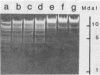
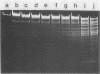
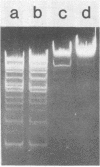
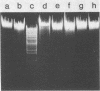
Abstract
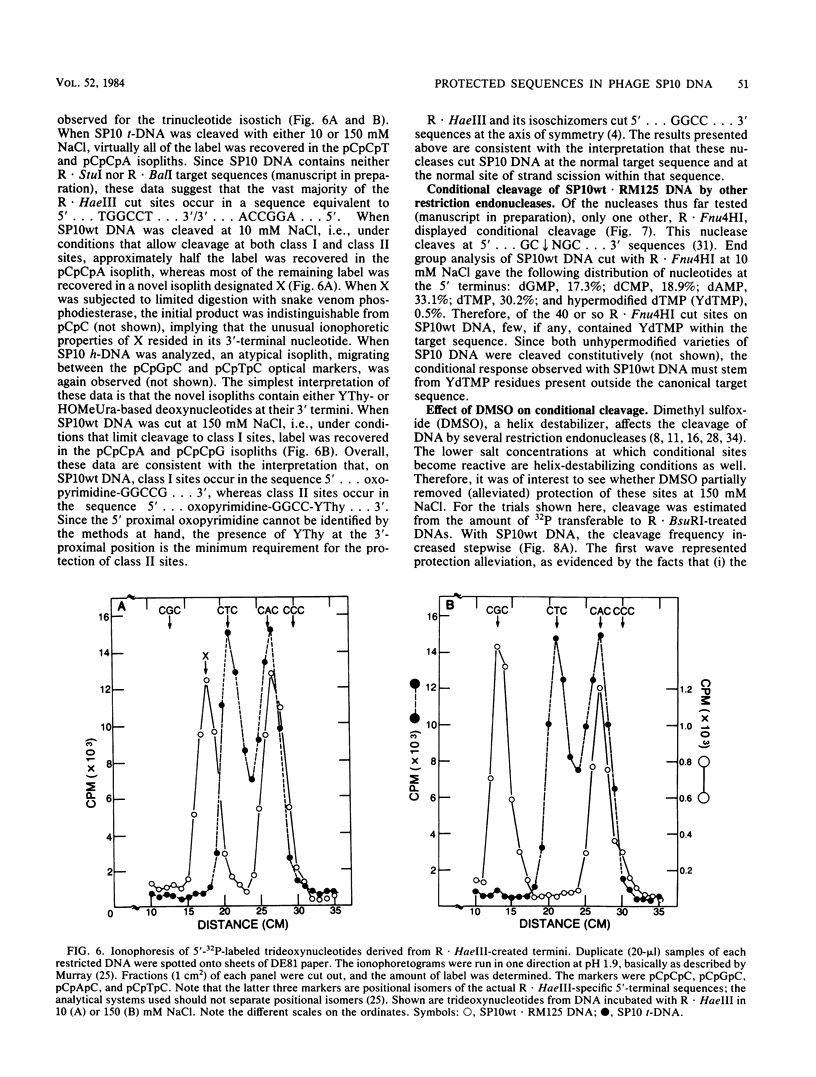
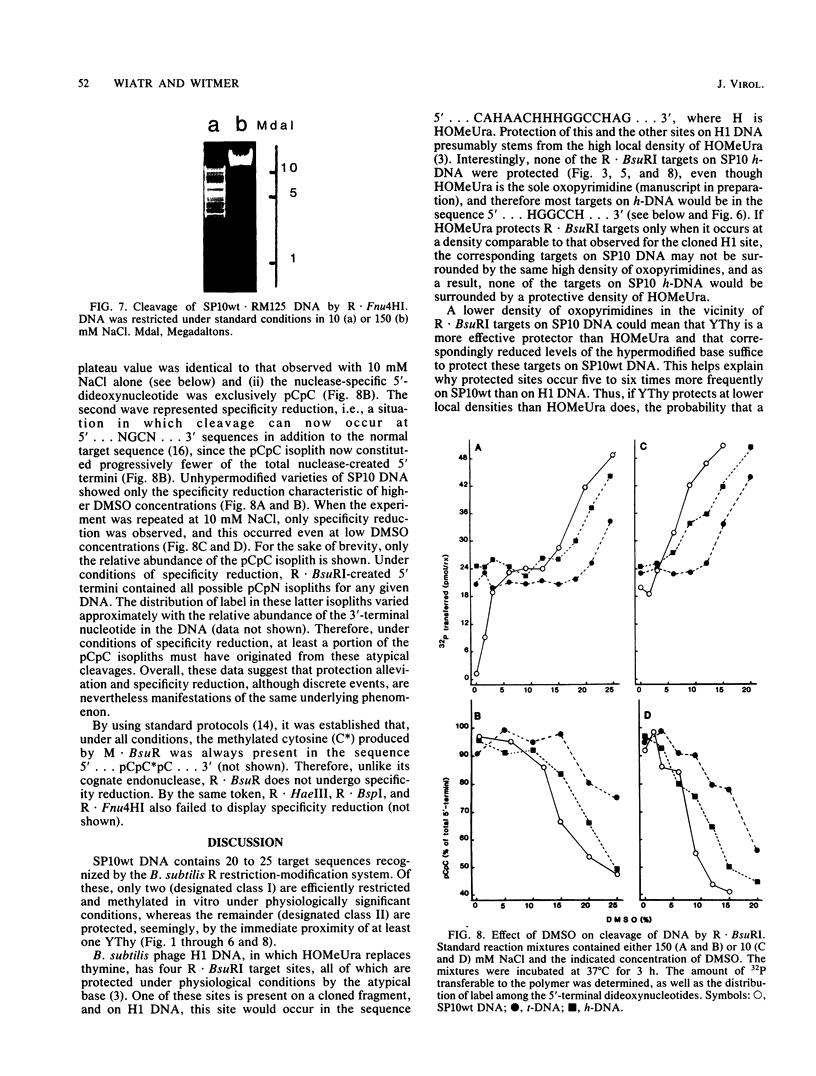
The DNA of Bacillus subtilis bacteriophage SP10 is partially resistant to cleavage and methylation in vitro by restriction enzyme R . BsuRI and its cognate methylase even though greater than 20 copies of the target sequence, 5' ... GGCC ... 3', are present on the phage genome. YThy, a hypermodified oxopyrimidine that replaces a fraction of the thymine residues in SP10 DNA, was responsible for this protection, since YThy-free DNA was no longer resistant. Sites that were normally resistant could nevertheless be cleaved or methylated in vitro if the salt concentration was reduced or dimethyl sulfoxide was added to the reaction buffer. Analysis of the termini produced by cleavage suggested that resistant sites occurred in the sequence 5' ... GGCC-YThy ... 3', whereas sensitive sites, of which there were only two per genome, occurred in the sequence 5' ... GGCCG ... 3'. These in vitro results provide an explanation for the in vivo resistance of SP10 to restriction-modification by B. subtilis R. They also suggest ways in which the presence of the atypical base YThy in regions that flank the target might upset critical DNA-enzyme interactions necessary to locate and recognize the specific site of cleavage or methylation. YThy also strongly protected 5' ... GCNGC ... 3' (R . Fnu4HI) sequences on SP10 DNA, but the biological relevance of this protection is unclear.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Bukhari A. I. Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol. 1975 Mar 15;92(4):529–540. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- Brandon C., Gallop P. M., Marmur J., Hayashi H., Nakanishi K. Structure of a new pyrimidine from Bacillus subtilis phage SP-15 nucleic acid. Nat New Biol. 1972 Sep 20;239(90):70–71. doi: 10.1038/newbio239070a0. [DOI] [PubMed] [Google Scholar]
- Bron S., Luxen E., Venema G. Resistance of bacteriophage H1 to restriction and modification by Bacillus subtilis R. J Virol. 1983 Jun;46(3):703–708. doi: 10.1128/jvi.46.3.703-708.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron S., Murray K. Restriction and modification in B. subtilis. Nucleotide sequence recognised by restriction endonuclease R. Bsu R from strain R. Mol Gen Genet. 1975 Dec 30;143(1):25–33. doi: 10.1007/BF00269417. [DOI] [PubMed] [Google Scholar]
