Abstract
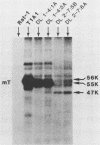
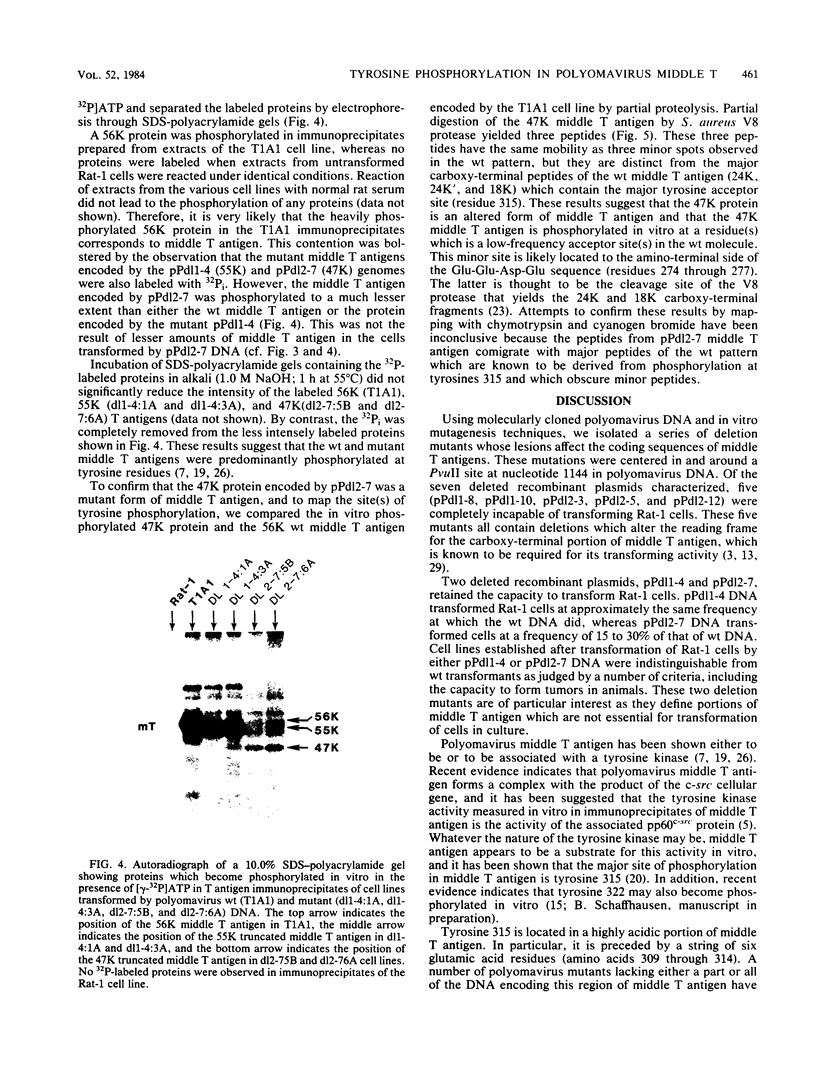
The induction of tumors and cellular transformation mediated by polyomavirus requires the action of middle T antigen. Accordingly, we have begun to define the domains of the viral protein important for these processes to learn more about its site and mechanism of action. One of the domains of middle T antigen which is thought to be important for its function includes a stretch of acidic amino acids and a vicinal tyrosine residue (tyrosine 315), the major site of tyrosine phosphorylation in vitro. To determine whether these acidic amino acids and tyrosine 315 are required to maintain the transforming activity of middle T antigen, we constructed deletions within the DNA sequences encoding these amino acids and measured the capacity of the resulting mutants to transform Rat-1 cells in culture. This was accomplished by using in vitro mutagenesis techniques with molecularly cloned polyomavirus DNA. Seven mutants were isolated. Five of these proved incapable of transforming Rat-1 cells and were found to contain deletions which altered the reading frame for middle T antigen. However, two mutants, pPdl1-4 and pPdl2-7, retained the capacity to transform Rat-1 cells at high frequencies. The middle T antigen encoded by one of these mutants, pPdl1-4, lacks part of the acidic string of amino acids but not tyrosine 315 (amino acids 304 through 310 are deleted), whereas the middle T antigen encoded by the other mutant, pPdl2-7, lacks the entire acidic amino acid stretch as well as tyrosine 315 (amino acids 285 through 323 are deleted). Rat-1 cells transformed by one or the other mutant DNA displayed a fully transformed phenotype, including the capacity to form tumors in animals. These results prove that the major site of tyrosine phosphorylation in middle T antigen and the acidic amino acids which precede it are not essential for its transforming activity.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asselin C., Gelinas C., Bastin M. Role of the three polyoma virus early proteins in tumorigenesis. Mol Cell Biol. 1983 Aug;3(8):1451–1459. doi: 10.1128/mcb.3.8.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. M. Cellular oncogenes and retroviruses. Annu Rev Biochem. 1983;52:301–354. doi: 10.1146/annurev.bi.52.070183.001505. [DOI] [PubMed] [Google Scholar]
- Carmichael G. G., Schaffhausen B. S., Dorsky D. I., Oliver D. B., Benjamin T. L. Carboxy terminus of polyoma middle-sized tumor antigen is required for attachment to membranes, associated protein kinase activities, and cell transformation. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3579–3583. doi: 10.1073/pnas.79.11.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael G., Schaffhausen B. S., Mandel G., Liang T. J., Benjamin T. L. Transformation by polyoma virus is drastically reduced by substitution of phenylalanine for tyrosine at residue 315 of middle-sized tumor antigen. Proc Natl Acad Sci U S A. 1984 Feb;81(3):679–683. doi: 10.1073/pnas.81.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtneidge S. A., Smith A. E. Polyoma virus transforming protein associates with the product of the c-src cellular gene. Nature. 1983 Jun 2;303(5916):435–439. doi: 10.1038/303435a0. [DOI] [PubMed] [Google Scholar]
- Ding D., Dilworth S. M., Griffin B. E. mlt Mutants of polyoma virus. J Virol. 1982 Dec;44(3):1080–1083. doi: 10.1128/jvi.44.3.1080-1083.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart W., Hutchinson M. A., Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979 Dec;18(4):925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- Eckhart W., Hutchinson M. A., Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979 Dec;18(4):925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T., Hutchinson M. A., Eckhart W. Translation of polyoma virus T antigens in vitro. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5917–5921. doi: 10.1073/pnas.75.12.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Brocklehurst J. R., Dulbecco R. Virus-specific proteins in the plasma membrane of cells lytically infected or transformed by pol-oma virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4666–4670. doi: 10.1073/pnas.74.