Abstract
The cytoskeleton plays an important role in neuronal morphogenesis. We have identified and characterized a novel actin-binding protein, termed Mayven, predominantly expressed in brain. Mayven contains a BTB (broad complex, tramtrack, bric-a-brac)/POZ (poxvirus, zinc finger) domain-like structure in the predicted N terminus and “kelch repeats” in the predicted C-terminal domain. Mayven shares 63% identity (77% similarity) with the Drosophila ring canal (“kelch”) protein. Somatic cell-hybrid analysis indicated that the human Mayven gene is located on chromosome 4q21.2, whereas the murine homolog gene is located on chromosome 8. The BTB/POZ domain of Mayven can self-dimerize in vitro, which might be important for its interaction with other BTB/POZ-containing proteins. Confocal microscopic studies of endogenous Mayven protein revealed a highly dynamic localization pattern of the protein. In U373-MG astrocytoma/glioblastoma cells, Mayven colocalized with actin filaments in stress fibers and in patchy cortical actin-rich regions of the cell margins. In primary rat hippocampal neurons, Mayven is highly expressed in the cell body and in neurite processes. Binding assays and far Western blotting analysis demonstrated association of Mayven with actin. This association is mediated through the “kelch repeats” within the C terminus of Mayven. Depolarization of primary hippocampal neurons with KCl enhanced the association of Mayven with actin. This increased association resulted in dynamic changes in Mayven distribution from uniform to punctate localization along neuronal processes. These results suggest that Mayven functions as an actin-binding protein that may be translocated along axonal processes and might be involved in the dynamic organization of the actin cytoskeleton in brain cells.
INTRODUCTION
Numerous studies have shown that the actin-based cytoskeleton, in conjunction with microtubules and intermediate filaments, provides an internal architectural framework, which regulates the structure and functions of all eukaryotic cells. The actin-based cytoskeleton is responsible for the generation and maintenance of cell polarity and cellular motility (Lauffenburger and Horwitz, 1996) and regulates organelle and protein distribution as well as mRNA transportbetween the nucleus and cytoplasm (Agutter, 1991). It controls the formation of neuronal cell processes, axons, and dendrites in developing neuronal tissues (Riederer, 1990), secretion from neurons and other secretory cells (Trifaro and Vitale, 1993), and regulates gated channels (Undrovinas et al., 1995). Significantly, the actin-cytoskeleton is often altered in malignant, metastatic, and senescent cells (Rao and Cohen, 1991) and in some diseases (Janmey and Chaponnier, 1995; Towbin, 1998). Dynamic changes in the architecture of the cell occur in response to extracellular signals during embryogenesis, and during metastasis, mitogenesis, or secretion. These events lead not only to changes in cell shape, but also to changes in gene expression, presumably by activation and/or transport of regulatory proteins into the nucleus and their subsequent interaction with the nuclear matrix (Ben-Ze’ev, 1991; Rosette and Karin, 1995).
The assembly and disassembly of actin filaments are regulated by a large number of actin-binding proteins (ABPs). ABPs are widely conserved across phylogeny, in both primary structure and biochemical properties, which implies their functional importance. Approximately 70 different ABPs have been characterized, but new members are still being discovered (Pollard, 1993). Recently, a new family of actin-binding proteins with sequences homologous to the Drosophila kelch protein has emerged (Xue and Cooley, 1993; Cooley and Theurkauf, 1994). The Drosophila kelch protein is localized to large, actin-rich intercellular ring canals, which regulate cytoplasmic transport from nurse cells to the developing oocyte within the egg chamber. The kelch protein is believed to be important for the maintenance of the ordered cytoskeleton, since Drosophila kelch mutants have ring canals that are occluded with disordered actin filaments that disrupt cytoplasmic transport and result in female sterility (Xue and Cooley, 1993; Knowles and Cooley, 1994). The kelch protein has two sequence motifs that are also found in other molecules. The first motif, which consists of ∼115 amino acids, has been named the BTB domain (for broad-complex, tramtrack, bric-a-brac) (Godt et al., 1993) or POZ domain (for poxvirus, zinc finger) (Bardwell and Treisman, 1994; Albagli et al., 1995). The second motif, composed of 50 amino acids repeated in tandem, is the kelch repeats (Cooley and Theurkauf, 1994). The BTB/POZ domain is found in diverse molecules: in several developmentally regulated zinc-finger-type transcription factors of Drosophila and mammals, in presumed modulators of chromatin structure (Albagli et al., 1995), in several open-reading frames (ORFs) of poxviruses (Koonin et al., 1992; Senkevich et al., 1993), in the cytoskeletal protein calicin derived from the sperm head (von Bulow et al., 1995), in the murine kelch-related gene Enc-1 (Hernandez et al., 1997), and in the human nuclear matrix protein NRP/B (for nuclear matrix protein expressed in brain) (Kim et al., 1998). The BTB/POZ domain has been proposed to function as a protein-protein interaction interface, and this domain organizes higher order structures involved in chromatin folding or cytoskeleton organization (Albagli et al., 1995). The kelch repeats have been identified in poxvirus ORFs, in calicin, in Enc-1, and NRP/B, all of which also encode the BTB/POZ domain. In addition, kelch repeats were found in the mouse placental-specific murine IAP-promoted placenta protein cDNA of unknown function (Chang-Yeh et al., 1991), in the Spe-26 gene that is believed to encode a cytoskeletal protein necessary for the spermatid development of Caenorhabditis elegans (Varkey et al., 1995), in the scruin protein (Way et al., 1995) known to cross-link actin acrosomal filaments in the sperm of Limulus (Schmid et al., 1994), and in a novel type of protein tyrosine kinase that phosphorylates actin–fragmin complexes, named actin-fragmin kinase or AFK (Eichinger et al., 1996).
Here we report the identification and characterization in brain cells of a novel ABP, termed Mayven, with high homology to Drosophila kelch protein. In the brain, Mayven is expressed in both primary hippocampal neurons and astrocytes. It is colocalized with the actin-cytoskeleton in stress fibers and in patchy regions of the actin-rich ruffling margins of U373-MG astrocytoma/glioblastoma cells. Immunoprecipitation studies after transfection with green fluorescent protein (GFP)-tagged Mayven cDNA demonstrated a direct association between recombinant Mayven and actin. Depolarization of primary hippocampal neurons with KCl enhanced the association of Mayven with actin and resulted in dynamic changes in cell architecture including punctate localization of Mayven in neuronal processes, suggesting that Mayven is transported along axons. Our studies suggest that Mayven might play a role as an actin-binding protein in the cytoskeleton of neuronal and glial cells.
MATERIALS AND METHODS
Materials
The λ-ZapII human hippocampus and human heart cDNA libraries and RNA isolation kits were purchased from Stratagene (La Jolla, CA). Primers for PCR and sequencing were synthesized in an automated DNA synthesizer (model 394, Applied Biosystems, Foster City, CA). Other reagents for PCR were from Perkin Elmer-Cetus (Norwalk, CT). 32P-labeled probes were labeled with Prime-It II Random Primer Labeling Kit (Stratagene). Restriction endonucleases and modifying enzymes were obtained from Amersham Pharmacia (Piscataway, NJ) and New England BioLabs (Beverly, MA). Dithiobis succinimidyl propionate (DSP) was purchased from Pierce Chemical (Rockford, IL). Sequencing was performed using an automated sequencing kit and automated laser fluorescent sequencer (Amersham Pharmacia). All other chemical reagents were purchased from Sigma Chemical (St. Louis, MO).
Cloning and Sequencing of the Mayven cDNA
Standard molecular biological protocols and PCR techniques (Sambrook et al., 1989) were employed unless otherwise indicated. Mayven was cloned with a probe corresponding to the expressed sequence tag (EST) sequence, GenBank accession no. Z30075. A portion of this EST sequence was amplified by PCR using forward oligonucleotide 5′-GGAGTACAGGTTTGTCATCTGTGG-3′ and reverse oligonucleotide 5′-GCCATCATGACCTCCTACAGCAT-3′ from total DNA prepared from a human hippocampus cDNA library. An amplified fragment of 289 base pairs (bp) (corresponding to nucleotides 1334–1623 of the cDNA) was ligated into a TA cloning vector, PCR (Invitrogen, San Diego, CA), sequenced to confirm its identity and used as a probe to screen human hippocampus and human heart cDNA libraries constructed in λ-ZapII (Stratagene). Approximately 1 × 106 clones were screened twice in order to obtain the coding sequence of Mayven from overlapping clones. Several clones were purified and partially sequenced. Two clones were selected and sequenced in their entirety on both strands. Nucleic acid and protein data banks were searched with Mayven sequences using the BLAST program (Altschul and Lipman, 1990).
Chromosomal Localization of Murine Mayven
Genomic DNAs from C57BL/6J, Mus spretus mice, and the (C57BL/6JEi × SPRET/Ei)F1 × SPRET/Ei type interspecific backcross DNA panel (BSS) were obtained from the Jackson Laboratory (Bar Harbor, ME) (Rowe et al., 1994). Approximately 4 μg of genomic DNA from C57BL/6J and M. spretus were digested with eight different restriction enzymes and blotted onto a Hybond N+ Nylon membrane. The Southern blots were probed with a 32P-labeled Mayven-specific probe (1340–1690 bp of Mayven cDNA). A suitable restriction fragment length polymorphism (RFLP) banding pattern was found with BamHI. Approximately 2 μg of each of the 96 genomic DNAs from the BSS type backcross panel were digested, and the corresponding Southern blots were probed with the same Mayven-specific probe. Segregation of the Mayven alleles was compared with other loci from the mouse genome database (MGD) by the Jackson laboratory backcross DNA map service (Rowe et al., 1994).