- Bron S., Murray K., Trautner T. A. Restriction and modification in B. subtilis. Purification and general properties of a restriction endonuclease from strain R. Mol Gen Genet. 1975 Dec 30;143(1):13–23. doi: 10.1007/BF00269416. [DOI] [PubMed] [Google Scholar]
- Casella E., Markewych O., Dosmar M., Witmer H. Production and expression of dTMP-enriched DNA of bacteriophage SP15. J Virol. 1978 Dec;28(3):753–766. doi: 10.1128/jvi.28.3.753-766.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerný R., Mushynski W. E., Spencer J. H. Nucleotide clusters in deoxyribonucleic acids. 3. Separation of pyrimidine isostichs according to base composition. Biochim Biophys Acta. 1968 Dec 17;169(2):439–450. doi: 10.1016/0005-2787(68)90052-x. [DOI] [PubMed] [Google Scholar]
- Clarke C. M., Hartley B. S. Purification, properties and specificity of the restriction endonuclease from Bacillus stearothermophilus. Biochem J. 1979 Jan 1;177(1):49–62. doi: 10.1042/bj1770049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Ehrlich K. C. A novel, highly modified, bacteriophage DNA in which thymine is partly replaced by a phosphoglucuronate moiety covalently bound to 5-(4',5'-dihydroxypentyl)uracil. J Biol Chem. 1981 Oct 10;256(19):9966–9972. [PubMed] [Google Scholar]
- George J., Blakesley R. W., Chirikjian J. G. Sequence-specific endonuclease Bam HI. Effect of hydrophobic reagents on sequence recognition and catalysis. J Biol Chem. 1980 Jul 25;255(14):6521–6524. [PubMed] [Google Scholar]
- Günthert U., Storm K., Bald R. Restriction and modification in Bacillus subtilis. Localization of the methylated nucleotide in the BsuRI recognition sequence. Eur J Biochem. 1978 Oct 16;90(3):581–583. doi: 10.1111/j.1432-1033.1978.tb12638.x. [DOI] [PubMed] [Google Scholar]
- Hattman S., Keister T., Gottehrer A. Sequence specificity of DNA methylases from Bacillus amyloliquefaciens and Bacillus brevis. J Mol Biol. 1978 Oct 5;124(4):701–711. doi: 10.1016/0022-2836(78)90178-x. [DOI] [PubMed] [Google Scholar]
- Hattman S. Specificity of the bacteriophage Mu mom+ -controlled DNA modification. J Virol. 1980 Apr;34(1):277–279. doi: 10.1128/jvi.34.1.277-279.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., van Ormondt H., de Waard A. Sequence specificity of the wild-type dam+) and mutant (damh) forms of bacteriophage T2 DNA adenine methylase. J Mol Biol. 1978 Mar 5;119(3):361–376. doi: 10.1016/0022-2836(78)90219-x. [DOI] [PubMed] [Google Scholar]
