume must be easy for technologists to prescribe regardless of experience level.
Acquisition time for any series should be less than 10 minutes.
Artifact reduction is more important than reduction in acquisition time.
At 3 T the increased signal-to-noise ratio (SNR) compared with 1.5 T was used to increase spatial resolution while increasing the receiver bandwidth to help compensate for the increased chemical shift and the more rapid susceptibility variation (as measured in hertz) at 3 T.
The 3D T1-weighted protocol must operate successfully with major imaging analysis methods that have been employed in this field such as manual and atlas-based region of interest generation, boundary shift integral, voxel-based morphometry, and tensor-based morphometry.
ADNI must include phantom-based methods to monitor scanner calibration across all sites over the course of the study.
Post-acquisition correction of certain image artifacts would be implemented where applicable, such as 3D distortion correction for warping due to gradient nonlinearity.
Several basic decisions about the composition of the MRI protocol for the execution phase were made on the basis of the cumulative experience of members of the MRI core with multisite MRI-based studies, information gleaned from surveys at the participating ADNI clinical sites, and the protocol guidelines established above. Each subject will be enrolled in the study for a 3-year period, and multiple types of data will be collected at each sampling point, so we anticipate a high research burden on each participant. Moreover, ADNI is an observational study, which offers participants no experimental treatment. In order to maximize scientific value while minimizing patient burden, the MRI core targeted the duration of the patient scanning portion of the execution phase MRI protocol to approximately 30 minutes. We also decided to scan the ADNI phantom immediately after each patient exam, rather than decoupling phantom and human scanning. Site surveys revealed that the average time window allotted for a single MRI examination of the head was approximately 45 minutes. The overall structure of the ADNI MRI execution phase was therefore targeted to be 30 minutes of patient scan time and 15 minutes of phantom scan time.
The protocol at a minimum had to include at least one high-quality, high-resolution 3D T1-weighted sequence, as well as a second acquisition that provided T2-weighted information to ascertain brain pathology. A number of optional additional imaging sequences were considered for the MRI protocol, including MR spectroscopy, diffusion tensor imaging, arterial spin labeling, and fluid attenuated inversion recovery (FLAIR) sequences. The MRI core decided to limit the scope of the protocol to two domains—high-quality 3D T1-weighted morphometric information and a dual contrast proton density/T2-weighted sequence for pathology detection. This decision was based on balancing the various competing considerations outlined above. Options for this high-quality 3D T1-weighted sequence were magnetization prepared rapid gradient echo (MP-RAGE; and IR-FSPGR, which is a related technique available on the GE scanners) and spoiled gradient echo (SPGR) or equivalent (spoiled fast low angle shot [FLASH] for Siemens systems and T1 fast field echo [FFE] for Philips systems). We also considered an approach that had been adopted by the Biomedical Informatics Research Network (BIRN) study: generating synthetic T1 images by acquiring stand-alone SPGR or equivalent 3D volumetric data sets with high and low flip angles (13,14).
Development Phase
During the development phase a total of 29 sample human studies with T1-weighted 3D volumetric studies at 1.5 and 3 T were obtained from various ADNI sites and the MRI vendors. Based on protocol features that worked well across vendor platforms, a set of generic protocols was generated and then reviewed and revised by the MRI core group along with industry and external advisors until a consensus was reached. The generic protocols were adapted to yield vendor-specific protocols for the preparatory phase study based on availability of software options. Also, minor protocol differences were introduced at this stage due to vendor-specific implementation differences. A customized MP-RAGE pulse sequence was developed for the GE platform to minimize vendor-to-vendor differences. At this stage we also identified and implemented pulse sequence fixes required for the T1-weighted IR pulse sequences on some platforms, including increased RF bandwidth of the inversion pulse at 3 T and increased gradient area for the end-of-sequence spoiler. Note that these customization steps resulted in sequences that in some cases were non-product. This in turn meant that ongoing support throughout the study by the MRI vendors as systems were upgraded was required. At the conclusion of the development phase, vendor-specific protocols that included the list of 3D T1-weighted sequences above (where appropriate by vendor platform) were created. This protocol was then employed in the preparatory phase.