Chromosomal Localization of the Human Mayven Gene
Genomic DNAs from NIGMS Hybrid Mapping Panel 2 and from somatic cell hybrids 11687 (chromosome 4) and 11714 (chromosome 5) containing monochromosomal human DNA were obtained from NIGMS Genetic Mutant Cell Repository (Coriel Institute for Medical Research, Camden, NJ). In addition, several chromosome 4 derivatives, including 11436, 11439, 11447, and 11448, were used in this study. The chromosome 4 derivative DNA samples, 11436 and 11439, respectively, also contain portions of chromosome 5. Mapping panel 2 consists of DNAs isolated from human and rodent parental cell lines (mouse and Chinese hamster) and from 24 human–rodent somatic cell hybrids retaining one or two human chromosomes. All but two of the hybrids retain a single human chromosome. Rodent–human cell hybrids 11687, 11714, 11436, 11439, 11447, and 11448 contain Chinese hamster chromosomes and either intact human chromosome 4 and 5 or a segment of chromosome 4. Approximately 5 μg of genomic DNA from human, hamster, and mouse were digested with BamHI, HindIII, and PstI to locate a suitable RFLP with the 32P-labeled Mayven gene-specific probe. Subsequently, DNAs from Panel 2 were cut with HindIII, and a Southern blot was probed with the gene-specific Mayven probe. Concordance between restriction fragments of the human genomic DNA and chromosome 4, as well as specific portions of the same chromosome, was used to establish the chromosomal localization of Mayven.
Northern Blot Analyses of Mayven Expression
The mRNA blots containing human adult and fetal tissue mRNAs were obtained from CLONTECH Laboratories (Palo Alto, CA). Total mRNAs from various cell lines were prepared using an mRNA isolation kit (Invitrogen) according to the manufacturer’s protocol. All cell lines were purchased from and cultured according to the specifications of the American Type Culture Collection (ATCC, Manassas, VA). The 32P-labeled Mayven probe corresponded to the nucleotides from 1148 to 2358 bp of the Mayven cDNA sequence.
Expression of the N-Terminal Portion of Mayven in Escherichia coli Fused to Glutathione-S-transferase (GST)
A GST–N-terminal Mayven (GST-N-Mayven) construct encoding amino acids (aa) 1–306 of Mayven fused to GST protein was generated by PCR using Mayven-specific oligonucleotides containing restriction site linkers, and then subcloned into the BamHI and EcoRI sites of the pGEX 4T-2 vector (Amersham Pharmacia). This construct was used to transform E. coli DH5α and was analyzed by sequencing. GST-N-terminal Mayven fusion protein expression was induced using 10 mM isopropyl β-thiogalactopyranoside. Recombinant protein was affinity purified using Glutathione-Sepharose 4B beads, according to Pharmacia’s protocol (GST Gene Fusion System, Second Edition, 1996) with the exception of elution steps, which were performed on ice instead of at room temperature. Purified protein solubilized in Glutathione Elution Buffer (Pharmacia) was dialyzed against cold PBS and stored in 50% ultrapure glycerol (United States Biochemical, Cleveland, OH) in 1 × PBS at −20°C.
Generation of anti-Mayven-specific Antibodies
Soluble, purified GST-N-Mayven protein was used to generate anti-Mayven antibodies in rabbits by standard methods (East Acres Biological, Southbridge, MA). IgG fractions from preimmune and immune anti-Mayven sera (R-5147) were purified using the ImmunoPure Binding/Elution Buffer System (Pierce Chemical) according to the manufacturer’s instructions. Briefly, sera were mixed with 3 volumes of ImmunoPure binding buffer and filtered through a 0.8-μm filter. Diluted sera were applied onto Protein-A columns and passed through the columns three times. Columns were washed three times with binding buffer, and IgG was eluted with ImmunoPure elution buffer. Eluted IgG was dialyzed against cold PBS, and then against 50% ultrapure glycerol in 1× PBS, and stored at −20°C. The specificity of the anti-Mayven antiserum was determined by preabsorbing the purified anti-Mayven IgG with recombinant GST-N-Mayven fusion protein. GST-N-Mayven (10 μg) was combined with 100 μl of anti-Mayven IgG (purified as described above) and incubated for 1.5 h at 4°C. Glutathione-Sepharose 4B beads (Amersham Pharmacia) were added, and the sample was incubated for 45 min. Complexes of anti-Mayven IgG and GST-N-Mayven fusion protein bound to Glutathione-Sepharose were pelleted and discarded. The absorbed serum was used for Western blotting.
Cross-linking of GST-N-Mayven Fusion Protein
The GST peptide was removed from the GST-N-Mayven fusion protein by adding ∼2 U of thrombin directly to ∼300 μg of fusion protein after elution from Glutathione-agarose beads. The protein was then incubated in PBS (Pierce) with 10 mM dithiobis (succinimidyl propionate), a bifunctional protein cross-linking reagent, freshly prepared as a 450 mM stock in DMSO (Sigma Chemical). Cross-linking was carried out at room temperature for 30 min and was terminated by adding either 2× Laemmli buffer, or 2× Laemmli buffer with DTT (an S-S reducing agent). Proteins were boiled for 5 min, resolved on a 4–12% gradient gel (Bio-Rad, Hercules, CA), and visualized by staining with Coomassie blue.
Cell Culture
U373-MG astrocytoma/glioblastoma cells were purchased from ATCC and cultured in DMEM containing 10% FBS, according to ATCC specifications. Rat primary astrocytes were isolated from postnatal day 1 rats as described previously (Brewer et al., 1993). Rat primary hippocampal neurons were prepared from the hippocampus of Sprague Dawley rats at gestational day 18 and cultured in neurobasal medium (Life Technologies, Gaithersburg, MD) supplemented with B27 (Life Technologies) and 0.5 mM l-glutamine for 4–5 d, as described by Brewer et al. (1993). For depolarization of neurons, neurons were cultured as above for 7–10 d, refed with neurobasal medium without B27 and l-glutamine for 4 h, and then depolarized by transfer into the above medium supplement with 40 mM KCl.
Immunolocalization by Confocal Microscopy
Standard immunohistochemical methods were used as described (Kim et al., 1998). Briefly, U373-MG astrocytoma/glioblastoma cells were seeded onto sterile 18-mm glass coverslips (Fisher Scientific, Pittsburgh, PA) in 12-well plates at different densities. Primary neurons were plated onto 12-mm BioCoat Poly-d-lysine coverslips (Collaborative Research, Bedford, MA) at 5 × 105 cells/coverslip. All cells were processed for immunostaining as follows: cells were washed in room temperature PBS for 2–3 min, fixed in 4% paraformaldehyde in PBS for 15 min, permeabilized with 0.2% Triton-X 100 in PBS for 5 min, and blocked in 3% BSA in PBS for 1 h at room temperature or overnight at 4°C. Cells were incubated for 1 h with anti-Mayven IgG (at 1:20 ratio) in PBS containing 0.1% BSA and 3% normal goat and/or horse serum, washed, and then incubated with 1:100 dilution of Texas Red-conjugated goat anti-rabbit IgG (Vector Laboratories, Burlingame, CA). For double staining of Mayven and filamentous actin (F-actin), cells were first stained for Mayven as indicated above, and then incubated for 40 min with fluorescein phalloidin (Molecular Probes, Eugene, OR) at 0.2 U/ml. Immunostained cells were washed, mounted with ProLong antifade reagent (Molecular Probes), and examined using Sarastro 2000 confocal laser scanning microscope (CLSM) (Molecular Dynamics, Sunnyvale, CA) and Leica TCS-NT (Leica, Wetzlar, Germany) confocal-laser-scanning microscopes optimized for simultaneous dual fluorescent imaging.
Appropriate controls and filter sets were used to obtain confocal micrographs using a Sarastro 2000 CLSM (Li et al., 1996). In the case of the Leica TCS-NT CLSM, laser power was tuned to minimize emission spectra cross-talk. Fields of adherent cells were brought into focus using a 60 × /1.4 numeric aperture PlanApo objective under bright-field conditions and briefly examined. A plane of focus 0.3–0.5 μm above the glass surface was selected, and optical sections were then recorded under fluorescent confocal microscopic conditions to reveal the distribution of Mayven and phalloidin-labeled actin. Image pairs were subjected to a two-dimensional (2-D) median filter to reduce background noise, and then examined as color composite images, with Mayven appearing red and phalloidin-labeled actin as green. Cell structures expressing both Mayven and phalloidin-labeled actin appeared yellow-orange.
Image Analysis.
To demonstrate the colocalization of Mayven with phalloidin-labeled actin, micrograph pairs were subjected to a quantitative image analysis procedure (Sugita et al., 1996; Liu et al., 1997) in which 2-D pixel intensity histograms from pairs of images containing Mayven or phalloidin-labeled actin were compared (Image-Space software, Molecular Dynamics). Image analysis was performed on pairs of images to determine the area occupied by Mayven, phalloidin-labeled actin, and those structures containing both Mayven and phalloidin-labeled actin. The pixel dimensions in all micrographs were 0.17 μm, and pixel intensities ranged from 0 to 255 intensity units.
