Abstract
ADP-ribosylation factor (ARF) proteins in Saccharomyces cerevisiae are encoded by two genes, ARF1 and ARF2. The addition of the c-myc epitope at the C terminus of Arf1 resulted in a mutant (arf1-myc arf2) that supported vegetative growth and rescued cells from supersensitivity to fluoride, but homozygous diploids failed to sporulate. arf1-myc arf2 mutants completed both meiotic divisions but were unable to form spores. The SPO14 gene encodes a phospholipase D (PLD), whose activity is essential for mediating the formation of the prospore membrane, a prerequisite event for spore formation. Spo14 localized normally to the developing prospore membrane in arf1-myc arf2 mutants; however, the synthesis of the membrane was attenuated. This was not a consequence of reduced PLD catalytic activity, because the enzymatic activity of Spo14 was unaffected in meiotic arf1-myc arf2 mutants. Although potent activators of mammalian PLD1, Arf1 proteins did not influence the catalytic activities of either Spo14 or ScPld2, a second yeast PLD. These results demonstrate that ARF1 is required for sporulation, and the mitotic and meiotic functions of Arf proteins are not mediated by the activation of any known yeast PLD activities. The implications of these results are discussed with respect to current models of Arf signaling.
INTRODUCTION
ADP-ribosylation factors (Arfs) are 21-kDa proteins of the Ras superfamily of GTP-binding proteins. Arf proteins were first identified as activators of cholera toxin-catalyzed ADP-ribosylation of the Gsα subunit of heterotrimeric G-proteins (Kahn and Gilman, 1986). A substantial amount of evidence has accumulated indicating that Arf proteins can regulate the formation of vesicles involved in both secretory and endocytic pathways (Serafini et al., 1991; Ktistakis et al., 1996; Chen et al., 1997; Faundez et al., 1997; West et al., 1997). Recently Arf proteins have been shown to directly activate phosphatidylinositol-4,5-bisphosphate (PIP2)-dependent mammalian phospholipase D (PLD) enzymes (Brown et al., 1993; Cockcroft et al., 1994; Brown et al., 1995; Hammond et al., 1995, 1997; Park et al., 1997). PLDs catalyze the hydrolysis of phosphatidylcholine (PC) to generate phosphatidic acid (PA) and choline. PLD-mediated changes in the lipid composition of membranes are thought to promote the assembly and release of clathrin coated vesicles at the trans-Golgi complex and plasma membrane (Ktistakis et al., 1996; Chen et al., 1997; West et al., 1997).
The earliest evidence that Arfs function in membrane traffic came from studies in Saccharomyces cerevisiae (Stearns et al., 1990a,b). Yeast have two ARF genes, ARF1 and ARF2, which together are required for viability (Stearns et al., 1990a). ARF1 and ARF2 are 96% identical and functional homologues, although ARF1 produces ∼90% of the Arf protein in the cell (Stearns et al., 1990a). Deletion of ARF1 results in a defect in the secretory pathway, as evidenced by an altered glycosylation of secreted proteins (Stearns et al., 1990b). Furthermore, arf1, but not arf2 mutants, grow slowly, are cold sensitive, and are supersensitive to fluoride (Stearns et al., 1990a). The pleiotropic phenotypes associated with arf mutants suggest that Arf proteins function in multiple signaling pathways. Recently, a family of proteins which mediate Arf-dependent mitotic growth have been identified; however, these proteins do not constitute the full array of Arf effectors (Zhang et al., 1998).
S. cerevisiae undergo sporulation when starved of nitrogen in the presence of a nonfermentable source of carbon (reviewed in Kupiec et al., 1997). Sporulation consists of a single round of DNA replication, followed by two meiotic divisions, within a single intact nuclear envelope. The meiotic divisions generate four haploid nuclei that are ultimately packaged into individual spores contained within a single ascus. Spore formation requires the de novo synthesis of a new double layered intracellular membrane, termed the prospore membrane (Byers, 1981). The prospore membrane develops as meiosis II proceeds. Once the nucleus divides at the end of meiosis II, the prospore membrane fuses with itself and in so doing encapsulates each of the four nuclei separately. The inner layer of the prospore membrane becomes the plasma membrane of the spore, whereas the luminal space between the two membranes serves as the site of spore wall synthesis.
