Abstract
A genetic hierarchy of interactions, involving myogenic regulatory factors of the MyoD and myocyte enhancer-binding 2 (MEF2) families, serves to elaborate and maintain the differentiated muscle phenotype through transcriptional regulation of muscle-specific target genes. Much work suggests that members of the cysteine-rich protein (CRP) family of LIM domain proteins also play a role in muscle differentiation; however, the specific functions of CRPs in this process remain undefined. Previously, we characterized two members of the Drosophila CRP family, the muscle LIM proteins Mlp60A and Mlp84B, which show restricted expression in differentiating muscle lineages. To extend our analysis of Drosophila Mlps, we characterized the expression of Mlps in mutant backgrounds that disrupt specific aspects of muscle development. We show a genetic requirement for the transcription factor dMEF2 in regulating Mlp expression and an ability of dMEF2 to bind, in vitro, to consensus MEF2 sites derived from those present in Mlp genomic sequences. These data suggest that the Mlp genes may be direct targets of dMEF2 within the genetic hierarchy controlling muscle differentiation. Mutations that disrupt myoblast fusion fail to affect Mlp expression. In later stages of myogenic differentiation, which are dedicated primarily to assembly of the contractile apparatus, we analyzed the subcellular distribution of Mlp84B in detail. Immunofluorescent studies revealed the localization of Mlp84B to muscle attachment sites and the periphery of Z-bands of striated muscle. Analysis of mutations that affect expression of integrins and α-actinin, key components of these structures, also failed to perturb Mlp84B distribution. In conclusion, we have used molecular epistasis analysis to position Mlp function downstream of events involving mesoderm specification and patterning and concomitant with terminal muscle differentiation. Furthermore, our results are consistent with a structural role for Mlps as components of muscle cytoarchitecture.
INTRODUCTION
Myogenesis involves a series of discrete processes beginning with specification and proliferation of the mesoderm, subdivision of functionally distinct muscle lineages, and ultimately, muscle differentiation. In vertebrates, commitment to a skeletal muscle fate requires the action of myogenic regulatory factors including members of the MyoD basic helix–loop–helix (bHLH) family and the myocyte enhancer-binding 2 (MEF2) proteins that influence muscle development through transcriptional regulation of muscle-specific target genes (for review, see Molkentin and Olson, 1996; Yun and Wold, 1996). A third family of proteins, the LIM domain-containing cysteine-rich proteins (CRPs), have also been implicated in promoting muscle differentiation (Arber et al., 1997; Louis et al., 1997). The crucial involvement of CRPs in myogenic differentiation has been confirmed by genetic studies in mice. Eliminating the function of one CRP family member, CRP3 (also called the muscle LIM protein MLP) results in postnatal lethality caused by heart failure that is associated with dilated cardiomyopathy and severe disruptions of cardiac muscle architecture (Arber et al., 1997).
Despite the dramatic consequences associated with loss of CRP3/MLP expression, the mechanistic details of CRP function in muscles remain speculative. Controversy over the precise role of CRPs stems in part from the fact that CRP isoforms have been observed in cell nuclei and in association with the actin-based cytoskeleton (Arber et al., 1994; Arber and Caroni, 1996; Crawford et al., 1994; Kong et al., 1997; Louis et al., 1997). Consistent with the hypothesis that CRP family members contribute to muscle differentiation by regulating the transcription of muscle structural genes, one study describes an interaction between CRP3/MLP and the bHLH transcription factor MyoD (Kong et al., 1997). Evidence for functionally relevant interactions between bHLH factors and LIM domain proteins has been established in other systems. For example, the hematopoietic bHLH transcription factor SCL (Tal1) controls erythroid differentiation in part through interactions with the nuclear LIM protein Lmo-2 (RBTN-2) (Valge-Archer et al., 1994; Wadman et al., 1997).
Other data are consistent with a structural role for CRP isoforms in muscle. In particular, CRPs are known to distribute along the actin cytoskeleton and at integrin-rich sites of adhesion in cultured fibroblasts and muscle cells (Sadler et al., 1992; Arber et al., 1994; Crawford et al., 1994). Moreover, two cytoskeletal proteins, zyxin and α-actinin, have been shown to interact directly with CRPs in a variety of biochemical assays (Sadler et al., 1992; Pomies et al., 1997). Zyxin, a protein displaying proline-rich sequences and three LIM domains, has been implicated in the control of microfilament dynamics (Golsteyn et al., 1997). α-Actinin, an actin cross-linking protein, is prominent in both nonmuscle and muscle cells; in the myofibril, α-actinin localizes to Z-bands that define the ends of each sarcomeric unit (McKenna et al., 1986). Thus, via interactions with cytoskeletal components, CRPs may contribute to muscle differentiation as an integral part of muscle cytoarchitecture. In this context, it is noteworthy that the terminal phenotype of CRP3/MLP null mice is disorganization of cardiac muscle myofibrils (Arber et al., 1997). Clearly, additional work is necessary to define more precisely the molecular mechanism by which CRPs act in myogenesis.
The availability of a genetic system for defining the pathways that require CRP function could provide significant insight into the underlying mechanisms by which CRPs participate in myogenesis. Toward that end, two Drosophila CRP family members, termed muscle LIM proteins Mlp60A and Mlp84B, have been identified (Arber et al., 1994; Stronach et al., 1996). Drosophila Mlps exhibit muscle-specific expression, accumulating in all of the body wall and visceral and pharyngeal muscles in the embryo. Analysis of the temporal expression profiles of Mlp60A and Mlp84B transcripts during development pointed to a potential requirement for the proteins late in the muscle differentiation process. Here, we have used molecular epistasis analysis to position the Mlps within the regulatory hierarchy that controls muscle development in Drosophila. Specifically, we have examined the expression and distribution of Mlps in the context of mutations that are known to disrupt specific processes in the myogenic pathway. For instance, we have evaluated the role of dMEF2, an essential myogenic transcription factor (Bour et al., 1995; Lilly et al., 1995), in Mlp expression. We have also determined whether myoblast fusion, a key event in somatic muscle differentiation, is required for Mlp accumulation. Finally, we analyzed at higher resolution the localization of Drosophila Mlp84B in various muscles at different stages of development and show that its localization is not dependent on two structural muscle proteins, α-actinin or PS2 integrin receptors.
MATERIALS AND METHODS
Drosophila Stocks and Crosses
All flies were reared at 25°C unless otherwise noted on standard cornmeal agar food plus yeast. Canton-S served as wild-type stock. For analysis of dMEF2 mutant embryos, we used the mutation mef22–21, an ethyl methane sulfonate-induced null allele (Bour et al., 1995). This allele was crossed into a w1118 background and balanced over a CyO chromosome carrying the lacZ gene under the control of the actin5C promoter (CyO-actinlZ), a gift from David VanVactor (Harvard Medical School, Boston, MA). The CyO-actinlZ balancer chromosome has been used previously to distinguish embryos carrying that chromosome from those that are homozygous for recessive lethal loci (Bourgouin et al., 1992; Lundgren et al., 1995). In our experiments, embryos that inherit the balancer chromosome show expression of β-galactosidase during most of embryogenesis; by immunocytochemistry, the protein is detected within the nuclei of many cells that are distributed throughout the internal space of the embryo. This particular staining pattern was absent in approximately one-fourth of embryos derived from the mef22–21/CyO- actinlZ stock, thus allowing for the identification of homozygous dMEF2 null mutant embryos. Aiding our identification of dMEF2 null embryos was the observation that only the portion of the population in which β-galactosidase immunoreactivity was absent also showed morphological and muscle differentiation defects consistent with loss of dMEF2 function.
For dMEF2 overexpression studies, we used w;GawB 69B, a homozygous viable enhancer trap line that expresses GAL4 in the epidermis at germ band extended stages (Brand and Perrimon, 1993). Males from this stock were crossed to virgins from the homozygous viable stock yw; UAS-MEF 1, which carries the dMEF2 cDNA under the control of GAL4 target upstream activating sites (UASs) (Lin et al., 1997). All embryos derived from this cross expressed dMEF2 in the epidermis. For analysis of myoblast fusion, a severe rollingstone allele was used, rost23, balanced over the CyO chromosome (Paululat et al., 1995). Mutant embryos were recognized by fusion defects. α-Actinin mutant alleles used in this study include ethyl methane sulfonate-induced l(1)EA43 and l(1)EA82 and x-ray-induced l(1)HC288, l(1)HC207, and l(1)C212 (Perrimon et al., 1985; Flybase, 1994). All of these alleles are larval lethal. Larvae from each stock were picked for midgut dissection. The integrin mutant stocks used were mys1/FM4, carrying a null allele of the βPS subunit (Flybase, 1994), and y v ifB4 f/FM6, carrying a null allele of the αPS2 subunit (Brown, 1994). These mutant embryos were recognized by morphological criteria including their specific muscle defects.