Preparatory Phase Study
The ADNI MRI preparatory phase consisted of a mini clinical trial the purpose of which was to select the 3D T1-weighted volume sequence that would be employed in the execution phase of the main ADNI study for morphometric assessment. Two endpoints were evaluated: first, the ability of the imaging sequence to appropriately differentiate normal control subjects from AD subjects cross-sectionally; and second, test-retest precision on serial scan pairs obtained in normal elderly control subjects (i.e., a situation in which no systematic biologic change is expected). Six different sites, representing the relevant MRI vendor platforms at 1.5 and 3 T, participated in the ADNI MRI preparatory phase. Over these six sites, 73 normal elderly subjects and 64 AD subjects were scanned once for cross-sectional comparison purposes. The control subjects were scanned again 2 weeks later in order to evaluate precision. Scanning sessions were performed on both phased-array receive and single-channel T/R birdcage RF coils for some platforms. The data acquired above were submitted for quantitative evaluation using the techniques that will be employed in the main ADNI data analyses: boundary shift integral, voxel-based morphometry, tensor-based morphometry, atlas-based hippocampal volumetrics, and also a measure of gray-white matter contrast to noise. In addition, scans were graded qualitatively by a trained individual for artifacts and general image quality on a four-point scale: none, mild, moderate, and severe. Each scan was graded on several separate criteria: blurring/ghosting; flow artifact; intensity and homogeneity; SNR; susceptibility artifacts; and gray-white CSF contrast.
RESULTS
A total of 208 unique MRI studies in 137 subjects were acquired during the preparatory phase. Plans for analysis of this data as initially outlined were compromised by the discovery of additional image imperfections on some system platforms while the preparatory phase was in progress. These included inconsistent polarity of the readout gradient between vendors that resulted in undesirable cephalic shift of fat over the brain at the base of the skull (Fig. 1) for sagittal acquisitions. Also, with transmit receive (T/R) head coils, the images were unacceptably noisy (Fig. 2a), which was corrected (at the expense of increased imaging time) by increasing the number of slices and the MP-RAGE repetition time, as indicated in Table 1 and illustrated in Fig. 2. Finally, some older systems imposed a 128 limit on the maximal number of slices. This not only degraded the SNR but also precluded sufficient coverage. Both the change in chemical shift direction and the lifting of the 128 slice limit required further pulse sequence modification. The protocol change to increase SNR for T/R birdcage coils and pulse sequence modification for chemical shift direction were made while the preparatory phase was in progress, resulting in undesirable discontinuities in this data.
Figure 1.
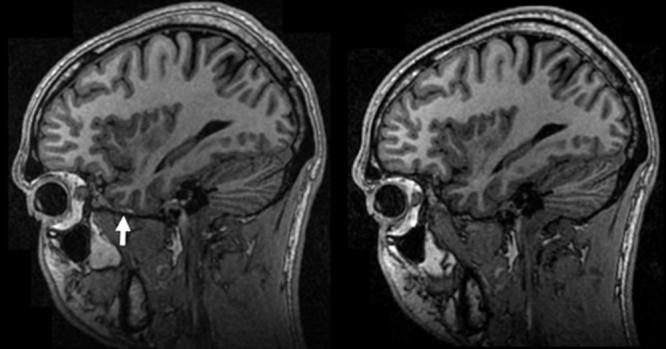
Undesirable chemical shift. The manufacturer's default polarity of the readout gradient in the SI direction for sagittal acquisitions shifted fat over the base of the brain (arrow, left) in the first version of the protocol. This hinders automated brain extraction algorithms. Reversal of this shift (right) required custom alteration of the manufacturer's product imaging sequence.
Figure 2.
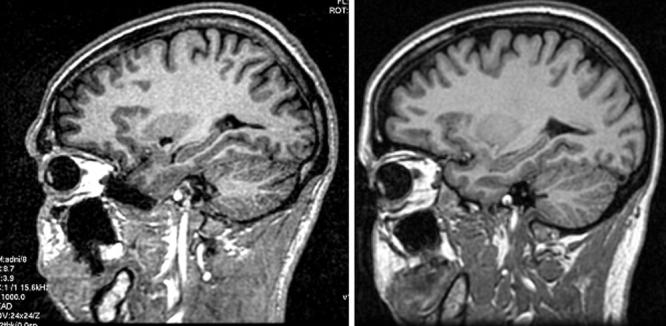
Poor SNR with single-channel birdcage coils in first version of protocol. As indicated in Table 1, the protocol using a single-channel birdcage coil differs from the phased array protocol. Left: When 1.5 T images are acquired using the phased array protocol with a birdcage coil, poor SNR results. Right: Making the parameter adjustments listed in Table 1 resolves the problem without increasing chemical shift.