The S. cerevisiae SPO14 gene product encodes a PIP2-dependent, PC-specific, PLD (Rose et al., 1995; Ella et al., 1996; Waksman et al., 1996). spo14 diploids are defective in the completion of the meiotic divisions and are unable to form spores (Honigberg et al., 1992; Rose et al., 1995). The assembly of the prospore membrane has been shown to require both localized PLD activity (Rudge et al., 1998) and a sporulation-specific branch of the secretory pathway (Neiman, 1998). Fusion of vesicles derived from the trans-Golgi complex are thought to be responsible for the formation of the prospore membrane (Neiman, 1998).
In this study we sought to investigate the role that Arf and Spo14 proteins play in the formation of the prospore membrane during meiosis and to directly test whether Arf proteins activate yeast PLD.
MATERIALS AND METHODS
Yeast Strains and Media
Routine growth and manipulation of S. cerevisiae strains were performed as described in (Rose et al., 1990). Strains used in this study are shown in Table 1.
Table 1.
Genotypes of yeast strains
| Strain | Genotype | Source |
|---|---|---|
| C163 | MATa arf1-3 arf2-Δ1 his3-Δ200 leu2-3, 112 lys2-801 ura3-52 | This study |
| MATα arf1-3 arf2::LEU2 his3-Δ200 leu2-3, 112 lys2 ura3-52 | ||
| RT321 | MATa arf1-myc his3-Δ200 leu2-3, 112 lys2-801 ura3-52 | This study |
| 511.4B | MATα arf2::LEU2 his3-Δ200 leu2-3, 112 ura3-52 | This study |
| C134 | MATa arf1-myc ARF2 his3-Δ200 leu2-3, 112 lys2-801 ura3-52 | This study |
| MATα ARF1 arf2::LEU2 his3-Δ200 leu2-3, 112 LYS2 ura3-52 | ||
| C134.1B | MATa arf1-myc arf2::LEU2 his3-Δ200 leu2-3, 112 ura3-52 | This study |
| C134.8A | MATα arf1-myc arf2::LEU2 his3-Δ200 leu2-3, 112 lys2-801, ura3-52 | This study |
| C135 | MATa arf1-myc arf2::LEU2 his3-Δ200 leu2-3, 112 LYS2 ura3-52 | This study |
| MATα arf1-myc arf2::LEU2 his3-Δ200 leu2-3, 112 lys2-801, ura3-52 | ||
| C138 | MATa ARF1 arf2::Δ12 ADE2 his3-Δ200 leu2-3, 112 lys2-801 ura3-52 | This study |
| MATα ARF1 arf2::LEU2 ade2-101 his3-Δ200 leu2-3, 112 lys2-801 ura3-52 | ||
| Y963 | C135 + HA-SPO14 CEN4 URA3 | This study |
| Y1172 | C138 + HA-SPO14 CEN4 URA3 | This study |
| Y980 | C135 + GFP-SPO14 2μ URA3 | This study |
| Y979 | C138 + GFP-SPO14 2μ URA3 | This study |
| Y494 | MATa spo14::URA3 leu2 thr1-4 trp1-4 ura3-1 arg4-8 ade2-1 | Rose et al., 1995 |
| MATα spo14::URA3 leu2 thr1-4 trp1-4 ura3-1 arg4-8 ade2-1 | ||
| Y568 | Y494 + HA-SPO14 CEN4 URA3 | Rudge et al., 1998 |
| PSY315 | MATa his3-Δ200 leu2-3, 112 lys2-801 ura3-52 | Kahn et al., 1995 |
| PSY316 | MATα his3-Δ200 leu2-3, 112 lys2-801 ura3-52 ade2-101 | This study |
| TT104 | MATa arf1::HIS3 his3-Δ200 leu2-3, 112 lys2-801 ura3-52 | This study |
| TT139 | MATα arf2::LEU2 his3-Δ200 leu2-3, 112 lys2-801 ura3-52 ade2-101 | This study |
| RT143 | PSY315 + GAL-ARF1 CEN4URA3 | Kahn et al., 1995 |
| RT143 | PSY315 + GAL-arf1Q71L CEN4URA3 | Kahn et al., 1995 |
| RT150 | PSY315 + GAL-arf1N126I CEN4URA3 | Kahn et al., 1995 |
| Y1125 | PSY315 spo14::URA3 | This study |
| Y1126 | PSY316 spo14::URA3 | This study |
| Y1127 | TT104 spo14::URA3 | This study |
| Y1128 | TT139 spo14::URA3 | This study |
| Y1129 | RT143 spo14::LEU2 | This study |
| Y1130 | RT149 spo14::LEU2 | This study |
| Y1131 | RT150 spo14::LEU2 | This study |
The expression of ARF alleles under the control of the GAL promoter on low copy number centromere (CEN) vectors (Table 1), was achieved by growing cells in liquid media containing 2% galactose instead of 2% glucose for 2 h.