Larval Midgut Preparation
Wild-type first or third instar larvae for dissection were picked out of Canton-S stock bottles. Hemizygous α-actinin mutant larvae were identified by their progressively paralyzed, flaccid phenotype among a population of developing larvae. Larvae were dissected, and midguts were processed as described (Saide et al., 1989). Primary antibodies were B50 (rabbit anti-Mlp84B) preabsorbed against early stage fixed embryos and diluted to 1:200 (Stronach et al., 1996) and 3A1 (mouse anti-α-actinin) culture supernatant diluted 1:2 (Saide et al., 1989). Secondary antibodies were affinity-purified Texas Red–conjugated goat anti-mouse IgG and fluorescein-conjugated goat anti-rabbit IgG used at 1:250 (Cappel Laboratories, Durham, NC). Samples were viewed using confocal microscopy.
Fluorescent Embryo Staining and Confocal Analysis
For immunofluorescence, embryos were collected and fixed according to standard procedures (Patel, 1994) except the Triton X-100 concentration was raised to 0.3% in all wash and blocking buffers. In all cases, egg collections were performed for no more than 15 h to minimize the presence of older embryos with impermeable cuticles. The following antibodies and dilutions were used in this study: rabbit anti-Mlp60A (B49) at 1:250, rabbit anti-Mlp84B (B50) at 1:250–500, rabbit anti-β-galactosidase at 1:5000 (Cappel Laboratories), mouse anti-βPS (CF6G11) at 1:1000 (Brower et al., 1984), and rabbit anti-muscle myosin at 1:500 (Kiehart and Feghali, 1986). In the analysis of Mlp expression in dMEF2 mutant embryos, both primary antibodies (anti-Mlp and anti-β-galactosidase) were derived from rabbit and thus were visualized using a single secondary antibody. Because the distributions of Mlps and β-galactosidase are nonoverlapping (our unpublished results), genotypes can be unambiguously assigned based on the presence or absence of β-galactosidase staining. All fluorochrome-conjugated secondary antibodies were obtained from either Cappel Laboratories or Jackson ImmunoResearch Laboratories (West Grove, PA) and used at 1:250–500. Images of embryos were captured as described previously (Stronach et al., 1996). Images of visceral muscle were obtained using a 60× objective and represent 1-μm-thick optical sections. For assessment of protein colocalization, we were cognizant of the alignment of tissue and degree of distortion in the Z dimension with respect to the axis of the laser during optical sectioning.
Genomic Sequence Analysis
For studying the genomic region comprising the Mlp84B gene, we first isolated clones from the Drosophila melanogaster genomic library “D.m. isox234” constructed in λEMBL3, a kind gift from John Tamkun (University of California, Santa Cruz, CA). Approximately 400,000 plaque-forming units were screened with the entire Mlp84B cDNA (Stronach et al., 1996) using standard techniques (Sambrook et al., 1989). Genomic sequence was obtained primarily from phage clone 4B2, because it fully spans the noncoding and coding portions of the Mlp84B transcript. Sequencing was performed by the DNA Sequencing Core Facility at the University of Utah using Mlp84B gene-specific primers. Either ABI dRhodamine dye terminators or ABI Prism BigDye terminators were used during cycle sequencing with Taq FS DNA polymerase. DNA sequence was collected and analyzed on an ABI Prism 377 automated DNA sequencer (Applied Biosystems, Foster City, CA). Approximately 5000 bp of sequence have been submitted to GenBank under accession number AF090832. Sequences were analyzed using DNASTAR software (DNASTAR, Madison, WI). The positions of each of the six regions that exactly match the MEF2 consensus binding site (Olson et al., 1995) are noted in the accession, as are four other sites that have a 9/10 match to the consensus.
The genomic region comprising the Mlp60A gene was sequenced by the Berkeley Drosophila Genome Project (Celniker et al., unpublished results) and obtained via GenBank accession number AC004642. A region of ∼5000 bp constituting the gene was analyzed for potential MEF2 binding sites using the same criteria.
Gel Mobility Shift Assays
Gel mobility shift assays were performed with dMEF2 protein synthesized using the Promega (Madison, WI) TNT rabbit reticulocyte lysate in vitro transcription and translation system (Lilly et al., 1994). For each 20-μl reaction, 3 μl of lysate containing PCite-dMEF2 or the control PCite vector alone were used. The lysates were incubated for 10 min at room temperature with 1.5 μg of poly(dI-dC), 1× binding buffer (40 mM KCL, 15 mM HEPES, pH 7.9, 1 mM EDTA, 0.5 mM DTT, and 5% glycerol), and the indicated competitor oligonucleotides. 32P-labeled probe (20,000 cpm) was then added and incubated for an additional 10 min before loading onto a 6% polyacrylamide gel in 0.5× Tris borate-EDTA buffer. The competitor oligonucleotides were added at 100-fold molar excess to the labeled probe. The sequences of the sense strand of the oligonucleotides used for probes and competitors were as follows (linker nucleotides added for end labeling are shown as lowercase): muscle creatine kinase (MCK) MEF2, gatcGCTCTAAAAATAACCCTGTCG; Mutant 6, gatcGCTCTAAACATAACCCTGTCG; Mlp60A-B, gatccGCCCCTCTATTTATAGATATG; and Mlp84B-D, gatcCACTATTATTAATAGATTCCG.
RESULTS
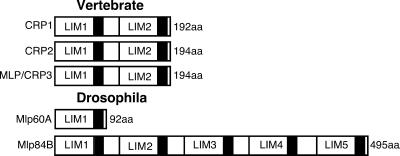
The CRP family of LIM domain proteins currently consists of three vertebrate isoforms, CRP1, CRP2, and CRP3/MLP, and two Drosophila members, Mlp60A and Mlp84B (Figure 1) (Weiskirchen and Bister, 1993; Weiskirchen et al., 1995; Arber et al., 1994; Crawford et al., 1994; Stronach et al., 1996; Louis et al., 1997). The vertebrate proteins share a common molecular architecture with two copies of the LIM domain, each followed by a small glycine-rich segment. Although the Drosophila family members diverge in the number of LIM-glycine repeats, they show significantly high sequence identity (46–60%) when compared with their vertebrate relatives. Much work suggests that CRP family members play an essential role in myogenic development by promoting muscle differentiation. Here, we address the potential requirements for Drosophila Mlps by positioning their function within a genetic regulatory hierarchy that controls myogenesis.
Figure 1.
Schematic representation of the CRP family of LIM domain proteins. In vertebrates, there are three conserved isoforms, CRP1, CRP2, and CRP3/MLP, encoded by unique genes. All share the same molecular architecture with two LIM domains, each followed by short glycine-rich regions (black box). In Drosophila, there are two proteins related to the vertebrate CRPs. Mlp60A exhibits a single LIM-glycine motif. Mlp84B comprises five tandem LIM-glycine modules.