- Heininger K., Hörz W., Zachau H. G. Specificity of cleavage by restriction nuclease from Bacillus subtilis. Gene. 1977 Jul;1(5-6):291–303. doi: 10.1016/0378-1119(77)90035-x. [DOI] [PubMed] [Google Scholar]
- Huang L. H., Farnet C. M., Ehrlich K. C., Ehrlich M. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 1982 Mar 11;10(5):1579–1591. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Roberts R. J. Unusual base sequence arrangement in phage phi 29 DNA. Gene. 1979 Jan;5(1):1–7. doi: 10.1016/0378-1119(79)90088-x. [DOI] [PubMed] [Google Scholar]
- Jack W. E., Terry B. J., Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D. A., Nierlich D. P. Cleavage of Nonglucosylated Bacteriophage T4 deoxyribonucleic acid by Restriction Endonuclease Eco RI. J Biol Chem. 1975 Mar 25;250(6):2395–2397. [PubMed] [Google Scholar]
- Kawamura F., Mizukami T., Shimotsu H., Anzai H., Takahashi H., Saito H. Unusually infrequent cleavage with several endonucleases and physical map construction of Bacillus subtilis bacteriophage phi 1 DNA. J Virol. 1981 Mar;37(3):1099–1102. doi: 10.1128/jvi.37.3.1099-1102.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Kiss A., Venetianer P. Biochemical characterization of the restriction-modification system of Bacillus sphaericus. Eur J Biochem. 1978 Sep 1;89(2):523–529. doi: 10.1111/j.1432-1033.1978.tb12557.x. [DOI] [PubMed] [Google Scholar]
- Markewych O., Boghosian A., Dosmar M., Ende D., Witmer H. SP-10 bacteriophage-specific nucleic acid and enzyme synthesis in Bacillus subtilis W23. J Virol. 1977 Jan;21(1):84–95. doi: 10.1128/jvi.21.1.84-95.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K. Nucleotide sequence analysis with polynucleotide kinase and nucleotide "mapping" methods. 5'-Terminal sequences of deoxyribonucleic acid from bacteriophages lambda and 424. Biochem J. 1973 Mar;131(3):569–582. doi: 10.1042/bj1310569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyer-Weidner M., Jentsch S., Pawlek B., Günthert U., Trautner T. A. Restriction and modification in Bacillus subtilis: DNA methylation potential of the related bacteriophages Z, SPR, SP beta, phi 3T, and rho 11. J Virol. 1983 May;46(2):446–453. doi: 10.1128/jvi.46.2.446-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C. M. Regulation of lambda exonuclease. I. Properties of lambda exonuclease purified from lysogens of lambda T11 and wild type. J Mol Biol. 1966 Jul;18(2):235–250. doi: 10.1016/s0022-2836(66)80243-7. [DOI] [PubMed] [Google Scholar]