Fluoride sensitivity was determined as previously described (Stearns et al., 1990a). YEPD plates containing 40 mM sodium fluoride were used within 5 d, and growth of cells was scored after 2–3 d.
Plasmids
Epitope tagged Arf1 consisted of the addition of a 6–amino acid thrombin recognition sequence (LVPRGS) and the 13–amino acid epitope, derived from c-myc and recognized by mAb 9E10 (SMEQKLISEEDLN; Evan et al., 1985), to the C-terminus of S. cerevisiae ARF1. The cDNA encoding this fusion protein was constructed by use of synthetic oligonucleotide primers that incorporated the indicated changes and unique restriction sites (NdeI and XbaI at the 5′ and 3′ ends, respectively) at each end of the open reading frame. Plasmid JCY1–85 was then constructed by insertion of the mutant arf1-myc into the YIp352-based plasmid, pJCB1–23, at unique NdeI and XbaI sites, which places it between the 5′ and 3′ untranslated regions of ARF1. The arf1-myc gene was then integrated into the genome at the ARF1 locus by gene replacement (Rothstein, 1991). The resultant strain, RT321, was mated with 511.4B to make the diploid, C134. Two segregants of C134 (C134.1B × C134.8A) were crossed to each other to get C135, a diploid that is homozygous for both arf1-myc and deletion of ARF2. Thus, C135 has two chromosomal copies of arf1-myc as its only source of Arf protein.
Spo14 was epitope-tagged with three copies of the hemagglutinin (HA) epitope and the Green Fluorescent Protein (GFP) as previously described by Rudge et al. (1998). The plasmids HA-SPO14 CEN4 URA3 and GFP-SPO14 2μ URA3 fully complemented the meiosis defect of spo14 diploids (Rudge et al., 1998).
Analysis of the Meiotic Divisions
Cells were grown and sporulated as previously described by Engebrecht et al. (1998). Aliquots from duplicate cultures were removed and fixed with 3.7% formaldehyde at the indicated times after transfer to sporulation medium. Fixed cells were then stained with the DNA-specific dye DAPI and examined using fluorescent microscopy (Rose et al., 1995). A minimum of 600 cells was examined for each time point.
Preparation of Nonidet-insoluble and -soluble Cell Fractions
Spheroplasts were prepared from mitotically dividing cells and from cells 15 hours after induction of meiosis as described by Rudge et al. (1998). Fifteen hours after induction of meiosis corresponds to the completion of the meiotic divisions and the initiation of prospore membrane biogenesis in this strain background (Rose et al., 1995). Consequently, we routinely isolated HA-Spo14 from cells at this time during meiosis.