Regulation of Muscle LIM Protein Expression by dMEF2
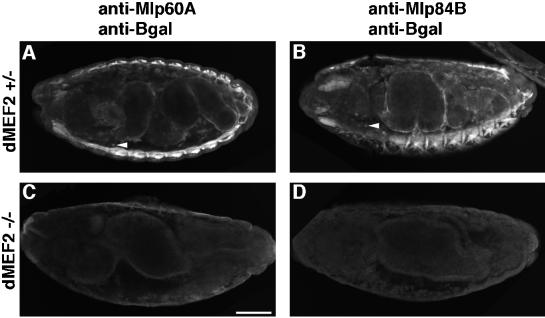
In Drosophila, the dMEF2 transcriptional regulator is essential for completion of myogenesis (Bour et al., 1995; Lilly et al., 1995). dMEF2 null mutant embryos exhibit a failure of muscle differentiation, although muscle specification and patterning occur fairly normally. Targets of dMEF2 activity include muscle structural genes that encode components of the myofibril, such as myosin and tropomyosin (Bour et al., 1995; Lin et al., 1996). To assess whether Mlps are downstream of dMEF2 activity, we used indirect immunofluorescence to evaluate the expression of Mlp60A and Mlp84B in dMEF2 mutant embryos. In heterozygous embryos, which express β-galactosidase encoded by a lacZ gene carried on the balancer chromosome, Mlps are expressed appropriately in the muscles (Figure 2, A and B; cf. Stronach et al., 1996). The Mlp expression pattern is quite distinct and exhibits no overlap with the expression of β-galactosidase. Mlps are found exclusively in muscle derivatives, whereas β-galactosidase is detected in many internal nonmuscle cells (Figure 2, A and B, arrowheads). Homozygous dMEF2 null embryos were identified by morphological criteria and by lack of staining for β-galactosidase; they constituted ∼25% of the total collected population. In the mutant embryos, neither Mlp60A nor Mlp84B expression was detected in any tissue (Figure 2, C and D). From the results presented here, we conclude that dMEF2 is an essential positive regulator of Mlp60A and Mlp84B expression. This lack of expression is notable given that it has been previously shown that mesoderm is formed and specified normally in dMEF2 mutants (Bour et al., 1995).
Figure 2.
The transcription factor dMEF2 is essential for Mlp expression. dMEF2 heterozygous embryos express Mlp60A (A) and Mlp84B (B) in somatic, visceral, and pharyngeal muscles. These embryos also carry the balancer chromosome from which the lacZ gene is expressed. Thus, by indirect immunofluorescence, we detect not only Mlp expression in these embryos but expression of β-galactosidase in single cells dispersed throughout the embryo (A and B, arrowheads). Mlp60A (C) and Mlp84B (D) fail to be expressed in the dMEF2 null mutant embryos. Null embryos display defective midgut morphology and no β-galactosidase–positive cells. All panels show lateral views of stage 16 (13- to 16-h) embryos oriented with dorsal up and anterior to the left. Bar, 50 μm.
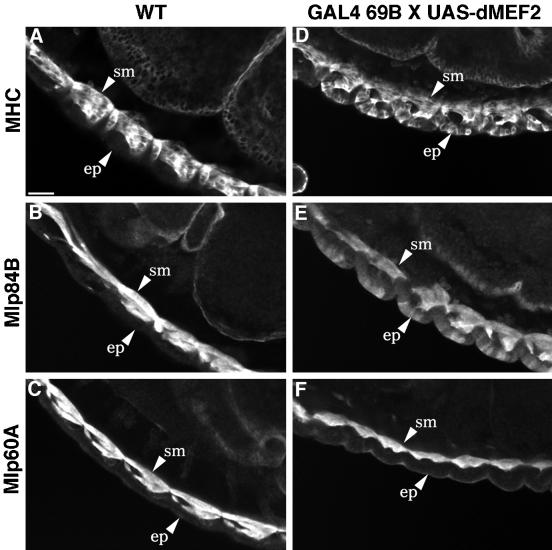
To investigate whether dMEF2 activity stimulates the expression of the Mlp genes, we used an ectopic tissue-specific expression system (Brand and Perrimon, 1993). Ectopic dMEF2 activity in the epidermis of germ band extended embryos is achieved using the GAL4 enhancer trap line 69B and a construct containing dMEF2 coding sequences downstream of the GAL4 recognition sites or UAS. dMEF2 is not normally found in this tissue, and ectopic expression leads to epidermal defects and subsequent embryonic lethality (Lin et al., 1997). We examined the possibility that ectopic expression of dMEF2 would result in up-regulation of muscle-specific targets such as the Mlps and myosin (Figure 3). In wild-type embryos undergoing muscle differentiation, myosin and the Mlps are observed in muscle tissues but not in epidermal cells (Figure 3, A–C); however, myosin expression is strongly induced in the epidermis by the presence of ectopic dMEF2 (Figure 3D). Mlp84B protein is also detected in the epidermis, concomitant with this ectopic expression of dMEF2 (Figure 3E). In contrast to both myosin and Mlp84B, Mlp60A protein is not detected at any appreciable level in epidermal cells of embryos that ectopically express dMEF2 (Figure 3F). We often observe a halo of staining in these embryos that is likely attributable to variable background associated with the anti-Mlp60A antibody. This staining does not appear cellular in nature, as that seen with the epidermal expression of myosin and Mlp84B, but rather as nonspecific surface staining.
Figure 3.
Ectopic dMEF2 expression stimulates expression of myosin and Mlp84B but not Mlp60A. In wild-type embryos, myosin (A), Mlp84B (B), and Mlp60A (C) are detected by indirect immunofluorescence in embryonic somatic muscles (sm) but not epidermal cells (ep). Myosin (D) and Mlp84B (E) expression can be induced in the epidermis by ectopic expression of dMEF2 under the control of the 69B GAL4 enhancer. Unlike myosin and Mlp84B, Mlp60A is not up-regulated by ectopic expression of epidermal dMEF2 (F). All panels display lateral views of 12- to 14-h embryos oriented with dorsal up and anterior to the left. Bar, 20 μm.
To assess whether the Mlp60A gene might require a higher threshold level of dMEF2 for its activation, we increased the dosage of dMEF2 in the epidermis by exploiting the temperature-sensitive nature of the GAL4 protein. At 29°C, GAL4 is reported to have higher activity resulting in greater accumulation of UAS-target gene product (Brand et al., 1994). Indeed, embryos raised at the higher temperature displayed more substantial phenotypic defects, suggesting increased dMEF2 expression in the epidermis, but still did not show ectopic up-regulation of Mlp60A protein (our unpublished results).
dMEF2 Binds Muscle LIM Protein Gene Sequences In Vitro
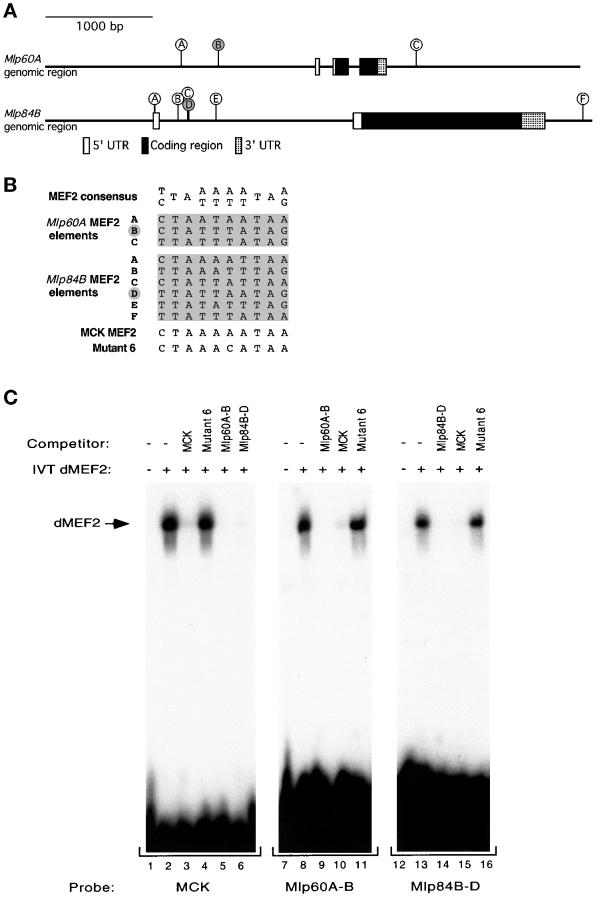
To begin to address whether Mlps may require dMEF2 activity directly for their expression, we analyzed the genomic sequences of the Mlp genes to identify potential dMEF2 binding sites. Figure 4A displays the genomic organization of the Mlp60A and Mlp84B genes denoting intron and exon structure. The Mlp60A gene exhibits three exons interrupted by two small introns, one in the 5′ untranslated region and another in the coding region. The Mlp84B gene contains one noncoding and one coding exon separated by a single large intron. Analysis of noncoding DNA within and surrounding the Mlp genes revealed the presence of multiple A/T-rich sequences matching exactly the reported MEF2 target binding consensus sequence (Olson et al., 1995). An alignment of the sites in the Mlp genes in comparison with the MEF2 consensus sequence and a bona fide mammalian MEF2 target sequence is displayed in Figure 4B. The Mlp60A gene contains three potential dMEF2 binding sites; two of these sites are located in the region 5′ to the start of gene, whereas the third is found 3′ to the coding sequence. Six putative dMEF2 binding sites are found in the Mlp84B gene. Four of the six are clustered in the intron, another is located 3′ to the coding region of the gene, and another is contained completely within the first exon.