The pixel intensity range corresponding to the cell cytoplasm was determined separately for each image in the pair. Pixel intensities corresponding to the cell ranged from 12 to 255 intensity units. The noncellular background composed of surrounding media and slide surface was determined to correspond to 0–11 pixel intensity units. Regions of the cell containing fluorescent-labeled Mayven were observed to have corresponding pixel intensities from 100 to 255, whereas pixel intensities for phalloidin-labeled actin ranged from 80 to 255 U. To compare the fluorochrome expression recorded in the image pair, pixels corresponding to Mayven or phalloidin-labeled actin were applied to a 2-D histogram of pixel intensities with Mayven (x-axis) plotted against phalloidin-labeled actin (y-axis) and mapped as binary images on a pixel-by-pixel basis. Pixels unique to either Mayven or actin were identified on the 2-D histogram and converted into binary sections. Binary sections were applied as a mask over the original image to produce area measurements on a per cell basis (Sugita et al., 1996; Liu et al., 1997). This procedure was repeated and used to measure the area occupied by Mayven, phalloidin-labeled actin, and those structures expressing both fluorescent probes.
Construction of GFP Epitope-tagged Mayven Expression Vectors
The full-length, N-terminal (aa 1–306) and C-terminal Mayven (aa 310–687) cDNAs were subcloned in the pGFP-C2 expression vector (CLONTECH), which contained the GFP epitope. The sequence and orientation were confirmed by sequencing both strands.
Transfection of Mayven
293T cells were grown in six-well plates with MEM or DMEM containing 10% FBS. Lipofectamine (Life Technologies, Gaithersburg, MD) was used for the transient transfections according to the protocol provided by the manufacturer, using GFP-Mayven constructs or the pGFP-C2 vector alone.
Immunoprecipitation, Immunoblotting, and Far Western Blotting
The cells were lysed in modified RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, leupeptin, and pepstatin, and 1 mM Na3VO4). Total cell lysates were clarified by centrifugation at 10,000 × g for 10 min, and protein concentrations were determined. Identical amounts of protein from each sample were precleared by incubation with protein G–Sepharose CL-4B (Sigma Chemical) for 10 min at 4°C. After the removal of protein G–Sepharose by brief centrifugation, the solution was incubated with Mayven antibodies overnight at 4°C. Immunoprecipitation of the antigen–antibody complex was accomplished by incubation for 1 h at 4°C with 30 μl of protein G–Sepharose. Bound proteins were solubilized in 20 μl of 2× Laemmli buffer. Samples were separated and analyzed by 7.5% or 10% SDS-PAGE, and then transferred to immobilon membranes. The membranes were blocked with 5% BSA (Boehringer Mannheim, Indianapolis, IN) and probed with primary antibody for 1 h at room temperature. Immunoreactive bands were visualized using horseradish peroxidase-conjugated secondary antibody and ECL reagents (Amersham Pharmacia, Piscataway, NJ).
Far Western Blot Analysis
For binding experiments, total cell lysates were immunoprecipitated with control antibody or Mayven antibodies. The immunoprecipitates were analyzed on 8% SDS-PAGE and transferred to membranes. The blots were incubated at 4°C overnight in 5% milk in PBS containing 0.1% Tween 20, followed by incubation with 10 μg of actin (Sigma Chemical) or purified Csk homologous kinase (CHK)-SH2 kinase (Price et al., 1997) as a control for 2 h at 4°C. The blots were washed, and rabbit anti-actin antibodies or rabbit anti-CHK antibodies were added for 1 h. After washing, horseradish peroxidase-conjugated anti-rabbit IgG (Amersham Pharmacia) was added for 1 h. Immunoreactive bands were visualized using ECL reagents.
RESULTS
Identification and Cloning of Mayven
In searching for proteins of potential importance in brain development, we have previously cloned a novel nuclear kelch-related gene, named NRP/B, predominantly expressed in brain (Kim et al., 1998). We were interested in NRP/B-related genes and have identified, under conditions of low-stringency hybridization, a human Mayven cDNA clone. In parallel, using an available public data bank, we have searched for an EST that is homologous to the NRP/B sequence. We have identified an EST sequence (GenBank accession number Z30075) with 50% similarity to NRP/B and have cloned this partial cDNA using PCR techniques. A probe corresponding to this EST was amplified and was used to screen the human hippocampus cDNA library. A full-length cDNA (2970 bp) was obtained from overlapping cDNA clones. A translation stop site is present in frame, 18 nucleotides from the 5′ of the putative initiator methionine (Figure 1). The 110 bp of 5′-untranslated region are rich in GC sequences, which is consistent with Kozak’s specifications for the translation initiation region (Kozak 1987, 1989). The 1779 bp of the predicted ORF encode a protein of 593 amino acids, with a predicted molecular mass of 66 kDa. The 3′-untranslated region of 1085 bp includes the polyadenylation signal (AATAAA), six nucleotides from the end of the clone.
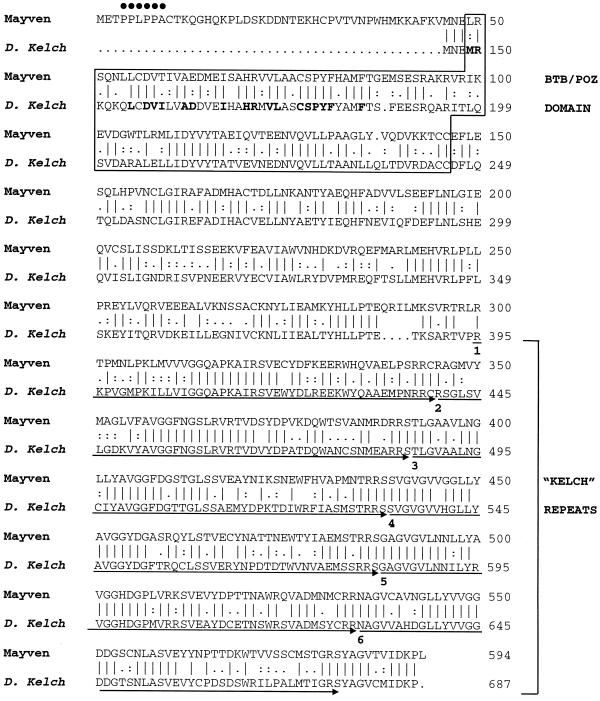
Figure 1.
Alignment of amino acid sequences of human Mayven and Drosophila kelch proteins. The PPXPPX sequence at the N terminus of Mayven, which is a putative ligand for SH3 domains, is marked with dots. The alignment starts at aa 46 of Mayven and aa 146 of kelch (based on the numbering of kelch by Xue and Cooley (1993). The BTB/POZ domain is boxed. Boldface type marks the residues conserved between Mayven and the bric-a-brac transcription factor. These residues of bric-a-brac are sufficient and necessary for its dimerization (Chen et al., 1995). The kelch repeats are indicated by arrows, and the beginning of each repeat is numbered.
Computer-assisted data bank searches revealed that Mayven shares 63% identity and 77% similarity with the Drosophila kelch protein and significant homologies with several other molecules with kelch-related sequences. Alignment between Mayven and kelch (shown in Figure 1) begins at aa 46 of Mayven and at aa 146 of Drosophila kelch, as the N termini of both of these proteins appear unique to them. Residues 3–8 possess a consensus sequence for the SH3 domain ligands, PPXPPX, where X is an aliphatic residue and P is a proline one. A secondary-structure-prediction algorithm predicts that the entire unique N-terminal region (aa 1–45) is likely to form a hydrophilic and highly flexible loop region (our unpublished data). Thus, Mayven may possibly interact via this sequence with other signaling molecules or other cytoskeletal proteins, which possess an SH3 domain. The remaining N-terminal portion of Mayven possesses a BTB/POZ domain predicted to be primarily α-helical (our unpublished data), which is consistent with the predicted structures of other BTB/POZ-containing proteins (Bardwell and Treisman, 1994). The C-terminal domain of Mayven encodes six tandem kelch repeats. It is predicted to form several hydrophobic and presumably buried β-strands, which are likely to form a superbarrel structure, based on the analysis of Bork and Doolittle (1994) of kelch repeats of the Drosophila kelch molecule.
Mapping of Mouse Mayven to Chromosome 8
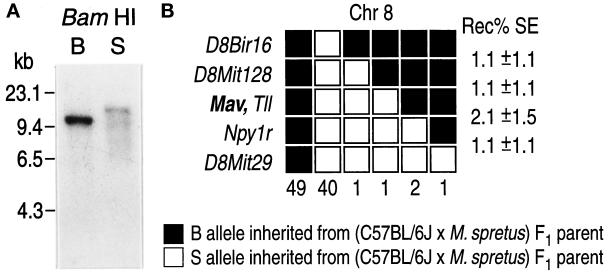
A human 32P-labeled PCR fragment of Mayven cDNA was used as a probe for BamHI RFLP analysis on genomic mouse DNA. It identified a fragment of 11 kilobases (kb) in C57BL/6J and of 14 kb in M. spretus, respectively (Figure 2A). There was no indication of cross-hybridization to other genes that are structurally related to Mayven. Allelic characterization was performed in 96 DNA samples from the (C57BL/6JEi × SPRET/Ei)F1 × SPRET/Ei type backcross panel (Rowe et al., 1994). Haplotype analysis indicated the mapping of the murine Mayven locus to the central region of chromosome 8 (Figure 2B). Perfect cosegregation with the mammalian tolloid-like protein (TLL) was observed. No recombinants were found between Mayven and TLL, indicating a distance of (cM) between the two loci.