Spheroplasts were suspended in 2 ml ice-cold immunoprecipitation (IP) lysis buffer (10 mM triethanolamine, pH 7.5, 150 mM sodium chloride, 5 mM EDTA, 5 mM EGTA, 50 mM sodium fluoride, 40 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 1 mM dithiothreitol (DTT), 2 mM PMSF, 2 mM benzamidine, 0.057 U/ml aprotinin and 10 μg/ml leupeptin) containing 1% (wt/vol) Nonidet P-40 (BDH Laboratory Supplies, Poole, Dorset, England), and incubated at 4°C for 40 min with gentle agitation. The lysate was cleared by centrifugation at 1000 × g for 6 min at 4°C to remove unlysed cells and large cellular debris. To accommodate for different efficiencies of spheroplasting and detergent lysis (especially between mitotic and meiotic samples), the total protein concentration of each supernatant was determined as described below. When necessary, samples were diluted with IP lysis buffer containing 1% Nonidet P-40. The supernatant was then prepared by centrifugation at 15,000 × g for 30 min at 4°C to yield the Nonidet P-40–insoluble (pellet) and –soluble (supernatant) fractions. The pellet was washed twice with IP lysis buffer and then suspended in a volume of IP lysis buffer (with 1% wt/vol Nonidet P-40) equal to the volume of supernatant collected. Fractions were then mixed with 4× Laemmli buffer (Laemmli, 1970) and boiled for 5 min for immunoblot analysis.
Immunoprecipitation of HA-Spo14
Spheroplasts, prepared from cells 15 h after induction for meiosis (Rudge et al., 1998), were suspended in 6 ml ice-cold IP lysis buffer containing 1% (wt/vol) Nonidet P-40 and lysed as described above. HA-Spo14 was immunoprecipitated directly from the soluble fraction using the 12CA5 mAb (BAbCo, Berkeley, CA), which recognizes the HA epitope. Briefly, 1-ml aliquots of the Nonidet P-40–soluble fraction were incubated for 1.5 h at 4°C in tubes containing 3 μg affinity-purified mAb 12CA5. Protein A-agarose (Life Technologies, Grand Island, NY) was then added (50 μl of a 50% suspension equilibrated in lysis buffer), followed by a further incubation for 1.5 h at 4°C. Immune complexes were washed three times in 1 ml IP lysis buffer (without PMSF, benzamidine, aprotinin, and leupeptin) containing 1% (wt/vol) Nonidet P-40, three times in 1 ml IP lysis buffer without detergent, and once in 1 ml Tris-buffered saline (TBS; 50 mM Tris, pH 7.5, 200 mM sodium chloride).
Immunoprecipitation of hPLD1
Human PLD1 (hPLD1) was immunoprecipitated from undifferentiated HL-60 cells. Cells (3.06 × 108 cells total) were lysed in IP lysis buffer containing 1% (wt/vol) Nonidet P-40 by probe sonication (three 5-s pulses at 10% output at 4°C). After 15 min on ice, the cell lysate was cleared by centrifugation at 1000 × g for 5 min at 4°C. The supernatant collected was then prepared by centrifugation at 100,000 × g for 30 min at 4°C. hPLD1 was immunoprecipitated directly from 200-μl aliquots of supernatant, diluted with 800 μl PBS (80 mM disodium hydrogen orthophosphate anhydrous, 20 mM sodium dihydrogen orthophosphate, and 100 mM sodium chloride, pH. 7.5), using 3 μg affinity-purified hPLD1 antipeptide polyclonal antibodies (Hammond et al., 1997). After 2 h of tumbling at 4°C, 40 μl of a 50% solution of protein A-Sepharose (Sigma, St. Louis, MO) in PBS was added to each immunoprecipitation. The tubes were tumbled for 1 h at 4°C. Immune complexes were sedimented by gravity for 30 min at 4°C and washed three times with Nonidet P-40 lysis buffer and four times with PBS.