Figure 4.
dMEF2 protein binds to consensus MEF2 sites found in the genomic regions of the Mlp genes. (A) Diagram of the genomic organization of the Mlp60A and Mlp84B genes. The boxed areas indicate the regions that correspond to cDNA sequences. The sticks indicate positions of 10-bp elements that exactly match the MEF2 consensus binding site. The two sites indicated by filled circles are those tested in the in vitro dMEF2 binding assay shown in C. The exact nucleotide range of the Mlp84B sites is noted in GenBank accession number AF090832. (B) The potential MEF2 binding elements found in the putative regulatory regions of Mlp60A and Mlp84B are shown aligned with the MEF2 consensus, the MEF2 regulatory site from the MCK enhancer, and the Mutant 6 form of the MCK site, which does not support MEF2 binding. Letters to the left correspond to the sites diagrammed in A, and the filled letters indicate those sites tested in C. (C) Mobility shift assays with in vitro–translated dMEF2 protein were performed with oligonucleotides corresponding to the MCK MEF2 site and one MEF2 element from each of the Mlp genes. The MEF2 elements used as probes were MCK MEF2 (lanes 1–6), Mlp60A-B (lanes 7–11), and Mlp84B-D (lanes 12–16). For each probe a control lysate lacking translated dMEF2 was included in parallel (lanes 1, 7, and 12). Competitor oligonucleotides were used at 100-fold molar excess of each probe. dMEF2 binds to each of the probes specifically (lanes 2, 8, and 13). Each of the Mlp MEF2 sites binds dMEF2 and competes for binding to the MCK element and their cognate site. A mutant form of the MCK element (Mutant 6) fails to compete for binding with any of the sites tested (lanes 4, 11, and 16).
One potential target site from each Mlp gene was chosen to test directly for dMEF2 binding activity in vitro, in parallel with a control site derived from the mammalian MCK gene (Gossett et al., 1989), which has been shown to bind dMEF2 in vitro (Lilly et al., 1994; Nguyen et al., 1994). Labeled oligonucleotides consisting of the core MEF2 binding site flanked by three to eight additional nucleotides derived from genomic sequence were generated and used in electrophoretic gel mobility shift assays with in vitro–translated dMEF2 protein (Lilly et al., 1994). Figure 4C shows a shifted complex with each of the three probes (MCK, Mlp60A-B, and Mlp84B-D) dependent on the addition of dMEF2 translation product. In addition, in all three cases, the bound probe could be efficiently competed by addition of excess unlabeled probe or an unlabeled control (MCK) probe. Furthermore, each of the shifted species was ineffectively competed by addition of excess unlabeled mutant probe, Mutant 6, in which a single base substitution within the MEF2 core binding site has been introduced (Cserjesi and Olson, 1991). Taken together, these results show that dMEF2 recognizes and binds specifically and directly to sequences derived from the Mlp60A and Mlp84B genes.
Myoblast Fusion Is Not Required for Muscle LIM Protein Expression
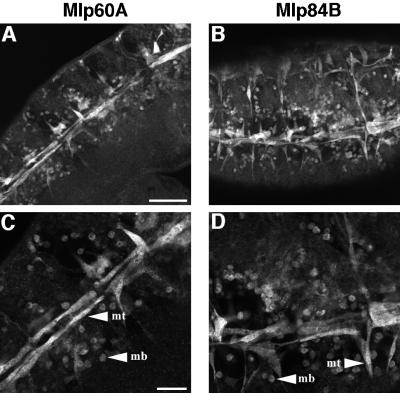
Fusion of myoblasts into syncytial muscle fibers is a key feature of somatic muscle development in Drosophila (Bate, 1990). Although the mechanism of fusion is not well understood, mutations that disrupt specific aspects of the process are beginning to be characterized. In Drosophila, embryos that harbor mutations in genes such as myoblast city (mbc) and rollingstone (rost) display severe defects in the process of myoblast fusion such that many single unfused myoblasts persist late into embryogenesis after they would normally be incorporated into a growing muscle fiber (Paululat et al., 1995; Rushton et al., 1995). Previously, we noted that myoblast fusion precedes Mlp protein accumulation by a few hours, suggesting that Mlps are not likely to be required for the fusion process (Stronach et al., 1996). However, the temporal relationship of fusion events and Mlp expression raises the possibility that Mlp accumulation in the somatic lineage might depend on cell fusion. Furthermore, because fusion defects are also associated with mutations in dMEF2, we assessed the expression of Mlps in the rost mutant background in which a failure of myoblast fusion is the primary defect. In embryos mutant for the rost gene, both Mlp60A and Mlp84B are observed in single unfused myoblasts (Figure 5), distributed in characteristic positions where, under normal circumstances, they would provide a pool of fusion competent cells for the developing syncytial myotubes (Bate, 1990). Thus, from these observations, we conclude that myoblast fusion is not required for Mlp expression, an observation that has also been noted for myosin and β3 tubulin (Paululat et al., 1995; Rushton et al., 1995).
Figure 5.
Myoblast fusion is not required for muscle LIM protein expression. Immunofluorescent detection of Mlp60A (A and C) or Mlp84B (B and D) in embryos mutant for the rollingstone gene is shown. These embryos display severe defects in myoblast fusion. Both Mlps are expressed in single unfused myoblasts (mb) as well as in myotubes (mt) that have formed (see higher magnification; C and D). All panels display ventral-lateral views of late stage embryos with anterior to the left. Bars: A and B, 50 μm; C and D, 20 μm.
Mlp84B Subcellular Distribution in Wild-Type and Mutant Visceral and Somatic Muscles
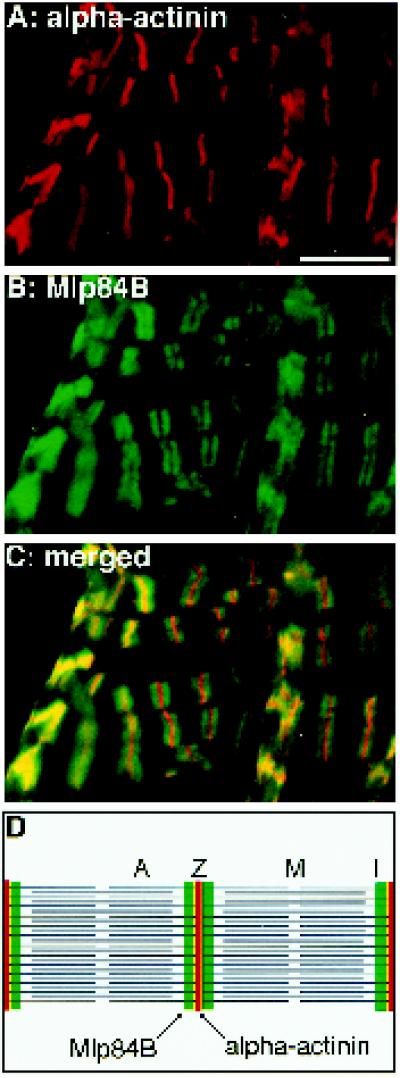
Knowledge of the subcellular distribution of a protein often contributes substantially to an understanding of its function. In embryonic somatic muscles, Mlp60A and Mlp84B are found in both the nuclear and cytoplasmic compartments, consistent with either a regulatory or structural role in differentiating muscle (Stronach et al., 1996). When expressed in rat embryo fibroblast cells, Drosophila Mlps showed a specific association with the actin cytoskeleton (Stronach et al., 1996). To determine the precise localization of Mlps within mature myofibrils at higher resolution, we double-labeled whole, third instar larval midguts using antibodies directed against the Mlps and α-actinin, which marks Z-bands (Figure 6).
Figure 6.