Figure 2.
(A) Mapping of the mouse Mayven gene to chromosome 8. Southern blot analysis of Mayven. C57BL/6J (B) and M. spretus (S) genomic DNAs were digested with BamHI. Probing with a [32P] Mayven-specific probe of human Mayven cDNA revealed an RFLP with bands of ∼11 and ∼14 kb for C57BL/6J and M. spretus, respectively. (B) Haplotype analysis of chromosome 8 genetic markers in (C57BL/6JEi × SPRET/Ei)F1 × SPRET/Ei type backcross mice (BSS type) shows linkage and the relative position of the Mayven gene. Black boxes indicate the inheritance of the C57BL/6JEi (B) allele, and the open boxes indicate the inheritance of the SPRET/Ei (S) allele from the (C57BL/6JEi × SPRET/Ei)F1 parent. The first two columns indicate the number of backcross progeny with no recombinants. The following columns show the recombinational events between adjacent loci (signified by the change from open to closed box). The number of recombinants are listed below each column, and the frequency of recombination (Rec %) between adjacent loci with the SE (SE) is indicated. Mapping data from this article have been deposited with the Mouse Genome Database under Accession No. MGD J:47396. Gene names and references to these loci can be found in the MGD (Mammalian Genome Database) on the World Wide Web at http://www.jax.org/recourses/documents/cmdata.
Mapping of the Human Mayven Gene to Chromosome 4
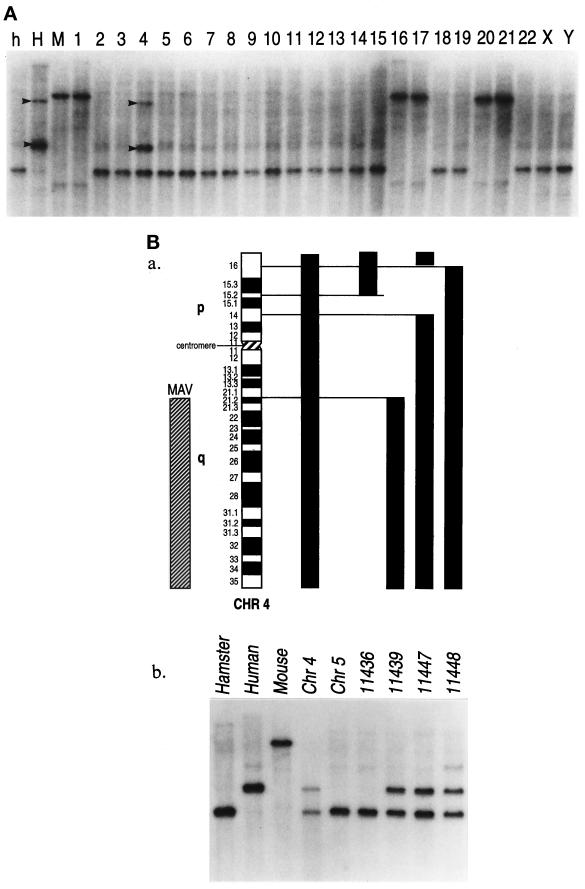
A somatic cell hybrid panel consisting of hamster, human, and mouse DNAs was digested with BamHI, HindIII, and PstI to identify a specific RFLP for the Mayven gene. Probing a Southern blot with a human Mayven-derived PCR probe revealed two specific bands in the human genomic DNA, and the somatic hybrid cell line 4, which contains human chromosome 4. All other hybrid cell lines were negative for the human-specific HindIII RFLP (Figure 3A). To further localize the Mayven locus on chromosome 4, several somatic cell lines representing fragments of human chromosomes 4 and 5 were analyzed. The human chromosome 5 somatic-cell-hybrid line was also analyzed since 11,436 and 11,439 chromosome 4 derivative cell lines carry fragments of both chromosomes 4 and 5. The DNAs were again digested with HindIII, and a Southern blot was hybridized with the 32P-labeled Mayven probe. The human chromosome 4-specific components are indicated in Figure 3B. No specific signal was detected in the hybrid cell line representing human chromosome 5, whereas the presence of the human-specific bands found in the chromosome 4 derivatives mapped the human Mayven to 4q21.2–4qter.
Figure 3.
Mapping of Mayven to human chromosome 4. (A) Genomic DNAs from hamster (h), human (H), and mouse (M) as well as 24 human-rodent somatic cell hybrids (lanes 1–22, X and Y) were digested with HindIII and probed with a human Mayven cDNA. The human-specific RFLP bands are indicated by arrowheads and are seen in the control lane, and in lane 4, which corresponds to human chromosome 4. (B) Mapping of Mayven to 4q21.2-qter. (a) Chromosome 4 and the somatic cell hybrids are indicated. The chromosome 4 content of each hybrid is shown as a solid bar. Mayven localization is indicated on the left. The human–mouse cell hybrid 11687 contains the entire human chromosome 4. (b) Autoradiography showing the Mayven human-specific band within the mapping panel. The cell lines from which the DNAs were extracted are shown above each lane.
Tissue and Cell Line Distribution of Mayven Expression
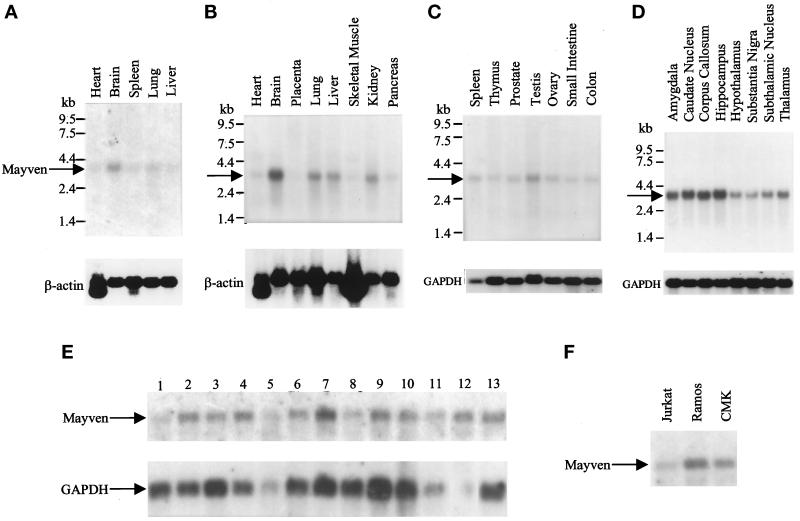
The expression of Mayven mRNA in various human fetal and adult tissues was determined by Northern blot analysis using a Mayven 3′-gene–specific probe. Mayven mRNA expression (3.4 kb) was found at different levels in the majority of human adult and fetal tissues examined (Figure 4). When normalized to either actin or GAPDH, the highest levels of expression were seen in adult and fetal brain (Figure 4, A and B, respectively). The adult brain, particularly the regions of amygdala, caudate nucleus, corpus callosum, and hippocampus, had the highest expression levels (Figure 4D). Lower mRNA levels were detected in the following tissues: lung, liver, kidney, and testis (Figure 4). Mayven expression was also detected at varying levels in transformed neuronal, glioblastoma, and astrocytoma cell lines (Figure 4E) and in several cell lines of hematopoietic lineage (such as Jurkat, Ramos, and CMK) (Figure 4F).
Figure 4.
Mayven mRNA expression in human tissues and cell lines. These blots were hybridized with a radiolabeled Mayven-specific probe and with control mRNA probes (β-actin or GAPDH). (A) Fetal tissue mRNAs. (B and C) Adult tissue mRNAs. (D) Adult brain regions. Blots A-C were obtained from CLONTECH. (E) mRNAs from cell lines. Cell lines were purchased from ATCC and mRNAs were prepared according to standard protocols. Lane 1, neuroglioma cells (H4); lanes 2–4, neuroblastoma cells (IMR32, SKN-MC, SKN-SH); lanes 5–10, glioblastoma cells (A-172, T-98G, U87-MG, U138-MG, U373-MG); lanes 11–12, astrocytoma cells (CCF-STG1 and SW 1983); lane 13, osteosarcoma cells (143B). (F) mRNAs from hematopoietic cell lines (Jurkat T cells, Ramos B cells, and CMK megakaryocytic cells).