PLD Assays of Immunoprecipitated HA-Spo14 and hPLD1
Immune complexes containing HA-Spo14, prepared as described above, were suspended in a mixture of hPLD1 assay buffer (Brown et al., 1993; 50 mM HEPES, pH 7.5, 3 mM EGTA, 80 mM potassium chloride, 1 mM DTT, 3 mM magnesium chloride, 2 mM calcium chloride) and lipid vesicles containing 50 μM 2-decanoyl-1-{O-[11-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionyl)amino]undecyl}-sn-glycero-3-phosphocholine (BODIPY-PC; Molecular Probes, Eugene, OR) and 5 μM PIP2 (purified from type I, Folch fraction I from Bovine brain; Sigma, as described by Morris et al., 1995). Immmunoprecipitated hPLD1 was suspended in hPLD1 assay buffer and lipid vesicles containing 3 μM BODIPY-PC, 92 μM phosphatidylethanolamine (Avanti Polar Lipids, Alabaster, AL), and 5 μM PIP2. Lipid vesicles were prepared by bath sonication of dry lipid films. In some reactions 100 μM GTPγS (Boeringer Mannheim, Indianapolis, IN), 6 μM bacterially expressed, purified yeast myristolyated Arf1 (Randazzo et al., 1992), or 6 μM purified recombinant human myristolyated ARF1 (Hammond et al., 1997) was included. In other reactions the myristolyated Arf proteins were first preactivated as described (Hammond et al., 1997). Six micromolar Arf protein was used, because it has previously been shown to provoke the maximal activation of immunopurified hPLD1 by ARF1 (Hammond et al., 1997). All assays were performed in triplicate.
After incubation for 30 min at 30°C in a final volume of 100 μl, the PLD reactions were terminated with the addition of 375 μl chloroform:methanol (1:2 vol/vol). Chloroform (125 μl) and 1 M MgCl2 (100 μl) were then added, and the lipid products of the lower phase were extracted and separated by TLC as described (Rose et al., 1995). The PLD reaction products were viewed by UV light, and the bands corresponding to BODIPY-PC and BODIPY-PA were scraped from the plates and extracted with methanol. The fluorescence of the methanol extracts was determined using a Packard (Downers Grove, IL) fluorometer at 485 nm excitation and 530 nm emission. Fluorescence of BODIPY-PA was quantified as a percentage of BODIPY-PC. This value was then converted into picomoles of BODIPY-PA formed per minute per IP. The fluorescence emission corresponding to BODIPY-PA for the lipid substrate control was always <5% of the value measured for PLD-generated BODIPY-PA.
PLD Assays of Total Yeast Cell Lysates
From each strain assayed, total cell lysates were prepared from three independent cultures using glass beads as previously described (Rose et al., 1995). Assays for PC-PLD activity were performed in a mixture of 100 μg protein, Spo14 assay buffer (25 mM HEPES, pH 7.0, 150 mM sodium chloride, 5 mM EGTA, 5 mM EDTA, 40 mM β-glycerophosphate, 1 mM DTT), and lipid vesicles containing 50 μM BODIPY-PC and 5 μM PIP2. The reaction mixture was incubated at 30°C for 30 min in a final volume of 100 μl. PLD assays were terminated, and the lipid products were extracted, separated, and quantitated as described above.
PE-PLD activity was assayed using 1-caproyl-2-{[6–7-(4-nitro-2–1,3-benzoxadiazol-4-yl)amino]caproyl}-sn-glycero-3-phosphatidylethanolamine (C6-NBD-PE; Avanti Polar Lipids) as a substrate (Waksman et al., 1997). These reactions were performed using 100 μg protein from spo14 mutant strains, in a mixture of Spo14 assay buffer, 12 mM CaCl2, and vesicles containing 50 μM C6-NBD-PE. Assays were incubated at 30°C for 30 min in a final volume of 100 μl. Reactions were terminated, and the lipid products were extracted as described above. C6-NBD-PE and C6-NBD-PA were separated by TLC using the solvent mixture chloroform:methanol:acetic acid (50:25:8) (Waksman et al., 1997). The spots corresponding to C6-NBD-PE and C6-NBD-PA were scraped from the plate, and the fluorescence of the methanol extracts was determined using a Packard fluorometer at 460 nm excitation and 530 nm emission. Fluorescence of C6-NBD-PA was quantified as a percentage of C6-NBD-PE; this value was converted into picomoles of C6-NBD-PA produced per minute per milligram of protein. The fluorescence emission corresponding to C6-NBD-PA for the lipid substrate control was always <5% of the value measured for PLD-generated C6-NBD-PA.