- Reeve J. N., Amann E., Tailor R., Günthert U., Scholz K., Trautner T. A. Unusual behaviour of SPO1 DNA with respect to restriction and modification enzymes recognizing the sequence 5'-G-G-C-C. Mol Gen Genet. 1980 Apr;178(1):229–231. doi: 10.1007/BF00267234. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1980 Jan 11;8(1):r63–r80. doi: 10.1093/nar/8.1.197-d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THORNE C. B. Transduction in Bacillus subtilis. J Bacteriol. 1962 Jan;83:106–111. doi: 10.1128/jb.83.1.106-111.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Tikchonenko T. I., Karamov E. V., Zavizion B. A., Naroditsky B. S. EcoRI activity: enzyme modification or activation of accompanying endonuclease? Gene. 1978 Nov;4(3):195–212. doi: 10.1016/0378-1119(78)90018-5. [DOI] [PubMed] [Google Scholar]
- Uozumi T., Hoshino T., Miwa K., Horinouchi S., Beppu T., Arima K. Restriction and modification in Bacillus species: genetic transformation of bacteria with DNA from different species, part I. Mol Gen Genet. 1977 Mar 28;152(1):65–69. doi: 10.1007/BF00264941. [DOI] [PubMed] [Google Scholar]
- Walker M. S., Mandel M. Biosynthesis of 5-(4'5'-dihydroxypentyl) uracil as a nucleoside triphosphate in bacteriophage SP15-infected Bacillus subtilis. J Virol. 1978 Feb;25(2):500–509. doi: 10.1128/jvi.25.2.500-509.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. A. Modified bases in bacteriophage DNAs. Annu Rev Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Blakesley R. W., Hardies S. C., Horn G. T., Larson J. E., Selsing E., Burd J. F., Chan H. W., Dodgson J. B., Jensen K. F. The role of DNA structure in genetic regulation. CRC Crit Rev Biochem. 1977;4(3):305–340. doi: 10.3109/10409237709102561. [DOI] [PubMed] [Google Scholar]
- Witmer H. Synthesis of deoxythymidylate and the unusual deoxynucleotide in mature DNA of Bacillus subtilis bacteriophage SP10 occurs by postreplicational modification of 5-hydroxymethyldeoxyuridylate. J Virol. 1981 Aug;39(2):536–547. doi: 10.1128/jvi.39.2.536-547.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead J. L., Malcolm A. D. Non-specific binding of restriction endonuclease EcoR1 to DNA. Nucleic Acids Res. 1980 Jan 25;8(2):389–402. doi: 10.1093/nar/8.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajko D. M., Valentine M. C., Weiss B. Mutants of Escherichia coli with altered deoxyribonucleases. II. Isolation and characterization of mutants for exonuclease I. J Mol Biol. 1974 May 15;85(2):323–343. doi: 10.1016/0022-2836(74)90367-2. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]