Expression of Mayven Protein
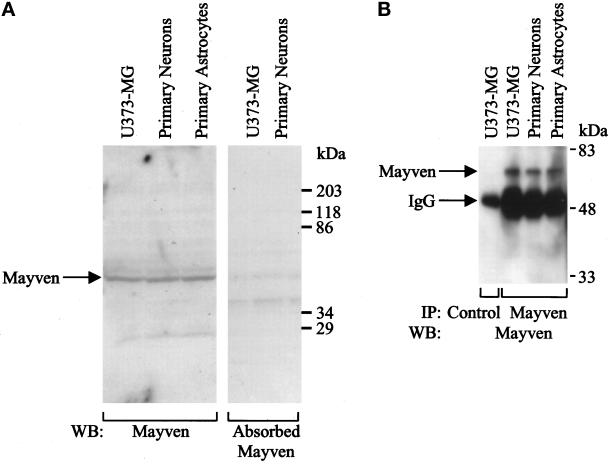
To examine the expression of Mayven protein, polyclonal antisera were produced in the rabbit against the N-terminal portion of Mayven (aa 1–306), which was fused in frame to GST protein in a pGEX 4T-2 vector (Pharmacia). Total cell lysates were prepared from confluent U373-MG glioblastoma cells, rat primary astrocytes, and primary hippocampal neurons. Proteins were resolved by 10% SDS-PAGE and transferred onto polyvinylidene fluoride membrane. Using anti-Mayven antibodies, a major protein of 66 kDa, which corresponds to the predicted molecular mass of Mayven protein, was observed (Figure 5A). The 66-kDa protein appeared to be competed in the presence of GST-Mayven fusion protein (10 μg/ml) (Figure 5A). The expression of Mayven protein in rat primary hippocampal neurons and astrocytes was also evaluated by immunoprecipitation analysis using Mayven polyclonal antibodies. Mayven protein (66 kDa) was expressed in both primary hippocampal neurons and in primary astrocytes (Figure 5, A and B). In addition, Mayven expression was observed in primary hippocampal neurons and astrocytes analyzed by immunofluorescent microscopy (our unpublished data) and confocal analysis (Figure 8).
Figure 5.
Expression of Mayven protein. (A) Total cell lysates were prepared from U373-MG astrocytoma/glioblastoma cells, rat primary hippocampal neurons, and rat primary astrocytes. Each lysate (200 μg) was analyzed by 8% SDS-PAGE and blotted with Mayven antibodies or with absorbed anti-Mayven antiserum (1:1000 dilution). The reactive proteins were detected using the ECL system (Amersham Pharmacia). (B) Total cell lysates (200 μg each) were immunoprecipitated with control antibody or with anti-Mayven antibodies and analyzed by 8% SDS-PAGE. The protein gel was blotted with Mayven antibodies (1:1000 dilution), and the reactive proteins were detected using the ECL system.
Figure 8.
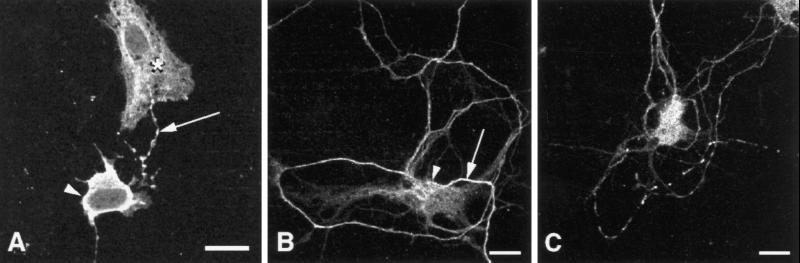
Confocal micrographs of cultured rat primary hippocampal neurons immunolabeled with Mayven (Texas Red secondary). (A) Mayven localizes to the cell body (arrowhead) and neuronal processes (arrow) in neurons cultured for 4 d. Cytoplasmic expression of Mayven was observed in primary astrocytes (*). Bar, 20 μm. (B) Confocal micrograph of cultured primary neuron immunolabeled with Mayven and stained with Texas Red-conjugated secondary antibody reveals Mayven distribution in the cell body, axon hillock (arrowhead), and neuronal processes (arrow). Bar, 10 μm. (C) Depolarized neuron 5 s after stimulation with 40 mM KCl reveals punctate Mayven redistribution along neuronal processes. Bar, 10 μm.
Recombinant BTB/POZ Domain of Mayven Dimerizes in Solution
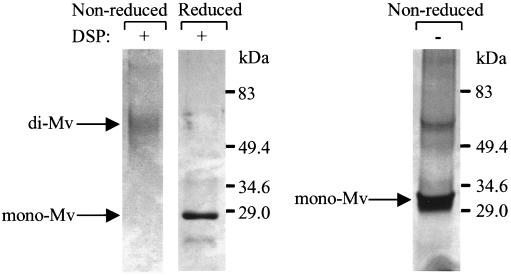
It has been established that the BTB/POZ domain of several proteins forms homodimers (Dhordain et al., 1995). Other BTB/POZ-containing transcription factors can form both homodimers and heterodimers, which have been demonstrated to regulate the transcriptional activity of zinc-finger proteins (Bardwell and Treisman, 1994). To determine whether Mayven can form multimers in solution, the Mayven protein was cleaved away from the GST sequence by thrombin and then cross-linked by a bifunctional protein cross-linking agent DSP. DSP forms stable linkages between free amine groups of interacting proteins, and these linkages are cleavable with S-S reducing reagents. We observed dimerization of Mayven in the nonreduced gel as compared with the reduced gel of the cross-linked Mayven and the non-cross-linked Mayven run under nonreducing conditions (Figure 6). Thus, the N-Mayven protein containing the BTB/POZ domain dimerized in solution.
Figure 6.
Dimer formation between BTB/POZ domains of Mayven revealed by protein cross-linking. GST sequence was proteolytically removed from recombinant Mayven-GST, and then recombinant Mayven was incubated in the presence (lanes 1 and 2) or absence (lane 3) of DSP. After quenching the residual DSP with Tris-HCl, pH 7.4, samples were run on reducing 8% SDS-PAGE (lane 2) or nonreducing 8% SDS-PAGE (lanes 1 and 3) and visualized by staining with Coomassie blue. Arrows indicate the monomeric (mono-Mv) and dimeric Mayven (di-Mv).
Mayven Colocalizes with F-Actin in U373-MG Astrocytoma/Glioblastoma Cells
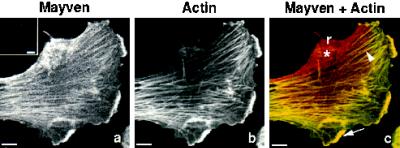
Since several proteins possessing kelch repeats have been shown to be either actin binding or associated with actin filaments, we anticipated that the Mayven protein would colocalize with the actin cytoskeleton. Using specific purified Mayven antibodies, Mayven expression was observed in these cells (Figure 7). Cells immunostained with preimmune serum did not reveal any specific staining patterns (inset, Figure 7a). Fluorescent-tagged phalloidin (FITC), a fungal toxin, was used to visualize filamentous actin (F-actin) in context with Mayven expression in these cells. Mayven (Figure 7a) and actin (Figure 7b) colocalize in fibrillar stress fibers (arrowhead) (Figure 7c) and in cortical patchy actin-rich locations (arrow) at the leading edge of the lamellipodia (Figure 7c). Quantitative 2-D analysis of this cell revealed that 43% of Mayven expression colocalized with fibrillar actin in stress fibers or in actin-rich patchy regions at the ruffled cell margin. Punctate basal-cell-surface expression of Mayven (*) near the retracting margins of the cell (r) coincides with the absence of actin stress fibers in this region. Mayven’s localization pattern in U373-MG cells suggests its involvement in cytoskeletal remodeling since Mayven protein is highly expressed at the retracting end/side of the cell and at the leading edge of the lamellipodia, where dynamic reorganization of actin occurs.
Figure 7.
Mayven colocalization with F-actin. Original confocal micrographs (a–c) of a cultured human U373-MG astrocytoma/glioblastoma cell immunostained for Mayven and actin. (a) Mayven expression only (preimmune serum control, inset a); (b) FITC phalloidin-labeled actin expression only; (c) color composite image resulting from the superimposition of (a) and (b) reveals colocalization of Mayven with fibrillar stress fibers (arrowhead) and the cortical actin-rich region of the ruffled cell margin (arrow). Mayven expression appears to be punctate at the basal cell surface (*). Mayven is highly expressed at the retracting margin of the cell (r). Bar, 10 μm.
We also examined, at varying time points after plating, whether Mayven in U373-MG cells associates with other filament systems, using anti-vimentin antibody as a probe for intermediate filaments, and anti-FAK antibody as a marker for focal adhesions. We observed that Mayven did not colocalize with intermediate filaments but did colocalize with focal adhesion kinase (our unpublished data).
Mayven’s Distribution Pattern Changes in Cultured Primary Neurons after Depolarization
Mayven localizes to the cell body (cytoplasm) of cultured rat primary hippocampal neurons and shows a punctate staining pattern along the processes (arrow) of neurons cultured for 4 d (Figure 8A). In the primary astrocyte marked with * in Figure 8A, Mayven has cytoplasmic expression, similar to that found in U373-MG astrocytoma/glioblastoma cells (Figure 7). In neurons cultured for 1 d, which are beginning to develop neuronal processes, intense and uniform Mayven staining was observed in the cell bodies and along the neuronal processes, mainly in the axons (our unpublished data). Neurons cultured for 4 d were rested for 4 h and were then depolarized with 40 mM KCl. Figure 8B shows that a resting neuron immunolabeled with Mayven has cytoplasmic expression (arrowhead) and intense, fairly uniform, axonal expression (arrow). Five seconds after depolarization of neurons with 40 mM KCl, Mayven distribution was changed from a relatively uniform to a punctate pattern on axons (Figure 8C), suggesting that Mayven is redistributed along neuronal processes in active neurons.