Total Protein Concentration Determination
Protein concentration was determined by the method of Bradford (1976), using a Bio-Rad (Hercules, CA) protein assay kit and bovine γ-globulin (Bio-Rad) as a standard.
Immunoblot Analysis
Nonidet P-40–soluble and –insoluble cell fractions prepared as described above were cleared by centrifugation at 16,000 × g for 10 min before being subjected to SDS-PAGE on 5% SDS-polyacrylamide gels. Proteins were electrophoretically transferred onto nitrocellulose membranes (pore size, 0.45 μm; Bio-Rad) for 18 h. Nitrocellulose membranes were blocked by incubation for 2 h at room temperature with 10% nonfat dry milk in TBS with 0.2% (vol/vol) Tween-20. Blots were then washed three times for 10 min with TBS with 0.1% (vol/vol) Tween-20 (TBS-T) and incubated with mAb 12CA5 diluted 1:3000 in TBS-T with 1% (wt/vol) fatty acid-free BSA for 2 h. After three washes with TBS-T, blots were incubated for 2 h with horseradish peroxidase–conjugated anti-mouse antiserum (Amersham International, Little Chalfont, Buckinghamshire, England) diluted 1:5000 in TBS-T with 1% (wt/vol) fatty acid-free BSA. After three final washes in TBS-T, proteins on immunoblots were visualized by ECL detection. As previously reported, 12CA5 mAb selectively recognizes HA-Spo14 (Rudge et al., 1998).
Cytology
Cytology was performed and documented as previously described (Rudge et al., 1998).
Determination of Dityrosine Content of Sporulating Yeast Cultures
Dityrosine was measured as described by Briza et al. (1986). Sporulating yeast cultures (1 × 108 cells) were collected by centrifugation, washed twice with sterile water, and suspended in 1 ml of 10% ammonium hydroxide. Cell suspensions were maintained at −80°C overnight, thawed, and lysed by repetitive vortexing (six times for 30 s each) with an equal volume of glass beads (425–600 μm, acid washed; Sigma) at 4°C.
Chloroform:methanol (1:1, 2.5 ml) was added, and the mixture was vortexed for 2 min at room temperature. The upper phase was collected after centrifugation, and the lower phase was washed with fresh 10% ammonium hydroxide. The two resultant upper phases were pooled, and 1.5 ml were prepared by centrifugation at 16,000 × g for 10 min at room temperature. Finally, 1 ml was removed, and the dityrosine content was determined by fluorescence spectrophotometry. Fluorescence was measured at room temperature with a Spex Industries (Edison, NJ) 212 Fluorolog spectrofluorometer operating in the ratio mode. Measurements were made in a semimicrocuvette. Intensity of emission was measured at 420 nm. The peak at 315 nm corresponds to dityrosine (Briza et al., 1986).
RESULTS
ARF1 Is Required for Sporulation
Partial or complete inability to sporulate has been observed with several mutant alleles of ARF1 (Kahn, unpublished data). Sporulation defects of arf1 alleles, and of mutations in yeast in general, are often found associated with poor growth on nonfermentable carbon sources, making clean resolution of these phenotypes difficult. Thus, we focused our investigations on an allele of ARF1, arf1-myc, which is completely defective in sporulation but otherwise wild type. However, the inability to sporulate is not unique to the arf1-myc allele as a conditional loss-of-function point mutant of ARF1, arf1–3, also displays a specific sporulation defect at the permissive temperature in arf2 diploids homozygous for the arf1–3 mutation (Table 2).
Table 2.
Sporulation in ARF1 arf2 and arf1-myc arf2 strains
| Relevant genotype | Sporulation (%) |
|---|---|
| ARF1 arf2a | 62 |
| arf1-3 arf2b | <0.1 |
| arf1-myc arf2c | <0.1 |
| arf1-myc arf2 + ARF1 CEN | 31 |
| arf1-myc arf2 + arf1-myc CEN | 0.7 |
| arf1-myc arf2 + SPO14 2μ | <0.1 |
Percent sporulation was calculated by counting a minimum of 600 cells from each of two independent cultures after 48 h in sporulation medium.
Strain C138.
Strain C163.
Strain C135.