Mlp84B localizes to discrete sites within sarcomeres of larval midgut visceral muscles. The mesoderm surrounding a third instar larval midgut has been double labeled for α-actinin (red), to visualize Z-bands, and Mlp84B (green). Most of the visceral muscles are positioned horizontally, but muscle cells can be seen on top of these positioned vertically in the figure. Mlp84B localizes as doublets within the muscles (B). In the merged image (C), Mlp84B is observed to flank α-actinin in a region adjacent to the Z-bands (see diagram of two adjacent sarcomeres with bands indicated in D). Bar, 10 μm.
Surrounding the midgut, elongated visceral mesodermal cells form a lattice of transverse and longitudinal fibers. Although these cells do not undergo myoblast fusion, they appear striated and display sarcomeric repeats (Saide et al., 1989; Tepass and Hartenstein, 1994). Within the midgut visceral mesoderm, α-actinin prominently localizes to Z-bands (Figure 6A). Z-bands demarcate the ends of individual sarcomeres (Figure 6D), where the barbed ends of actin thin filaments terminate. In the same tissue, Mlp84B distributes as a doublet that flanks each Z-band (Figure 6B). As seen in merged images (Figure 6C), α-actinin and Mlp84B are localized in adjacent regions. Mlp84B extends away from the periphery of the Z-band, whereas α-actinin is clearly more restricted (Figure 6, C and D). The localization of Mlp84B to discrete sites within the muscle sarcomere provides evidence for a specific association of Mlp84B with the microfilament cytoskeleton in vivo. No nuclear staining for Mlp84B in the visceral muscles was observed at this time in development. Although Western immunoblot analysis revealed that Mlp60A is present in isolated midgut preparations, we were unable to detect the protein using immunofluorescent methods. It is unclear why Mlp60A protein was not observed in situ, but perhaps within the mature myofibril, Mlp60A is complexed with protein partners such that the epitopes recognized by our antibodies are masked.
Effects of α-Actinin Mutations on Mlp84B Distribution.
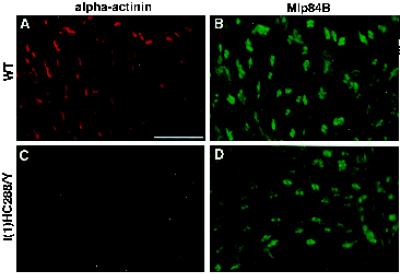
It has been reported that vertebrate CRP1 interacts with α-actinin (Pomies et al., 1997). This interaction may underlie the association of CRPs with the microfilament cytoskeleton. By inference, Drosophila Mlps, relatives of the vertebrate CRPs, may associate with the cytoskeleton via interactions with α-actinin. Although α-actinin and Mlp84B are not extensively colocalized, the distribution in adjacent domains may indicate that a subset of the molecules associate at the Z-band periphery. To evaluate whether Mlp84B localization within sarcomeres depends on the presence of α-actinin, we analyzed the distribution of Mlp84B in α-actinin mutant larval midguts. Mutant larvae become progressively paralyzed and flaccid between hatching and the second instar molt. These larvae were dissected, and midguts were double labeled for Mlp84B and α-actinin in parallel with similar staged wild-type larval midguts. Figure 7 illustrates comparable Mlp84B localization in α-actinin mutant versus wild-type midgut visceral muscles (Figure 7, compare B and D). Although mutant myofibers appear generally more disorganized than wild type, doublets of Mlp84B protein were observed in repeated arrays, reflecting some residual sarcomeric organization. Identical results were seen in four other α-actinin mutant backgrounds. Thus, in the absence of functional α-actinin protein, Mlp84B is still capable of localizing to discrete sites within the developing sarcomeres. Therefore, α-actinin is not absolutely essential for recruiting Mlp84B to its normal subcellular location within muscle tissue. We do observe some inappropriate localization of Mlp84B in α-actinin–deficient muscles (Figure 7D). This may result from the decaying muscle cytoarchitecture in the mutant or may reflect some contribution of α-actinin in restricting Mlp84B localization.
Figure 7.
Loss of α-actinin does not affect Mlp84B localization within visceral muscles. α-Actinin null mutant l(1)HC288/Y and wild-type midgut muscles have been double labeled for α-actinin (A and C) and Mlp84B (B and D). The null mutant lacks staining for α-actinin protein (C), and wild type is shown for comparison (A). Note that Mlp84B distribution is similar in the wild-type (B) and mutant (D) visceral muscles showing doublets flanking the Z-bands. In the mutant, Mlp84B still localizes to discrete sites within sarcomeres. Bar, 10 μm.
Localization of Mlp84B to Muscle Attachment Sites (MASs) Does Not Require PS2 Integrins.
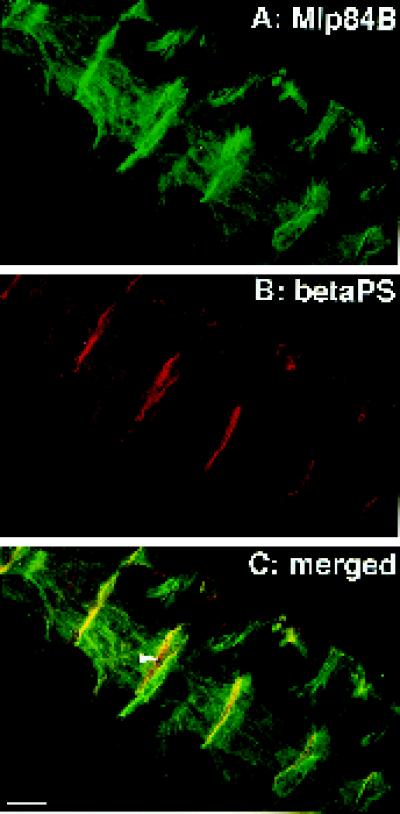
The identification of components required for normal Mlp84B localization may provide insight into its role in muscle differentiation. We noted previously that, in embryos, Mlp84B is enriched at the ends of somatic muscle fibers where they make attachments to the body wall. These attachment sites are thought to be analogous to the prominent focal adhesions of cultured fibroblast cells. Indeed, both focal adhesions and MASs are areas where actin filaments terminate at the membrane and become linked to extracellular matrix through a series of protein interactions culminating with the transmembrane integrin receptors (Burridge et al., 1988; Reedy and Beall, 1993; Tepass and Hartenstein, 1994). To confirm that the enrichment of Mlp84B at the ends of the somatic muscle fibers coincides with the location of the MAS, we double labeled embryos with antibodies directed against Mlp84B and the βPS integrin subunit (Figure 8). βPS forms a dimer with αPS2 in the muscle cell and with αPS1 in the neighboring tendon cell membrane (Bogaert et al., 1987; Leptin et al., 1989). As revealed in the merged image (Figure 8C), significant colocalization of Mlp84B and βPS was observed at the MASs. Thus, we conclude that Mlp84B colocalizes with muscle integrin complexes. Integrin staining that was not coincident with Mlp84B was also observed in between the neighboring longitudinal muscles (Figure 8C, arrow), which may reflect the presence of βPS complexes in the tendon cell membranes.
Figure 8.
Mlp84B colocalizes with βPS integrin at the MASs. Ventral-lateral longitudinal muscles of a stage 16 embryo are double labeled for Mlp84B (green) and the βPS subunit of integrin (red). The merged image (C) reveals significant colocalization of the two proteins (yellow) at the MASs. The arrow in C indicates βPS complexes in the tendon cells. Bar, 20 μm.