Mayven Associates In Vivo with Actin and Colocalizes with the Actin Cytoskeleton
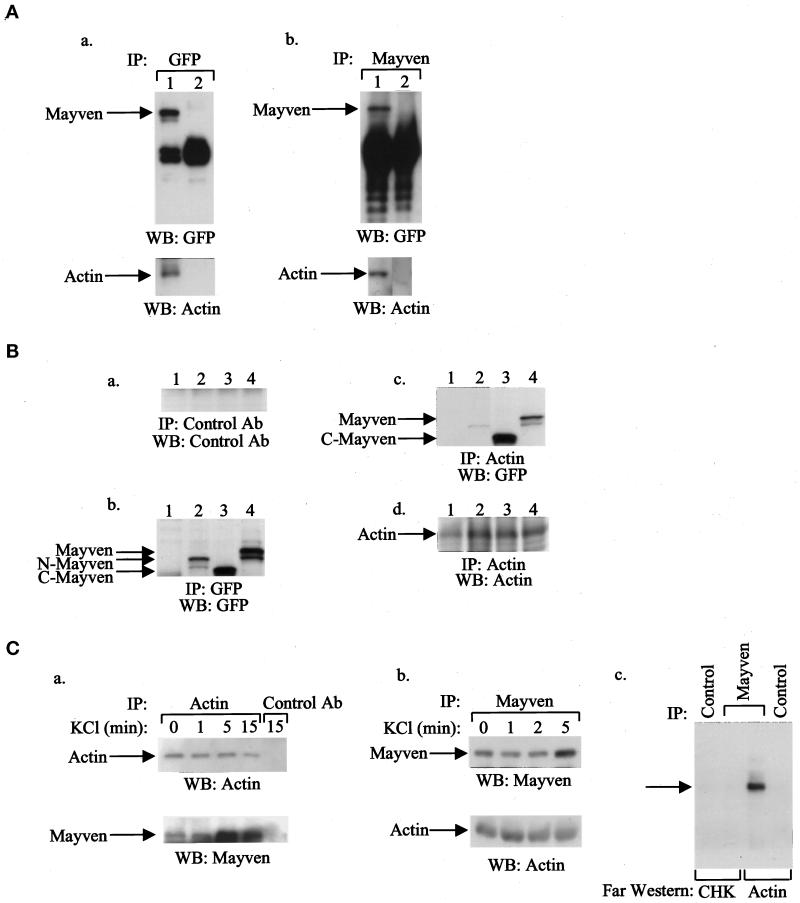
To study the potential association of Mayven with actin, we transfected 293T cells with an expression vector encoding GFP epitope-tagged Mayven cDNA (GFP-tag-Mayven). The GFP-tagged Mayven was detected by immunoprecipitation analyses using either GFP antibodies or Mayven-specific antibodies (Figure 9A). In addition, GFP-tagged Mayven was coimmunoprecipitated with actin, suggesting an in vivo association of Mayven and actin (Figure 9A). The association of Mayven to actin is mediated through the kelch repeats within the C terminus of Mayven (Figure 9B).
Figure 9.
In vivo association of Mayven with actin in transfected 293T cells and primary hippocampal neurons. (A) 293T cells were transfected with a GFP-Mayven cDNA expression vector (lane 1) or GFP vector alone (lane 2). After 48 h, total cell lysates were prepared, immunoprecipitated with GFP antibody (panel a) or with Mayven antibody (panel b), analyzed by 8% SDS-PAGE, and transferred to immobilon membranes. The membranes were blotted with the indicated antibodies (diluted 1:1000), and the reacted proteins were detected using the ECL system. (B) 293T cells were transfected with cDNA expression vectors containing either GFP full-length Mayven, GFP-N-terminus-Mayven (N-Mayven) or GFP-C-terminus-Mayven (C-Mayven) or with pGFP vector alone. After 48 h, total cell lysates were prepared, immunoprecipitated with control antibody (a), GFP antibody (b), or actin antibody (c and d), analyzed by 8% SDS-PAGE, and transferred to immobilon membranes. The membranes were blotted with the indicated antibodies (diluted 1:1000), and the reacted proteins were detected using the ECL system. Lane 1, 293T cells transfected with pGFP vector; lane 2, 293T cells transfected with N-Mayven cDNA; lane 3, 293T cells transfected with C-Mayven cDNA; and lane 4, 293T cells transfected with full-length Mayven cDNA. (C) Primary hippocampal neurons were grown for 7 d, and then KCl depolarization of these cells from 1–15 min was per formed, as indicated. Total cell lysates were prepared and immunoprecipitated with either specific antibodies for actin (a) or Mayven (b) as indicated. The immunoprecipitates were analyzed by 8% SDS-PAGE and blotted with the indicated antibodies (1:1000). The reacted proteins were detected using the ECL system. (c) Far Western blotting analysis: rat primary hippocampal neurons were grown for 7 d, and then KCl depolarization of these cells for 5 min was performed. Total cell lysates were prepared and immunoprecipitated with either specific antibodies for Mayven or control antibodies. The immunoprecipitates were analyzed by 8% SDS-PAGE, and far Western blottings were performed using actin (10 μg) or the GST-CHK-SH2 domain as a control.
The difference in Mayven’s localization pattern in 1- versus 4-d-old cultured primary neurons prompted an examination of this pattern related to the electrical activity of neurons, since neurons maintained in culture exhibit a gradual increase in spontaneous activity (Ling et al., 1990). In an attempt to explore the interactions between Mayven and actin in neuronal cells, the ability of Mayven to bind native actin was tested. Primary hippocampal neurons were treated with KCl for various time points, and coimmunoprecipitation studies were performed. Actin was coimmunoprecipitated with Mayven in depolarized neurons, indicating an association of Mayven with actin upon KCl depolarization (Figure 9C). In addition, in a reciprocal experiment, Mayven was coimmunoprecipitated with actin in primary hippocampal neurons (Figure 9C). Interestingly, the level of Mayven associated with actin was increased during KCl depolarization of primary hippocampal neurons. To further characterize the binding of Mayven with actin, we performed far Western blotting analysis. Total cell lysates were prepared from primary hippocampal neurons untreated or treated with KCl for 5 min and immunoprecipitated with anti-Mayven antibodies or control antibodies. The immunoprecipitates were analyzed by 8% SDS-PAGE and immunoblotted with either purified actin or CHK kinase as a control. In the presence of KCl, binding of Mayven to actin was observed (Figure 9C). In contrast, the CHK protein, as a negative control, did not bind actin. Taken together, these results indicate that Mayven binds to actin in neurons. Furthermore, Mayven’s increased association with actin upon KCl depolarization of primary hippocampal neurons suggests that Mayven is an actin-binding protein that might play a role in the dynamics of cytoskeletal processes in neurons.
DISCUSSION
In this study, we have characterized a novel actin-binding protein, termed Mayven. Mayven is predominantly expressed in human brain and is up-regulated during development. Mayven is colocalized with actin in the cytoskeleton-remodeling regions in stress fibers in U373-MG glioblastoma cells. In addition, Mayven is highly expressed in the cell body and in neurite processes. Depolarization of primary hippocampal neurons with KCl resulted in a rapid change in Mayven distribution from uniform to punctate expression in axons. Furthermore, association of Mayven with actin was observed, suggesting that Mayven may be involved in axonal transport.
The human and the mouse genes for Mayven have been mapped using backcross and somatic cell hybrid panels, respectively. The mouse Mayven gene maps to a region on chromosome 8, which has homology to human chromosome 4 and includes the TLL locus. The mammalian TLL protein was mapped in humans to 4q32–33 and shows strong expression in structures of the developing neonatal and adult brain. Loci for some mouse developmental abnormalities map to the same general chromosomal location as Mayven and TLL (Takahara et al., 1996).
Structural analysis of both the nucleotide sequences and the predicted secondary structure of Mayven revealed three major structural elements: the α-helix BTB domain-like structure; a 45-aa sequence that contains a potential motif (PPXPPX) capable of binding to an SH3 domain-containing ligand in the predicted N terminus; and a β-sheet with six repeats of kelch motif in the predicted C terminus with a striking homology to Drosophila kelch protein (ring canal) of 63% identity (and 77% similarity) on the protein level. BTB/POZ domains are found in actin-binding and in many nuclear DNA-binding proteins such as developmentally regulated transcription factors found in a wide range of metazoans, in regulators of chromatin structure (Dorn et al., 1993; Bardwell and Treisman, 1994; Zollman et al., 1994; Albagli et al., 1995), and in NRP/B, a nuclear matrix protein (Kim et al., 1998). BTB/POZ domains have been proposed to function as protein–protein interaction interfaces that organize higher order structures of the cytoskeleton or chromatin (Bardwell and Treisman, 1994). We have demonstrated that the BTB/POZ domain of Mayven forms multimers in vitro (Figure 6). All residues, shown to be sufficient and necessary for dimerization of bric-a-brac transcription factor (Chen et al., 1995), are conserved between Mayven and bric-a-brac (Figure 1). Therefore, it is likely that Mayven may also multimerize in vivo. Such Mayven homomultimers may enhance actin cross-linking, thus contributing to the stabilization of actin filaments or actin bundling.