Table 1.
Range of Parameters for MP-RAGE Acquisition
| B0 | Coil | Nx | Ny |
Nz No. of slices |
FoV (mm) | TRa (ms) | TI (ms) |
Flip (deg) |
Plane | Δz (mm) |
RBW (Hz/pixel) |
Time (min:s) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.5T | BC | 192 | 192 | 184–208 | 240 × 240 | 3000 | 1000 | 8 | Sagittal PEb = A/P | 1.2 | 162–180 | 9:36–9:38 |
| 1.5T | MA | 192 | 192 | 160–170 | 240 × 240 | 2300–2400 | 1000 | 8 | Sagittal PE = A/P | 1.2 | 162–200 | 7:11–7:42 |
| 3.0T | MA or BC | 256 | 256c | 160–170 | 256–260 × 240 | 2300 or 3000d | 853–900 | 8–9 | Sagittal PE = A/P | 1.2 | 240–244 | 9:14–9:22 |
TR is defined here as the repetition time for the inversion pulses.
PE is the phase-encoded direction.
256 is the base resolution. Because the field of view is rectangular at 3.0 T, the net number of acquired phase-encoding steps is approximately 0.94 × 256 = 240.
The longer value of TR = 3000 at 3 T is used only with pulse sequences where the train of gradient echo readouts represents the in-plane phase encoding. Because there are fewer encoded slices than in-plane phase encodings in the 3 T protocol, the net acquisition time is approximately equal to the TR = 2300 msec case.
MA = multicoil phased-array head coil; BC = birdcage or volume head coil.
The MRI core along with the external advisors met during the 2004 annual ISMRM meeting in Miami. The purpose of this meeting was to review data from the preparatory phase and select a final protocol for the execution phase of ADNI. A general summary of the qualitative ranking results from best to worst were: MP-RAGE > SPGR > synthetic T1. The different scan types were also graded on the basis of quantitative measures made by the individual image analysis groups in the MRI core. These data have appeared or will appear as separate independent publications (15).
Overall the results were mixed. There was no clear indication across the different analyses performed that one image type outperformed the other. In addition, where one image type was found to be better than another, differences were typically small. There was an overall consensus that the MP-RAGE and SPGR or equivalent sequences outperformed the synthetic T1 images. The primary advantages of the SPGR over MP-RAGE were superior SNR and superior performance on applications that placed a premium on brain-CSF segmentation. The advantages of the MP-RAGE sequence were superior gray/white contrast to noise, superior performance in some applications requiring cortical segmentation, and imaging times that were under 10 minutes for all vendor platforms at both field strengths. The evaluation group also noted that the SNR advantage for SPGR was to a large extent present on birdcage coil acquisitions at 1.5 T acquired with the initial unsatisfactory version of the preparatory phase MP-RAGE protocol, which was acquired with birdcage coils. The protocol changes illustrated in Fig. 2 resulted in a significant improvement in the performance of MP-RAGE in the various image processing algorithms. The evaluation group decided to extrapolate to what we would have seen had the higher SNR protocol been used throughout the entire preparatory phase when weighing the pros and cons of various MR sequences.
The evaluation group unanimously selected MP-RAGE as the 3D sequence for the ADNI execution phase (Fig. 3). The suggestion was also made to acquire back-to-back MP-RAGE sequences as opposed to the more traditional approach of a single acquisition per exam. The advantage of incorporating back-to-back acquisitions as a standard feature of the protocol is that the decision to repeat the scan on the basis of image quality will not be placed in the hands of individual technologists at the sites performing the scans. The ADNI MRI quality control center at the Mayo Clinic will select the better MP-RAGE sequence at each time point based on centralized and standardized criteria. Perhaps most importantly, requiring back-to-back MP-RAGE scans should reduce the number of examinations that must be repeated due to poor image quality. This in turn should minimize patient burden and enhance patient retention in the study. Finally, if both MP-RAGE scans are of equivalent high quality, the magnitude images could be combined (typically after spatial registration of the two image volumes) to produce a single image an improvement in SNR approaching
Figure 3.