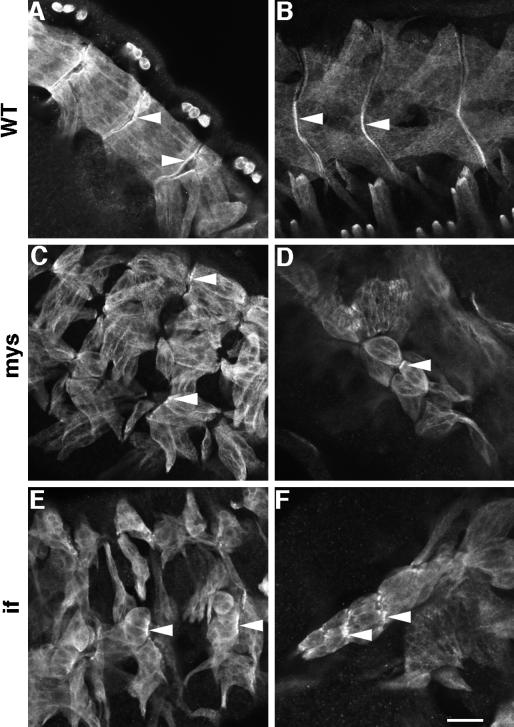
Colocalization of Mlp84B and βPS integrin at MASs raises the possibility that integrin receptors, through interactions with extracellular ligands, recruit and stabilize the association of Mlp84B with the junction and that this association is required for muscle development or attachment. We sought to address the role of integrin receptor engagement in the recruitment of Mlp84B by analyzing the distribution of Mlp84B in an integrin mutant background. PS2 integrin mutations are lethal in part because of detachment of the muscles late in embryogenesis and subsequent failure of larvae to hatch from the eggshell (Wright, 1960). Using null alleles for both the βPS subunit encoded by the myospheroid (mys) gene and the αPS2 subunit encoded by inflated (if) (Leptin et al., 1989; Wilcox et al., 1989; Brown, 1994), we looked specifically for the presence of Mlp84B at the MASs of muscles in the mutant embryos (Figure 9). In both mys and if mutants, Mlp84B was observed at the junctional attachment sites in muscles before complete detachment or at remnant sites after detachment (Figure 9, C and E). Often we detected Mlp84B enriched at the membrane of muscles that appeared to be adhering end-to-end with one another (Figure 9, D and F). Therefore, Mlp84B localizes to MASs independent of PS2 integrin-ligand interactions. By staining mutant embryos with the anti-βPS antibody, we detected no residual integrin complexes containing the βPS subunit that may have been contributed maternally (our unpublished results).
Figure 9.
Mlp84B is capable of associating with MASs in the absence of integrin–ligand interactions. Immunofluorescent detection of Mlp84B in somatic muscles of stage 16 wild-type (A and B), mys (C and D), or if (E and F) embryos is shown. Note the characteristic rounded muscles observed in embryos mutant for either integrin subunit. Arrows indicate Mlp84B enrichment at the MASs of wild-type embryos (A and B) or at remnant junctions in mutant embryos (C-F). Bar, 20 μm.
DISCUSSION
Key to understanding the process of muscle differentiation is the characterization of myogenic regulatory factors and their molecular targets. Presumably, it is the unique combination of these target proteins that ultimately defines the differentiated morphology and function of distinct muscle types. In this study, we have described the results of molecular epistasis analysis designed to unravel mechanisms responsible for regulating Mlp gene expression during the process of myogenic differentiation and for establishing the subcellular distributions of Mlps within muscles. This analysis confirms that Mlps lie downstream of events involving mesoderm specification and patterning. Our findings indicate that Mlps are likely targets of transcriptional regulation by a member of the MEF family of MADS (named for MCM-1, agamous, deficiens, and serum response factor) box myogenic transcription factors that regulate muscle differentiation. In addition, the process of myoblast fusion in the somatic muscle lineage is not required for Mlp expression. Furthermore, we demonstrate that Mlp84B associates specifically with the microfilament-based cytoskeleton within sarcomeres and that Mlp84B is enriched at the MASs in embryonic somatic muscles. The discrete localization of Mlp84B is not affected by loss of the prominent Z-band protein α-actinin or the transmembrane PS2 integrin receptor.
Mlps Require dMEF2 Activity during Myogenesis
Many lines of evidence point to a role for CRP family members in muscle differentiation. To understand at what point in the differentiation program Mlps may function, we examined the interplay between dMEF2 transcription factor function and Mlp expression. dMEF2 is an invertebrate member of a family of vertebrate myocyte enhancer binding factors that display a MADS box and MEF2 domain, which promote DNA binding and dimerization (Nguyen et al., 1994; Olson et al., 1995). Cell biological and biochemical studies using vertebrate systems have implicated the MEF family of transcription factors in myogenesis, specifically in regulating muscle differentiation (Olson et al., 1995). During Drosophila development, dMEF2 accumulates in all muscle subtypes and displays a biphasic expression pattern suggesting a requirement for the protein in larval and adult myogenesis (Nguyen et al., 1994; Bour et al., 1995; Ranganayakulu et al., 1995). Despite early expression of dMEF2 during gastrulation, genetic analysis revealed a role for the protein relatively late in the myogenic pathway. dMEF2 mutant embryos display defective myoblast fusion and a failure of muscle differentiation (Bour et al., 1995; Lilly et al., 1995), consistent with the postulated role of vertebrate MEF2 proteins. Identification of the targets of dMEF2 has provided substantial insight into why differentiation fails in mutant embryos. dMEF2 activity regulates expression of several late markers that contribute to muscle structure and function. These include myosin, tropomyosin I, and the αPS2 integrin (Ranganayakulu et al., 1995; Lin et al., 1996). Our findings demonstrate clearly that dMEF2 is also essential for the expression of both Mlp60A and Mlp84B in all of the muscle tissues in which they are normally expressed, because we observe no Mlp-positive cells in dMEF2 null mutant embryos. Indeed, in embryos homozygous for a severe hypomorphic allele of dMEF2 (mef113) in which some nuclear dMEF2 protein can be detected (Ranganayakulu et al., 1995), we observed very weak expression of Mlp60A and Mlp84B (our unpublished observations). This result supports the notion that Mlp genes may be sensitive to changes in the levels of dMEF2 transcriptional activity.
To investigate whether dMEF2 is capable of stimulating Mlp expression, we used the GAL4-UAS system to express dMEF2 ectopically in the epidermis, a tissue in which it is not normally found. As a consequence of ectopic dMEF2 expression, we noted a robust up-regulation of myosin protein, a target of dMEF2. Similarly, Mlp84B was induced in the epidermis. We did not observe Mlp60A protein accumulation in epidermal cells that were programmed to express dMEF2. It is possible that either transcription from the Mlp60A gene is not initiated in the context of epidermal cells expressing dMEF2 or that protein is produced but then rapidly degraded. Consistent with the former hypothesis, Lin and colleagues (1997), using a similar dMEF2 overexpression system, failed to detect Mlp60A transcripts by in situ hybridization in the embryonic epidermis, with the exception of several cells at the ventral midline. Perhaps the Mlp60A gene is subject to different regulatory constraints than myosin and Mlp84B. For instance, Mlp60A expression may be specifically repressed by an epidermal transcriptional regulator or may lack a required coactivator present in mesoderm but not ectoderm. Collectively, these results indicate that Mlps may be members of a group of dMEF2 target proteins that contribute to differentiated muscle cytoarchitecture and function. Interestingly, forced premature overexpression of dMEF2 in the mesoderm did not result in premature accumulation of Mlps in muscle tissue (our unpublished observations). This observation implies that there are likely to be additional transcriptional regulatory inputs that converge at the Mlp gene promotors during myogenic development.
To explore whether the regulation of Mlp gene expression by dMEF2 could be direct, we searched the noncoding regions of Mlp genomic DNA for potential dMEF2 binding sites. Indeed, we identified several such sequences in both Mlp genes that matched exactly the reported consensus binding sites for MEF2 transcription factors (Olson et al., 1995). These putative binding sites are located in regions of the genes where regulatory information, such as enhancer binding sites, are characteristically found. We chose one putative site from each gene to test for dMEF2 binding in an in vitro binding assay. Labeled oligonucleotides derived from the Mlp60A and Mlp84B genes were directly and specifically bound by dMEF2 protein. Demonstration of candidate dMEF2 sites in the regulatory regions of the Mlp genes, coupled with the ability of a site from each gene to support specific binding of dMEF2 protein in vitro, suggests that Mlps could be direct targets of dMEF2 activity in vivo. Definitive proof that Mlps are direct targets of dMEF2 will require further in vivo analysis of the MEF2 sites present in the Mlp genes.
Our analysis of the genomic organization of the Mlps genes revealed an interesting finding. Although the Mlp60A gene encodes a protein with a single LIM domain, sequences 3′ to the coding region appear to contain information that may have, at one time, encoded an additional four LIM domains. Perhaps an ancestral gene coding for a protein with five LIM domains gave rise to two Mlp genes, with the result that the Mlp84B gene was maintained to encode five domains, whereas the Mlp60A gene diverged and was corrupted to produce a truncated reading frame encoding a single LIM domain. It is also noteworthy that each of the two Mlp genes has a putative dMEF2 binding site located just downstream of the gene and that these two sites appear to be related to each other. Otherwise, the genomic structure of these two genes appears to have significantly diverged.