Mayven, like kelch protein, has six tandem kelch repeats. The possible function of kelch repeats may be obtained from cloning and ultrastructural studies of scruin, an actin-binding and bundling protein from the acrosomal process of the Limulus sperm (Way et al., 1995). Scruin has two sets of kelch repeats, and its N and C termini are separated with a spacer sequence. Scruin kelch repeats interact with helix-loop-β motifs in subdomain 1 and 3 along a single actin filament (Schmid et al., 1994). Tropomyosin also binds to subdomain 3 of actin (Schmid et al., 1994), and tropomyosin binding appears to protect actin filaments from depolymerization by actin-depolymerizing factor (ADF)/cofilin proteins (Bernstein and Bamburg, 1982). ADF/cofilin, like Mayven, localizes to the leading edges of cells and lamellipodia where rapid actin polymerization takes place.
In addition to the sequences found in the kelch family of proteins, Mayven has a unique domain at its extreme N terminus, and this domain possesses a proline-rich sequence with a potential motif similar to that found in ligands of the src-homology-3 (SH3) domain. SH3 domains are protein modules found in many signal-transducing molecules and in several cytoskeletal proteins. These proteins include the actin-binding proteins α-spectrin, myosin 1b and ABP-1, SLA-1, and BEM-1, molecules isolated from Saccharomyces cerevisiae (Pawson, 1995). In addition, several signaling molecules with SH3 domains associate with actin filaments, although they are not believed to bind to actin directly. Rather, binding is via the proline-rich sequences of actin-binding proteins that function as ligands for SH3 domains. Secondary structure prediction algorithms suggest that this domain of Mayven is highly flexible (our unpublished data) and is thus likely to participate in protein–protein interactions. Thus potentially, Mayven may interact via this sequence with other signaling molecules or other cytoskeletal proteins, which possess SH3 domains.
We have expressed the N-terminal portion of Mayven fused to the GST protein. When eluted at room temperature, this recombinant protein appeared to form large macroaggregates and was thus lost from the soluble pool. Robinson and Cooley (1997) also reported that recombinant kelch at concentrations >0.4 mg/ml tended to form aggregates of very large molecular mass (>500 kDa). Similarly, scruin has been reported to form amorphous aggregates (Way et al., 1995). This self-aggregation property appears to be intrinsic to the kelch family proteins.
Mayven mRNA is predominantly expressed in brain tissue (Figure 4, A, B, and C). The distribution of Mayven mRNA in brain regions showed high levels in the amygdala, caudate nucleus, hippocampus, and corpus callosum (Figure 4D). In addition to Mayven, two other kelch-related proteins, NRP/B and Enc-1, are highly expressed in the brain. NRP/B is a neuronal-specific nuclear matrix protein involved in neuronal differentiation (Kim et al., 1998). Enc-1 has been shown to colocalize with the actin cytoskeleton and to immunoprecipitate with actin (Hernandez et al., 1997).
Immunofluorescent staining and confocal microscopy were used to localize endogenous Mayven protein in situ. The Mayven localization pattern in U373-MG astrocytoma/glioblastoma cells suggests its involvement in cytoskeletal remodeling, since Mayven was highly localized to the retracting end of the cell, and to the leading edge of the lamellipodia (Figure 7), where dynamic reorganization of actin takes place. This actin reorganization includes local actin polymerization and bundling and rapid actin depolymerization and retrograde flow (Lauffenburger and Horwitz, 1996). The leading edges of the lamellipodia are especially rich in actin and actin filament-binding proteins. Other actin filament-binding proteins include fimbrin/α-actinin/filamin, villin, fascin, and scruin (Matsudaira, 1994). It is likely, based on its localization and shared kelch repeats with scruin, that Mayven is a member of the actin filament-binding proteins.
Mayven is also prominent in patchy actin-rich locations at the leading cell edge (Figure 7). This region consists of submembrane complexes of actin and adaptor proteins linked to integrin- or cadherin-type cell receptors (Yamada and Geiger, 1997). Mayven is also localized with the focal adhesion kinase (our unpublished data), thus suggesting localization of Mayven in focal adhesion plaques. Based on these results, Mayven might play either a structural or signal-transducing role. Mayven also appears to colocalize with actin in patterns similar to plectin, a known actin-binding protein, in actin stress fibers and focal adhesion contacts (Seifert et al., 1992).
In primary hippocampal neurons, resting neurons (Figure 8) had a uniform distribution of Mayven along axons, while depolarization of primary neurons with KCl induced changes in Mayven distribution from a uniform to punctate staining pattern (Figure 8). This suggests movement of Mayven along axons and dendrites, where various types of organelles, cytoskeletal proteins (Terada et al., 1996), and mRNAs (Davis et al., 1990) are transported bidirectionally. Both actin and tubulin filament systems are colinear and closely associated in neurons (Fath and Lasek, 1988). The high level of expression of Mayven in adult brain and its localization pattern in cultured primary neurons and astrocytes suggest that Mayven might play a role in the development and function of the nervous system. Cotransfection of 293T cells with GFP-tag-Mayven demonstrated that Mayven was coimmunoprecipitated with actin by using specific antibodies for GFP or Mayven. In addition, in a reciprocal experiment, actin was coimmunoprecipitated with Mayven using specific antibodies for actin (Figure 9). In transfection studies with 293T cells, we were able to show that the association of Mayven with actin was mediated by the kelch repeats in the C terminus of Mayven (Figure 9B). Coimmunoprecipitation of actin and Mayven from day 1 hippocampal neurons showed constitutive binding of Mayven and actin and, upon depolarization of these cells with KCl, the enhanced association of Mayven protein with actin after 5 min (Figure 9C). Furthermore, far Western blotting revealed binding of Mayven with actin (Figure 9C). Taken together, the confocal data along with biochemical analyses, transfection experiments, and far Western analysis indicated a dynamic association and interaction of actin with Mayven in neuronal cells.
Thus, our studies demonstrate that Mayven is an evolutionarily conserved actin-binding protein predominantly expressed in brain and is involved in axonal transport in cultured primary neurons.
ACKNOWLEDGMENTS
We thank Dr. Bijia Deng for help in establishing rat primary hippocampal cultures; Daniel J. Price, Roanna London, and Yigong Fu for their technical help; and Jean Lai for expert microscopy and image analysis. We thank Dr. Sheila Zrihan-Licht for her critical reading of the manuscript. We thank Janet Delahanty for her help in editing the manuscript, Nancy DesRosiers for preparation of the figures, and Peter Park and Tee Trac for their typing assistance. The authors thank Dr. Jerome E. Groopman for his overall support of the project. Sequence data for this article have been deposited with the GenBank/EMBL data libraries under accession No. AF059569, and chromosomal mapping data have been deposited in the mouse genome database under No. MGD J: 47396. This work was supported in part by National Institutes of Health grants R01 51456, R01 55445, CA-76226, P01 HL-43510, and P01 HL-33009. This paper is dedicated to Martin and Anne Peretz and family for their friendship and support for our research program, and to the memories of Dr. Dananagoud Hiregowdara and Diana Michaelis.
Abbreviations used:
- ABPs
actin-binding proteins
- ADF
actin-depolymerizing factor
- BTB domain
broad-complex, tramtrack, bric-a-brac
- CLSM
confocal laser scanning microscope
- CSK
Csk homologous kinase
- DSP
dithiobis succinimidyl propionate
- EST
expressed sequence tag
- F-actin
filamentous actin
- GFP
green fluorescent protein
- GFP-tag-Mayven
GFP epitope-tagged Mayven
- GST-N-Mayven
GST-N-terminal Mayven
- MGD
mouse genome database
- NRP/B
nuclear matrix protein expressed in brain
- ORF
open reading frame
- POZ
domain, poxvirus, zinc finger
- RFLP
restriction fragment length polymorphism
- SH3
src-homology-3
- TLL
tolloid-like protein
REFERENCES
- Agutter PS. Role of the cytoskeleton in nucleocytoplasmic RNA and protein distributions. Biochem Soc Trans. 1991;19:1094–1098. doi: 10.1042/bst0191094. [DOI] [PubMed] [Google Scholar]
- Albagli O, Dhordain. P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth & Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- Altschul SF, Lipman DJ. Protein database searches for multiple alignments. Proc Natl Acad Sci USA. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes & Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- Ben-Ze’ev A. Animal cell shape changes and gene expression. Bioessays. 1991;13:207–212. doi: 10.1002/bies.950130502. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- Bork P, Doolittle RF. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Chang-Yeh A, Mold DE, Huang RC. Identification of a novel murine IAP-promoted placenta-expressed gene. Nucleic Acids Res. 1991;19:3667–3672. doi: 10.1093/nar/19.13.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Zollman, S., Couderc, J.L., and Laski, F.A. (1995). The BTB domain of bric a brac mediates dimerization in vitro [retracted by Chen W, Zollman S, Couderc JL, Laski FA. In: Mol. Cell. Biol. 1997 Nov;17(11):6772]. Mol. Cell. Biol. 15, 3424–3429.