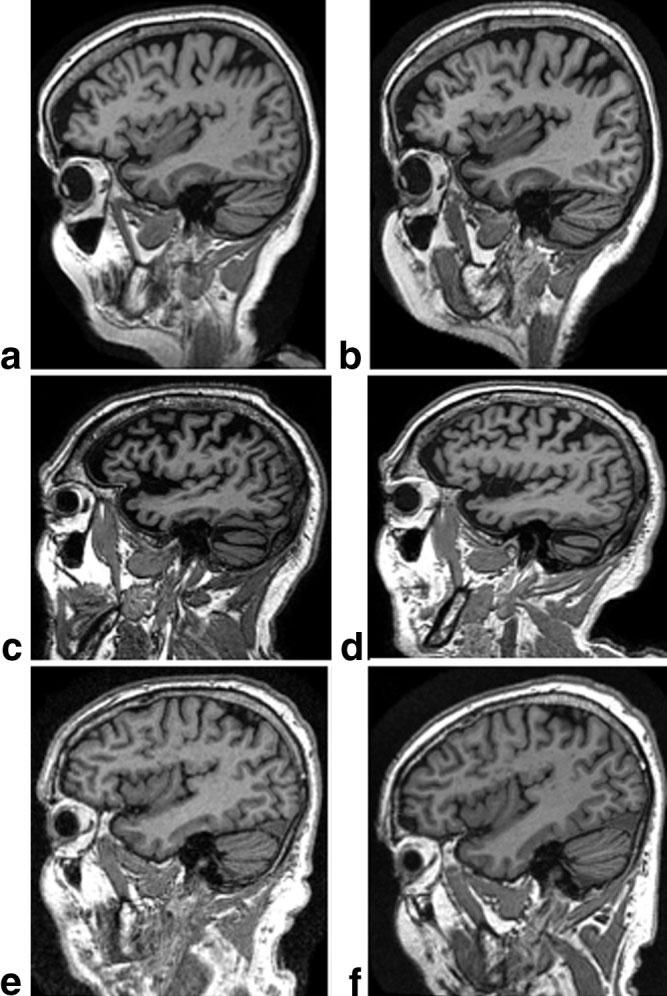
Example MP-RAGE images for each manufacturer at 1.5 T (left) and 3 T (right). a,b: GE. c,d: Philips. e,f: Siemens.
Discussion of the appropriate imaging sequence to employ for cerebral pathology detection centered on FLAIR vs. dual contrast fast spin echo. The 30 minutes allotted to patient scan time could accommodate only one of these additional sequences. Although the FLAIR sequence is highly useful, it was decided that some clinical groups would find a double fast spin echo sequence more in keeping with their general practice than a stand-alone FLAIR image. This was an important consideration because an on-site interpretation of all ADNI MRI studies by a local radiologist for medical alerts is a feature of the overall ADNI study design. The MR core therefore decided on the following for the final format of the ADNI execution phase protocol:
Standard prescan and scouting procedure recommended by the manufacturer
Sagittal 3D MP-RAGE
Sagittal 3D MP-RAGE repeat
Sagittal B1-calibration scan (phased array)
Sagittal B1-calibration scan (body coil)
Axial proton density T2 dual contrast FSE/TSE.
These six series are typically completed in 30 minutes or less. Series 4 and 5 are low-resolution maps used to correct for B1-intensity variation of the phased array receive coil. Consequently, they are omitted when only a single-channel, T/R birdcage head coil is available. In the case of the Philips scanners, a product B1-correction was available at the beginning of the study and was integrated into the protocol. The other two vendors introduced product B1-corrections later, but reference scans 4 and 5 were not replaced, to help ensure continuity of the longitudinal data. A list of parameters for series 2 and 3 is provided in Table 1, and representative images for each vendor at both field strengths are shown in Fig. 3. Detailed lists of parameters for each ADNI-supported vendor platform can be downloaded at http://www.loni.ucla.edu/ADNI/Research/Cores/.