Myoblast Fusion Is Not Required for Mlp Expression
In addition to a failure of muscle differentiation, the process of myoblast fusion is also affected in dMEF2 mutant embryos. To address whether the failure to accumulate Mlps is a result of defects in fusion, we analyzed Mlp expression in rost mutant embryos. In these embryos, unfused myoblasts persist well beyond the normal period of fusion and eventually express several differentiation markers including myosin (Paululat et al., 1995). We demonstrate that Mlps can also be expressed in single myoblasts in rost mutants. Thus, the defects in myoblast fusion resulting from mutations in dMEF2 do not appear to be the primary cause of the observed failure of Mlp expression in dMEF2 mutant embryos. Moreover, myoblast fusion does not appear to be a crucial checkpoint for ensuing muscle differentiation. Late in embryogenesis, these myoblast levels decline, presumably through an apoptotic mechanism coupled with macrophage-mediated phagocytosis (Rushton et al., 1995). We have never detected Mlp expression in macrophages under normal conditions. However, it is possible that some Mlp-positive cells observed in the rost mutant embryos represent phagocytic cells that have engulfed Mlp-expressing myoblasts.
Mlps Are Components of the Contractile Apparatus in Myofibrils
Vertebrate CRPs have been localized to the actin cytoskeleton in certain cell types; CRP3/MLP gene disruption in the mouse leads to defects in cardiac muscle cytoarchitecture; and Drosophila Mlps are able to colocalize with actin fibers in fibroblast cells (Arber et al., 1994, 1997; Stronach et al., 1996; Louis et al., 1997). These observations point to a potential role for CRP family members in establishing muscle structure. In previous studies we noted the appearance of a linear staining pattern for Mlps in somatic myotubes in the embryo; however, it was not possible to assess clearly the association of Mlps with subdomains of the microfilament cytoskeleton. Therefore, we turned to a later stage of development to examine the distribution of Mlps in relation to a mature sarcomeric pattern of striated muscle tissue. In the larva, a layer of striated visceral muscle cells encases the midgut and provides contractile activity to move food down the alimentary canal (Skaer, 1993). In these cells, we observed a highly localized distribution of Mlp84B protein in association with the sarcomeric cytoskeleton. Specifically, Mlp84B was concentrated in double stripes flanking the Z-bands, which are rich in α-actinin protein. To determine whether α-actinin influences the localization of Mlp84B, we assessed the distribution of Mlp84B in muscles that fail to express α-actinin. In α-actinin mutant larval midguts, sarcomeres appear somewhat disorganized, but Mlp84B is localized normally. These data suggest that α-actinin is not absolutely essential for the proper recruitment of Mlp84B to the sarcomere. Because Mlp84B displays five copies of a protein-binding LIM-glycine motif, each of which may direct interactions with additional proteins within the contractile apparatus, it is possible that the lack of redistribution of Mlp84B in α-actinin mutant muscles may reflect the involvement of multiple components for Mlp84B localization. Interestingly, a similar protein distribution has been noted for the actin capping protein tensin in cultured myotubes (Bockholt et al., 1992), as well as for the vertebrate CRP3/MLP isoform in mouse cardiomyocytes (Arber et al., 1997). The comparable subcellular distributions of Mlp84B and CRP3/MLP in muscle cells points to a potential functional conservation among vertebrate and invertebrate CRP family members. Although the mechanism underlying Mlp84B localization is still unknown, our data illustrate the specific association of Mlp84B with the microfilament cytoskeleton in vivo and lend support for a structural role for Mlp84B in mature differentiated muscles.
MAS Formation and Function
In striated muscles, Mlp84B localizes near regions enriched in the barbed, fast-growing ends of actin filaments, such as the Z-band. In the developing embryo, Mlp84B accumulates at MASs where the barbed ends of actin filaments terminate at the membrane and associate with transmembrane integrin receptors. Although Mlp84B and βPS proteins are colocalized in vivo, in the absence of PS2 integrins, Mlp84B localizes appropriately. Both the normal muscle patterning and polarity seen in the integrin mutant embryos before muscle detachment and the ability of Mlp84B to localize to the attachment sites in the absence of integrins suggest that integrin-ligand interactions are not instructive for defining the ends of the muscle fiber and assembling the attachment site. In further support of this notion, it is noteworthy that βPS molecules, incapable of binding extracellular ligands, also localize appropriately to MASs, results that place further emphasis on the importance of intracellular mechanisms for the organization of muscle termini (MartinBermudo and Brown, 1996). βPS complexes appear to be necessary only for the continued maintenance of the attachment site once it has been assembled, an observation previously noted by careful analysis of the myospheroid phenotype (Wright, 1960).
Concluding Remarks
Using molecular epistasis methods, we have examined the relationships between Mlps and several gene products necessary for Drosophila myogenesis. This analysis revealed the dependence of Mlp gene expression on the transcriptional regulator dMEF2 and places Mlp function late in the terminal stages of muscle differentiation. We show that myoblast fusion per se is not necessary for expression of the Mlp proteins and, furthermore, that neither α-actinin nor PS2 integrin is required to direct Mlp84B to its proper position in muscle cells. Future work will continue to address the mode of Mlp gene regulation and their functional significance in the process of myogenesis. Genetic analysis of Mlp function is likely to provide important insights into their physiological roles in muscle. Thus far, no mutations in the Mlp60A gene have been identified, and deficiency analysis has been hampered by an apparent haploinsufficient locus in the region. However, we have begun to characterize phenotypes associated with loss of Mlp84B function. For this analysis, we have used a set of overlapping deficiencies that remove a small region of genomic DNA including the coding region of the Mlp84B gene. The Mlp84B-deficient animals die as larvae and early pupae, exhibiting locomotor and morphological defects consistent with abnormal muscle function (Clark and Beckerle, unpublished observations). Although additional work needs to be done to characterize fully the Mlp84B null phenotype, preliminary results suggest that gene function is essential for viability and proper muscle function.
ACKNOWLEDGMENTS
We thank Kathleen Clark for recognizing the additional LIM coding sequences in the vicinity of the Mlp60A gene and for helpful comments during the course of this work. We also thank the Berkeley Drosophila Genome Project for genomic sequence in the Mlp60A region and the DNA Sequencing Core Facility at the University of Utah, supported in part by National Cancer Institute grant CA-42014, for genomic sequence in the Mlp84B region. We are grateful to Bob Schackmann and the University of Utah Oligonucleotide Synthesis Facility for the synthesis of all sequencing primers used in this study as well as the Mlp60A and Mlp84B MEF2 elements. Additional thanks to Ed King and the Biology Department Imaging Facility for assistance with confocal microscopy and image processing. We thank N. Perrimon, N. Brown, R. Schulz, A. Paululat, D. Van Vactor, and C. Keller for graciously providing fly stocks and J. Saide, D. Kiehart, and D. Brower for antibodies. This work was supported by National Institutes of Health grants GM-50877 and HL-60591. M.C.B. is the recipient of a faculty research award from the American Cancer Society.