The arf1-myc allele is the full-length ARF1 coding region with an additional 19 residues containing the c-myc epitope, added to the C-terminus as described in MATERIALS AND METHODS. To use the arf1-myc allele in studies of sporulation, it was necessary to establish that the allele behaved like wild type in all other respects. Expression of arf1-myc on a centromere plasmid (low copy number) rescued arf1 haploid cells from the lethal effects of 40 mM sodium fluoride and overcame the slow growth defect on YEPD plates, indicating that arf1-myc produced a functional protein.
One genomic copy of ARF1 was replaced with arf1-myc in the diploid strain, C134 (Table 1), and a strain homozygous for both arf1-myc1 and arf2 was generated, C138 (Table 1). Deletion of both ARF1 and ARF2 is lethal (Stearns et al., 1990a); however, arf1-myc arf2 strains were viable. Furthermore arf1-myc arf2 mutants were not supersensitive to fluoride ions or cold sensitive and grew at wild-type rates on YEPD and nonfermentable carbon sources. Taken together, these results indicate that strains harboring arf1-myc as their only source of Arf behaved like wild type in mitotically dividing cells.
When arf1-myc arf2 diploids were induced to undergo meiosis, they were unable to form spores (Table 2). ARF1 expressed on a centromere plasmid rescued the sporulation defect of arf1-myc arf2 mutants (Table 2). In contrast, arf1-myc on a centromere plasmid failed to restore sporulation to a significant extent (Table 2). ARF2 on a centromere plasmid also rescued the sporulation defect of arf1-myc arf2 diploids to a similar extent as ARF1.
Diploids Homozygous for arf1-myc and arf2 Complete Meiosis I and II but Do Not Form Spores
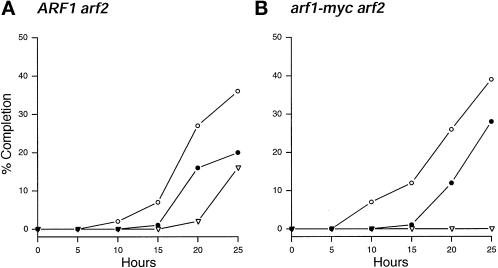
Meiotic nuclear division and asci formation were followed in arf1-myc arf2 diploids induced to undergo meiosis. The percentage of cells completing meiosis I and meiosis II was determined by staining fixed cells with the DNA-specific dye DAPI. In the arf1-myc arf2 mutant, binucleate and tetranucleate cells appeared at the same time as in the ARF1 arf2 strain (Figure 1). Moreover, the percentages of cells completing meiosis I and II were comparable in the two strains. However, in contrast to ARF1 arf2, arf1-myc arf2 mutants failed to form spores (Figure 1 and Table 2).
Figure 1.
Effect of arf1-myc arf2 on the meiotic divisions and sporulation. (A) ARF1 arf2::LEU2 diploids (strain C138). (B) arf1-myc arf2::LEU2 diploids (strain C135). The percentages of cells completing meiosis I (open circles) and meiosis II (filled circles) were monitored by DAPI-fluorescence microscopy, and ascus formation (open triangles) was monitored by phase-contrast microscopy. The percentage of cells having completed meiosis I was calculated by dividing the sum of bi-, tri-, and tetranucleated cells and asci by the total number of cells. The percentage of cells having completed meiosis II was similarly calculated by dividing the sum of bi-, tri-, and tetranucleated cells and asci by the total number of cells. At least 600 cells were counted at each time point.
Sporulation in S. cerevisiae is regulated by a temporal sequence of gene expression (reviewed in Mitchell, 1994). Genes are classified as either early, middle, or late, depending on the time they are either transcribed or their transcription is induced (Mitchell, 1994). Northern blot analysis was performed on RNA extracted from ARF1 arf2 and arf1-myc arf2 diploids in mitosis and at various times throughout meiosis. The expression of HOP1 (Hollingsworth and Byers, 1989) and SPS1 (Friesen et al., 1994), which correspond to early and middle meiosis-specific genes, respectively, were monitored. The timing and relative expression levels of HOP1 and SPS1 were indistinguishable between ARF1 arf2 and arf1-myc arf2 diploids. These results are consistent with the microscopic observations of DAPI-stained cells and demonstrate that the initiation and execution of the meiotic divisions are unaffected in arf1-myc arf2 mutants.