DISCUSSION
The requirements for the ADNI MRI protocol differ from those used to generate a routine clinical protocol. For example, while an experienced radiologist often can “read through” minor artifacts, the automated software programs used for the analysis typically cannot. In fact, as a general rule, the more fully automated the analysis algorithm is, the less tolerant it is to image imperfections. Therefore, smaller fields of view that can lead to wraparound artifacts (e.g., the nose onto the back of the head) were avoided, as well as parallel imaging methods like sensitivity encoding (SENSE) that can sometimes result in residual aliasing when there is imperfect calibration. Also, partial k-space acquisition was avoided because of the associated artifacts that can result in regions of rapid susceptibility variation. Consequently, the acquisition time of the MP-RAGE series is somewhat longer than a corresponding protocol typically used for diagnostic purposes. The ADNI MRI protocol was selected in a data-driven manner with considerable deliberation by a group of experts in the field of MRI. Because of the large multicenter nature of ADNI, MRI techniques that were widely available in the 2004–2005 time frame were given preference, although important pulse sequence changes (like the fat-water chemical shift direction) were allowed. The protocol follows a set of principles (outlined by the MRI core, ADNI executive committee, and external advisory committee) that are meant to maximize scientific utility while minimizing research burden for participants in ADNI.
In addition to imaging study subjects, ADNI is also acquiring images of a phantom in the same examination period as each human MRI exam (Fig. 4). The phantom was specifically designed for ADNI by a subset of the ADNI MRI core along with partners in industry (16-18). The ADNI phantom and accompanying analysis software will measure gradient calibration, residual nonlinearity after 3D distortion (i.e., “gradwarp”) correction, SNR, and contrast. These measurements will be used to monitor scanner performance over time for each scanner involved in the ADNI study. Tolerance specifications for each parameter will be established when sufficient longitudinal data from all participating sites have been acquired and analyzed. These specifications will be used to inform sites of any deviation from tolerance limits detected on individual scanners. In addition, the gradient calibration measures acquired at each imaging time point will be linked with its corresponding human scan. This will permit retrospective rescaling of human images to correct for drift or discontinuities in gradient calibration.
Figure 4.
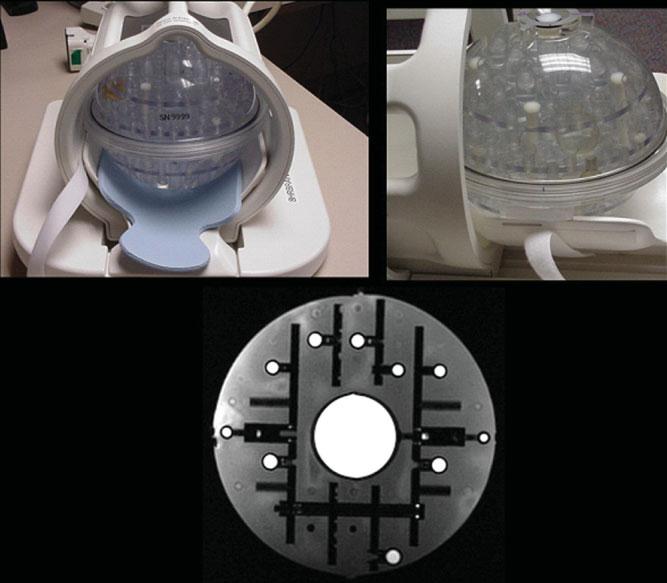
ADNI phantom. The ADNI phantom is spherical, with multiple inclusions that are used both for fiducial purposes and for SNR and contrast measurements. Image at the bottom is an MRI illustrating the central SNR inclusion as well as smaller spheres for fiducial measurements.