- Cooley L, Theurkauf WE. Cytoskeletal functions during Drosophila oogenesis. Science. 1994;266:590–596. doi: 10.1126/science.7939713. [DOI] [PubMed] [Google Scholar]
- Davis L, Burger B, Banker GA, Steward O. Dendritic transport: quantitative analysis of the time course of somatodendritic transport of recently synthesized RNA. J Neurosci. 1990;10:3056–3068. doi: 10.1523/JNEUROSCI.10-09-03056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhordain P, Albagli O, Ansieau S, Koken MH, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert JP, Leprince D. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene. 1995;11:2689–2697. [PubMed] [Google Scholar]
- Dorn R, Krauss V, Reuter G, Saumweber H. The enhancer of position-effect variegation of Drosophila, E(var)3–93D, codes for a chromatin protein containing a conserved domain common to several transcriptional regulators. Proc Natl Acad Sci USA. 1993;90:11376–11380. doi: 10.1073/pnas.90.23.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L, Bomblies L, Vandekerckhove J, Schleicher M, Gettemans J. A novel type of protein kinase phosphorylates actin in the actin-fragmin complex. EMBO J. 1996;15:5547–5556. [PMC free article] [PubMed] [Google Scholar]
- Fath KR, Lasek RJ. Two classes of actin microfilaments are associated with the inner cytoskeleton of axons. J Cell Biol. 1988;107:613–621. doi: 10.1083/jcb.107.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt D, Couderc JL, Cramton SE, Laski FA. Pattern formation in the limbs of Drosophila: bric a brac is expressed in both a gradient and a wave-like pattern and is required for specification and proper segmentation of the tarsus. Development. 1993;119:799–812. doi: 10.1242/dev.119.3.799. [DOI] [PubMed] [Google Scholar]
- Hernandez MC, Andres-Barquin PJ, Martinez S, Bulfone A, Rubenstein JL, Israel MA. ENC-1: a novel mammalian kelch-related gene specifically expressed in the nervous system encodes an actin-binding protein. J Neurosci. 1997;17:3038–3051. doi: 10.1523/JNEUROSCI.17-09-03038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA, Chaponnier C. Medical aspects of the actin cytoskeleton. Curr Opin Cell Biol. 1995;7:111–117. doi: 10.1016/0955-0674(95)80052-2. [DOI] [PubMed] [Google Scholar]
- Kim T-A, Lim J, Ota S, Raja S, Rogers R, Rivnay B, Avraham H, Avraham S. NRP/B, a novel nuclear matrix protein, associates with p110RB and is involved in neuronal differentiation. J Cell Biol. 1998;141:553–566. doi: 10.1083/jcb.141.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles BA, Cooley L. The specialized cytoskeleton of the Drosophila egg chamber. Trends Genet. 1994;10:235–241. doi: 10.1016/0168-9525(94)90170-8. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Senkevich TG, Chernos VI. A family of DNA virus genes that consists of fused portions of unrelated cellular genes. Trends Biochem Sci. 1992;17:213–214. doi: 10.1016/0968-0004(92)90379-n. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eukaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Li J, Avraham H, Rogers RA, Raja S, Avraham S. Characterization of RAFTK, a novel focal adhesion kinase, and its integrin-dependent phosphorylation and activation in megakaryocytes. Blood. 1996;88:417–428. [PubMed] [Google Scholar]
- Ling DS, Petroski RE, Chou W, Geller HM. Development of spontaneous electrical activity by rat hypothalamic neurons in dissociated culture. Brain Res Dev Brain Res. 1990;53:276–282. doi: 10.1016/0165-3806(90)90018-t. [DOI] [PubMed] [Google Scholar]
- Liu J, Oh P, Horner T, Rogers RA, Schnitzer JE. Organized cell surface signal transduction in caveolae distinct from glycosyl-phosphatidylinositol-anchored protein microdomains. J Biol Chem. 1997;272:7211–7222. doi: 10.1074/jbc.272.11.7211. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Actin cross-linking proteins at the leading edge. Semin Cell Biol. 1994;5:165–174. doi: 10.1006/scel.1994.1021. [DOI] [PubMed] [Google Scholar]
- Pawson T. Protein modules and signaling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Actin and actin binding proteins. In: Kreis T, Vale R, editors. Guidebook to the Cytoskeletal and Motor Proteins. Oxford, UK: Oxford University Press; 1993. [Google Scholar]
- Price DJ, Rivnay B, Fu Y, Jiang S, Avraham S, Avraham H. Direct association of Csk homologous kinase (CHK) with the diphosphorylated site Tyr568/570 of the activated c-KIT in megakaryocytes. J Biol Chem. 1997;272:5915–5920. doi: 10.1074/jbc.272.9.5915. [DOI] [PubMed] [Google Scholar]
- Rao KM, Cohen HJ. Actin cytoskeletal network in aging and cancer. Mutat Res. 1991;256:139–148. doi: 10.1016/0921-8734(91)90007-x. [DOI] [PubMed] [Google Scholar]
- Riederer BM. Some aspects of the neuronal cytoskeleton in development. Eur J Morphol. 1990;28:347–378. [PubMed] [Google Scholar]
- Robinson DN, Cooley L. Drosophila kelch is an oligomeric ring canal actin organizer. J Cell Biol. 1997;138:799–810. doi: 10.1083/jcb.138.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosette C, Karin M. Cytoskeletal control of gene expression: depolymerization of microtubules activates NF-kappa B. J Cell Biol. 1995;128:1111–1119. doi: 10.1083/jcb.128.6.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe LB, Nadeau JH, Turner R, Frankel WN, Letts VA, Eppig JT, Ko MS, Thurston SJ, Birkenmeier EH. Maps from two interspecific backcross panels available as a community genetic mapping resource. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmid MF, Agris JM, Jakana J, Matsudaira P, Chiu W. Three-dimensional structure of a single filament in the Limulus acrosomal bundle: scruin binds to homologous helix-loop-beta motifs in actin. J Cell Biol. 1994;124:341–350. doi: 10.1083/jcb.124.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Lawson D, Wiche G. Immunolocalization of the intermediate filament-associated protein plectin at focal contacts and actin stress fibers. Eur J Cell Biol. 1992;59:138–147. [PubMed] [Google Scholar]
- Senkevich TG, Muravnik GL, Pozdnyakov SG, Chizhikov VE, Ryazankina OI, Shchelkunov SN, Koonin EV, Chernos VI. Nucleotide sequence of XhoI O fragment of ectromelia virus DNA reveals significant differences from vaccinia virus. Virus Res. 1993;30:73–88. doi: 10.1016/0168-1702(93)90017-h. [DOI] [PubMed] [Google Scholar]
- Sugita M, Jackman RM, van Donselaar E, Behar SM, Rogers RA, Peters PJ, Brenner MB, Porcelli SA. Cytoplasmic tail dependent localization of CD1b antigen-presenting molecules to MIICs. Science. 1996;273:349–352. doi: 10.1126/science.273.5273.349. [DOI] [PubMed] [Google Scholar]
- Takahara K, Brevard R, Hoffman GG, Suzuki N, Greenspan DS. Characterization of a novel gene product (mammalian tolloid-like) with high sequence similarity to mammalian tolloid/bone morphogenetic protein-1. Genomic. 1996;34:157–165. doi: 10.1006/geno.1996.0260. [DOI] [PubMed] [Google Scholar]
- Terada S, Nakata T, Peterson AC, Hirokawa N. Visualization of slow axonal transport in vivo. Science. 1996;273:784–788. doi: 10.1126/science.273.5276.784. [DOI] [PubMed] [Google Scholar]
- Towbin JA. The role of cytoskeletal proteins in cardiomyopathies. Curr Opin Cell Biol. 1998;10:131–139. doi: 10.1016/s0955-0674(98)80096-3. [DOI] [PubMed] [Google Scholar]
- Trifaro JM, Vitale ML. Cytoskeleton dynamics during neurotransmitter release. Trends Neurosci. 1993;16:466–472. doi: 10.1016/0166-2236(93)90079-2. [DOI] [PubMed] [Google Scholar]
- Undrovinas AI, Shander GS, Makielski JC. Cytoskeleton modulates gating of voltage-dependent sodium channel in heart. Am J Physiol. 1995;269:H203–214. doi: 10.1152/ajpheart.1995.269.1.H203. [DOI] [PubMed] [Google Scholar]
- Varkey JP, Muhlrad PJ, Minniti AN, Do B, Ward S. The Caenorhabditis elegans spe-26 gene is necessary to form spermatids and encodes a protein similar to the actin-associated proteins kelch and scruin. Genes & Dev. 1995;9:1074–1086. doi: 10.1101/gad.9.9.1074. [DOI] [PubMed] [Google Scholar]
- von Bulow M, Heid H, Hess H, Franke WW. Molecular nature of calicin, a major basic protein of the mammalian sperm head cytoskeleton. Exp Cell Res. 1995;219:407–413. doi: 10.1006/excr.1995.1246. [DOI] [PubMed] [Google Scholar]
- Way M, Sanders M, Garcia C, Sakai J, Matsudaira P. Sequence and domain organization of scruin, an actin-cross-linking protein in the acrosomal process of Limulus sperm. J Cell Biol. 1995;128:51–60. doi: 10.1083/jcb.128.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue F, Cooley L. Kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- Yamada KM, Geiger B. Molecular interactions in cell adhesion complexes. Curr Opin Cell Biol. 1997;9:76–85. doi: 10.1016/s0955-0674(97)80155-x. [DOI] [PubMed] [Google Scholar]
- Zollman S, Godt D, Prive GG, Couderc JL, Laski FA. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]