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y. Polyoma virus-specific 55K protein isolated from plasma membrane of productively infected cells is virus-coded and important for cell transformation. Virology. 1979 Oct 15;98(1):261–266. doi: 10.1016/0042-6822(79)90545-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mes A. M., Hassell J. A. Polyoma viral middle T-antigen is required for transformation. J Virol. 1982 May;42(2):621–629. doi: 10.1128/jvi.42.2.621-629.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S. V., Tyndall C., Magnusson G. Deletion mapping of a short polyoma virus middle T antigen segment important for transformation. J Virol. 1983 Apr;46(1):284–287. doi: 10.1128/jvi.46.1.284-287.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra B. A., Harvey R., Ely B. K., Markham A. F., Smith A. E. Transforming activity of polyoma virus middle-T antigen probed by site-directed mutagenesis. Nature. 1983 Aug 4;304(5925):456–459. doi: 10.1038/304456a0. [DOI] [PubMed] [Google Scholar]
- Patschinsky T., Hunter T., Esch F. S., Cooper J. A., Sefton B. M. Analysis of the sequence of amino acids surrounding sites of tyrosine phosphorylation. Proc Natl Acad Sci U S A. 1982 Feb;79(4):973–977. doi: 10.1073/pnas.79.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M., Cowie A., Carr A., Glaichenhaus N., Kamen R., Cuzin F. The roles of individual polyoma virus early proteins in oncogenic transformation. Nature. 1982 Dec 23;300(5894):713–718. doi: 10.1038/300713a0. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M., Naghashfar Z., Cowie A., Carr A., Grisoni M., Kamen R., Cuzin F. Expression of the large T protein of polyoma virus promotes the establishment in culture of "normal" rodent fibroblast cell lines. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4354–4358. doi: 10.1073/pnas.80.14.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B. S., Dorai H., Arakere G., Benjamin T. L. Polyoma virus middle T antigen: relationship to cell membranes and apparent lack of ATP-binding activity. Mol Cell Biol. 1982 Oct;2(10):1187–1198. doi: 10.1128/mcb.2.10.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B. S., Silver J. E., Benjamin T. L. Tumor antigen(s) in cell productively infected by wild-type polyoma virus and mutant NG-18. Proc Natl Acad Sci U S A. 1978 Jan;75(1):79–83. doi: 10.1073/pnas.75.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B., Benjamin T. L. Comparison of phosphorylation of two polyoma virus middle T antigens in vivo and in vitro. J Virol. 1981 Oct;40(1):184–196. doi: 10.1128/jvi.40.1.184-196.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K., Ito Y. Differential subcellular localization of in vivo-phosphorylated and nonphosphorylated middle-sized tumor antigen of polyoma virus and its relationship to middle-sized tumor antigen phosphorylating activity in vitro. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6812–6816. doi: 10.1073/pnas.79.22.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J., Schaffhausen B., Benjamin T. Tumor antigens induced by nontransforming mutants of polyoma virus. Cell. 1978 Oct;15(2):485–496. doi: 10.1016/0092-8674(78)90018-1. [DOI] [PubMed] [Google Scholar]
- Smith A. E., Smith R., Griffin B., Fried M. Protein kinase activity associated with polyoma virus middle T antigen in vitro. Cell. 1979 Dec;18(4):915–924. doi: 10.1016/0092-8674(79)90204-6. [DOI] [PubMed] [Google Scholar]
- Smolar N., Griffin B. E. DNA sequences of polyoma virus early deletion mutants. J Virol. 1981 Jun;38(3):958–967. doi: 10.1128/jvi.38.3.958-967.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Griffin B. E. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979 Jun;17(2):357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- Templeton D., Voronova A., Eckhart W. Construction and expression of a recombinant DNA gene encoding a polyomavirus middle-size tumor antigen with the carboxyl terminus of the vesicular stomatitis virus glycoprotein G. Mol Cell Biol. 1984 Feb;4(2):282–289. doi: 10.1128/mcb.4.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Novak U., Favaloro J., Kamen R. Transformation of rat cells by an altered polyoma virus genome expressing only the middle-T protein. Nature. 1981 Aug 13;292(5824):595–600. doi: 10.1038/292595a0. [DOI] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978 Jul;14(3):725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]