To enhance standardization across sites and platforms of images acquired in the ADNI study, post- acquisition correction of certain image artifacts has been implemented. These include corrections in image geometry for gradient nonlinearity, i.e., 3D gradwarp (19,20); corrections for intensity nonuniformity due to nonuniform receiver coil sensitivity (21); and correction of image intensity nonuniformity due to other causes such as wave effects at 3 T. These corrections are system specific. For example, only systems with apparent gradient nonlinearity undergo this correction; and the implementation of 3D gradwarp (or equivalent) is specific for each gradient configuration (Fig. 5). Similarly, correction for intensity inhomogeneity due to nonuniform sensitivity of multiarray receiver coils was implemented for those systems in which the manufacturer did not provide this feature as a product at the inception of the study (Fig. 6). In addition to the uncorrected original image files, the images with all the corrections and some with intermediate steps will be available to the general scientific community, as described at http://www.loni.ucla.edu/ADNI. These data correction procedures as well as image quality control procedures are performed at a single site (Mayo Clinic). Image data quality control includes inspection of each incoming image file for protocol compliance, clinically significant medical abnormalities, and image quality. The results of image quality control analysis are uploaded to the ADNI central data base, where this information is linked to the relevant image file and is available to the general scientific community. The actual image processing analyses for ADNI are performed at five separate sites.
Figure 5.
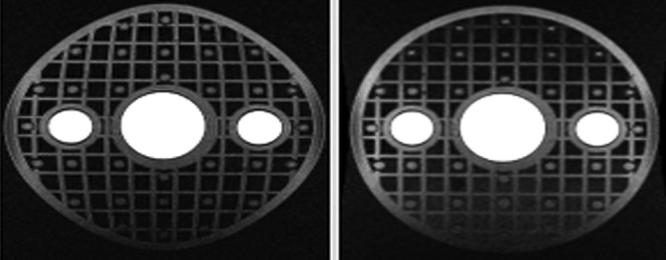
Effect of gradwarp. Spherical phantom with rectilinear grid inclusion before (left) and after (right) gradwarp correction.
Figure 6.
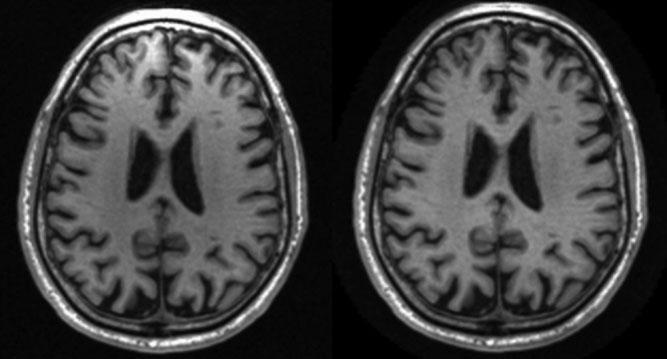
Intensity in-homogeneity correction. Phased array coil acquisition at 1.5 T before (left) and after (right) intensity nonuniformity correction. Images have been reformatted from the sagittal into the axial plane to illustrate the intensity in-homogeneity anteriority prior to correction.
The approach to standardization across sites and platforms of the MRI protocol and acquisition parameters for each imaging sequence, post-acquisition correction of image artifacts, and phantom-based monitoring of the instruments themselves could be extended to other multisite trials, including those in other research areas. This approach will minimize variation in the data collected due to technical nonuniformity and thus maximize across-site sensitivity to true biologic variation. In large multisite studies where the role of MRI is to capture relevant phenotypic information in all study participants, the importance of standardization supersedes the natural impulse by MR scientists to employ the most cutting edge MR methods that usually will be unique to each platform and can introduce undesirable technical variability.
ACKNOWLEDGMENTS
The authors dedicate this manuscript to the memory of Dr. Leon Thal. Dr. Thal devoted his career to the goal of finding a cure(s) for Alzheimer's disease. He was universally respected and admired and was instrumental in establishing the ADNI study. See the following web site: http://www.alzforum.org/spotlight/Thaltribute.asp#barrett.
Contract grant sponsor: the Alzheimer's Disease Neuroimaging Initiative (ADNI; Principal Investigator: Michael Weiner; National Institutes of Health grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and the Foundation for the National Institutes of Health, through generous contributions from the following companies and organizations: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, the Alzheimer's Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging (ISOA), with participation from the U.S. Food and Drug Administration.