REFERENCES
- Arber S, Caroni P. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes & Dev. 1996;10:289–300. doi: 10.1101/gad.10.3.289. [DOI] [PubMed] [Google Scholar]
- Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- Arber S, Hunter JJ, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriad J-C, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- Bockholt SM, Otey CA, Glenny JRJ, Burridge K. Localization of a 215 kDa tyrosine phosphorylated protein that cross-reacts with tensin antibodies. Exp Cell Res. 1992;203:39–46. doi: 10.1016/0014-4827(92)90037-9. [DOI] [PubMed] [Google Scholar]
- Bogaert T, Brown N, Wilcox M. The Drosophila PS2 antigen is an invertebrate integrin that, like the fibronectin receptor, becomes localized to muscle attachments. Cell. 1987;51:929–940. doi: 10.1016/0092-8674(87)90580-0. [DOI] [PubMed] [Google Scholar]
- Bour BA, O’Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes & Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- Bourgouin C, Lundgren SE, Thomas JB. apterous is a Drosophila LIM domain gene required for the development of a subset of embryonic muscles. Neuron. 1992;9:549–561. doi: 10.1016/0896-6273(92)90192-g. [DOI] [PubMed] [Google Scholar]
- Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. Vol. 44. San Diego: Academic Press; 1994. pp. 635–654. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brower DL, Wilcox M, Piovant M, Smith RJ, Reger LA. Related cell surface antigens expressed with positional specificity in Drosophila imaginal discs. Proc Natl Acad Sci USA. 1984;181:7485–7489. doi: 10.1073/pnas.81.23.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NH. Null mutations in the αPS2 and βPS integrin subunit genes have distinct phenotypes. Development. 1994;120:1221–1231. doi: 10.1242/dev.120.5.1221. [DOI] [PubMed] [Google Scholar]
- Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu Rev Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- Crawford AW, Pino JD, Beckerle MC. Biochemical and molecular characterization of the chicken cysteine-rich protein, a developmentally regulated LIM-domain protein that is associated with the actin cytoskeleton. J Cell Biol. 1994;124:117–127. doi: 10.1083/jcb.124.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserjesi P, Olson E. Myogenin induces muscle-specific enhancer binding factor MEF2 independently of other muscle-specific gene products. Mol Cell Biol. 1991;11:4854–4862. doi: 10.1128/mcb.11.10.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flybase The Drosophila genetic database. Nucleic Acids Res. 1994;22:3456–3458. doi: 10.1093/nar/22.17.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golsteyn RM, Beckerle MC, Koay T, Friederich E. Structural and functional similarities between the human cytoskeletal protein zyxin and the ActA protein of Listeria monocytogenes. J Cell Sci. 1997;110:1893–1906. doi: 10.1242/jcs.110.16.1893. [DOI] [PubMed] [Google Scholar]
- Gossett LA, Kelvin DJ, Sternberg EA, Olson EN. A new myocyte-specific enhancer binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol Cell Biol. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Feghali R. Cytoplasmic myosin from Drosophila melanogaster. J Cell Biol. 1986;103:1517–1525. doi: 10.1083/jcb.103.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Flick MJ, Kudla AJ, Konieczny SF. MLP promotes myogenesis by enhancing the activity of MyoD. Mol Cell Biol. 1997;17:4750–4760. doi: 10.1128/mcb.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptin M, Bogaert T, Lehmann R, Wilcox M. The function of PS integrins during Drosophila embryogenesis. Cell. 1989;56:401–408. doi: 10.1016/0092-8674(89)90243-2. [DOI] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN. d-MEF2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci USA. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor d-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Lin M-H, Bour BA, Abmayr SM, Storti RV. Ectopic expression of MEF2 in the epidermis induces epidermal expression of muscle genes and abnormal muscle development in Drosophila. Dev Biol. 1997;182:240–255. doi: 10.1006/dbio.1996.8484. [DOI] [PubMed] [Google Scholar]
- Lin MH, Nguyen HT, Dybala C, Storti RV. Myocyte-specific enhancer factor 2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosin gene muscle expression. Proc Natl Acad Sci USA. 1996;93:4623–4628. doi: 10.1073/pnas.93.10.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis HA, Pino JD, Schmeichel KL, Pomies P, Beckerle MC. Comparison of three members of the cysteine-rich protein (CRP) family reveals functional conservation and divergent patterns of gene expression. J Biol Chem. 1997;272:27484–27491. doi: 10.1074/jbc.272.43.27484. [DOI] [PubMed] [Google Scholar]
- Lundgren SE, Callahan CA, Thor S, Thomas JB. Control of neuronal pathway selection by the Drosophila LIM homeodomain gene. apterous. Development. 1995;121:1769–1773. doi: 10.1242/dev.121.6.1769. [DOI] [PubMed] [Google Scholar]
- Martin-Bermudo MD, Brown NH. Intracellular signals direct integrin localization to sites of function in embryonic muscles. J Cell Biol. 1996;134:217–226. doi: 10.1083/jcb.134.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N, Johnson C, Wang Y. Formation and alignment of Z lines in living chick myotubes microinjected with rhodamine-labeled alpha-actinin. J Cell Biol. 1986;103:2163–2171. doi: 10.1083/jcb.103.6.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J, Olson E. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Bodmer R, Abmayr SM, McDermott JC, Spoerel NA. d-mef2: a Drosophila mesoderm-specific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proc Natl Acad Sci USA. 1994;91:7520–7524. doi: 10.1073/pnas.91.16.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, Perry M, Schulz RA. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. Vol. 44. San Diego: Academic Press; 1994. pp. 446–487. [DOI] [PubMed] [Google Scholar]
- Paululat A, Burchard S, Renkawitz-Pohl R. Fusion from myoblasts to myotubes is dependent on the rolling stone gene (rost) of Drosophila. Development. 1995;121:2611–2620. doi: 10.1242/dev.121.8.2611. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Engstrom L, Mahowald AP. Developmental genetics of the 2C-D region of the Drosophila X-chromosome. Genetics. 1985;111:23–41. doi: 10.1093/genetics/111.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomies P, Louis HA, Beckerle MC. CRP1, a LIM domain protein implicated in muscle differentiation, interacts with alpha-actinin. J Cell Biol. 1997;139:157–168. doi: 10.1083/jcb.139.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. A series of mutations in the d-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev Biol. 1995;171:169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- Reedy MC, Beall C. Ultrastructure of developing flight muscle in Drosophila. II. Formation of the myotendon junction. Dev Biol. 1993;160:466–479. doi: 10.1006/dbio.1993.1321. [DOI] [PubMed] [Google Scholar]
- Rushton E, Drysdale R, Abmayr SM, Michelson AM, Bate M. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophila muscle development. Development. 1995;121:1979–1988. doi: 10.1242/dev.121.7.1979. [DOI] [PubMed] [Google Scholar]
- Sadler I, Crawford AW, Michelsen JW, Beckerle MC. Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J Cell Biol. 1992;119:1573–1587. doi: 10.1083/jcb.119.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saide JD, Chin-Bow S, Hogan-Sheldon J, Busquets-Turner L, Vigoreaux JO, Valgeirsdottir K, Pardue ML. Characterization of components of Z-bands in the fibrillar flight muscle of Drosophila melanogaster. J Cell Biol. 1989;109:2157–2167. doi: 10.1083/jcb.109.5.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Skaer H. The alimentary canal. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Vol. 2. New York: Cold Spring Harbor Laboratory Press; 1993. pp. 941–1012. [Google Scholar]
- Stronach BE, Siegrist SE, Beckerle MC. Two muscle-specific LIM proteins in Drosophila. J Cell Biol. 1996;134:1179–1195. doi: 10.1083/jcb.134.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U, Hartenstein V. The development of cellular junctions in the Drosophila embryo. Dev Biol. 1994;161:563–596. doi: 10.1006/dbio.1994.1054. [DOI] [PubMed] [Google Scholar]
- Valge-Archer VE, Osada H, Warren AJ, Forster A, Li J, Baer R, Rabbitts TH. The LIM protein RBTN2 and the basic helix-loop-helix protein TAL1 are present in a complex in erythroid cells. Proc Natl Acad Sci USA. 1994;91:8617–8621. doi: 10.1073/pnas.91.18.8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman I, Osada H, Grutz G, Agulnick A, Westphal H, Forster A, Rabbitts T. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskirchen R, Bister K. Suppression in transformed avian fibroblasts of a gene (crp) encoding a cysteine-rich protein containing LIM domains. Oncogene. 1993;8:2317–2324. [PubMed] [Google Scholar]
- Weiskirchen R, Pino JD, Macalma T, Bister K, Beckerle MC. The cysteine-rich protein family of highly related LIM domain proteins. J Biol Chem. 1995;270:28946–28954. doi: 10.1074/jbc.270.48.28946. [DOI] [PubMed] [Google Scholar]
- Wilcox M, DiAntonio A, Leptin M. The function of PS integrins in wing morphogenesis. Development. 1989;107:891–897. doi: 10.1242/dev.107.4.891. [DOI] [PubMed] [Google Scholar]
- Wright TRF. The phenogenetics of the embryonic mutant, lethal myospheroid, in Drosophila melanogaster. J Exp Zool. 1960;143:77–99. doi: 10.1002/jez.1401430107. [DOI] [PubMed] [Google Scholar]
- Yun K, Wold B. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr Opin Cell Biol. 1996;8:877–889. doi: 10.1016/s0955-0674(96)80091-3. [DOI] [PubMed] [Google Scholar]