REFERENCES
- 1.Fox NC, Warrington EK, Freeborough PA, et al. Presymptomatic hippocampal atrophy in Alzheimer's disease. A longitudinal MRI study. Brain. 1996;119:2001–2007. doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- 2.Fox NC, Crum WR, Scahill RI, Stevens JM, Janssen JC, Rossor MN. Imaging of onset and progression of Alzheimer's disease with voxel-compression mapping of serial magnetic resonance images. Lancet. 2001;358:201–205. doi: 10.1016/S0140-6736(01)05408-3. [DOI] [PubMed] [Google Scholar]
- 3.Jack CR, Jr, Petersen RC, Xu Y, et al. Prediction of AD with MRI- based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jack CR, Jr, Shiung MM, Weignad SD, et al. Brain atrophy rates predict subsequent clinical conversion in normal elderly and amnestic MCI. Neurology. 2005;65:1227–1231. doi: 10.1212/01.wnl.0000180958.22678.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y, Jack C, Jr, O'Brien PC, et al. Usefulness of MRI measures of entorhinal cortex vs hippocampus in AD. Neurology. 2000;54:1760–1767. doi: 10.1212/wnl.54.9.1760. [DOI] [PubMed] [Google Scholar]
- 6.Dickerson BC, Goncharova I, Sullivan MP, et al. MRI-derived entorhinal and hippocampal atrophy in incipient and very mild Alzheimer's disease. Neurobiol Aging. 2001;22:747–754. doi: 10.1016/s0197-4580(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 7.Dickerson BC, Salat D, Bates JF, et al. Medial temporal lobe function and structure in mild cognitive impairment. Ann Neurol. 2004;56:27–35. doi: 10.1002/ana.20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Killiany R, Gomez-Isla T, Moss M, et al. Use of structural magnetic resonance imaging to predict who will get Alzheimer's disease. Ann Neurol. 2000;47:430–439. [PubMed] [Google Scholar]
- 9.Killiany RJ, Hyman BT, Gopmez-Isla T, et al. MRI measures of entorhinal cortex vs hippocampus in preclinical AD. Neurology. 2002;58:1188–1196. doi: 10.1212/wnl.58.8.1188. [DOI] [PubMed] [Google Scholar]
- 10.Du AT, Schuff N, Amend D, et al. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2001;71:441–447. doi: 10.1136/jnnp.71.4.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller SG, Weiner MW, Thal LJ, et al. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dementia. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller SG, Weiner MW, Thal LJ, et al. The Alzheimer's Disease Neuroimaging Initiative. Neuroimaging Clin North Am. 2005;15:869–877. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischl B, Salat DH, van der Kouwe A, et al. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 2004;23:S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 14.Deoni SC, Peters TM, Rutt BK. High-resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2. Magn Reson Med. 2005;53:237–241. doi: 10.1002/mrm.20314. [DOI] [PubMed] [Google Scholar]
- 15.Leow AD, Klunder AD, Jack CR, Jr, et al. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. For the ADNI Preparatory Phase Study (2006) NeuroImage. 2006;31:627–40. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunter JL, Bernstein MA, Borowski B, et al. Validation testing of the MRI calibration phantom for the Alzheimer's Disease Neuroimaging Initiative Study. ISMRM 14th Scientific Meeting and Exhibition; Seattle, WA. 2006. [Google Scholar]
- 17.Mallozzi RP, Blezek DJ, Gunter JL, Jack CR, Jr, Levy JR. Phantom- based evaluation of gradient non-linearity for quantitative neurological MRI studies. ISMRM 14th Scientific Meeting and Exhibition; Seattle, WA. 2006. [Google Scholar]
- 18.Mallozzi RP, Blezek DJ, Ward CP, Gunter JL, Jack CR., Jr Phantom-based geometric distortion correction for volumetric imaging of Alzheimer's disease. ISMRM 12th Annual Scientific Meeting and Exhibition; Kyoto, Japan. 2004. [Google Scholar]
- 19.Hajnal JV, Hill DLG, Hawkes DJ. Medical image registration. CRC Press; New York: 2001. [Google Scholar]
- 20.Jovicich J, Czanner S, Greve DN, et al. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. NeuroImage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 21.Narayana PA, Brey WW, Kulkarni MV, Sievenpiper CL. Compensation for surface coil sensitivity variation in magnetic resonance imaging. Magn Reson Imaging. 1988;6:271–274. doi: 10.1016/0730-725x(88)90401-8. [DOI] [PubMed] [Google Scholar]