The Failure of arf1-myc arf2 Mutants to Form Spores Is Not a Consequence of Reduced PLD Activity
SPO14 encodes a major PC-PLD activity in yeast as no PLD activity is detected in spo14 deletion strains under the assay conditions used (Rose et al., 1995). Although Spo14 is expressed and active in mitotically dividing cells, the critical function of Spo14 is in the formation of the prospore membrane during meiosis (Rose et al., 1995; Rudge et al., 1998). Because mammalian PLD1 is directly activated by Arf proteins (Brown et al., 1993; Cockcroft et al., 1994; Singer et al., 1996; Hammond et al., 1997; Park et al., 1997), we tested the hypothesis that the sporulation defect of arf1-myc arf2 mutants was the result of reduced PC-PLD activity. Total cell lysates were made from ARF1 arf2 and arf1-myc arf2 diploids dividing mitotically and at various stages of meiosis. PC-PLD activity in arf1-myc arf2 mutants was equivalent to that measured in ARF1 arf2 cells (Table 3).
Table 3.
Time course of Spo14 activity during mitosis and meiosis
| Spo14 activity (pmol/min/mg protein) Time course of meiosis (h) | ||||||
|---|---|---|---|---|---|---|
| 0 | 5 | 10 | 15 | 20 | 25 | |
| ARF1 arf2 diploidsa | 46 ± 3 | 42 ± 4 | 65 ± 2 | 58 ± 2 | 82 ± 6 | 94 ± 8 |
| arf1-myc arf2 diploidsb | 52 ± 4 | 46 ± 4 | 61 ± 4 | 59 ± 2 | 73 ± 9 | 83 ± 5 |
Total cell lysates were prepared from mitotically dividing cells (0 h) and at various times after induction of meiosis (5, 10, 15, 20 and 25 h). PLD activity was assayed in triplicate as described in MATERIALS AND METHODS. SDs were calculated with PLD values obtained from individual experiments.
Strain C138.
Strain C135.
The requirement for Arf in coatomer-coated vesicle formation can be overcome by the addition of increased PLD activity (Ktistakis et al., 1996). To determine whether the requirement for Arf in sporulation can likewise be overcome by increasing PLD activity, arf1-myc arf2 diploids were transformed with SPO14 on a 2μ plasmid (for review, see Broach and Volkert, 1991). Expression of SPO14 on a 2μ plasmid results in an approximate 10-fold increase in PC-PLD activity (Rudge et al., 1998); however, the presence of increased PLD activity did not overcome the sporulation defect of the arf1-myc arf2 mutants (Table 2).
Spo14 Relocalization Occurs Normally in arf1-myc arf2 Diploids
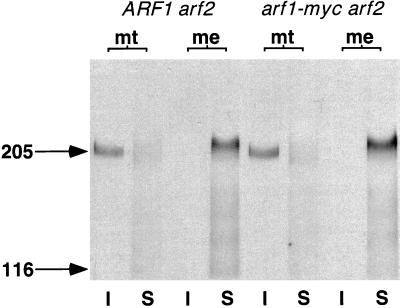
Spo14 relocalizes to the developing membrane during meiosis; relocalization is essential for membrane formation and correlates with phosphorylation of the Spo14 protein (Rudge et al., 1998). To monitor protein movement, we performed cell fractionation studies using a functional HA-tagged derivative of Spo14 (HA-Spo14; Rudge et al., 1998). In mitotically dividing ARF1 arf2 and arf1-myc arf2 mutant cells, <5% of the total HA-Spo14 was solubilized by the nonionic detergent Nonidet P-40. Consequently, HA-Spo14 was detected predominantly in the detergent-insoluble cell fraction (Figure 2). During meiosis, HA-Spo14 is readily solubilized by such treatment in both ARF1 arf2 and arf1-myc arf2 mutants and was detected predominantly in the soluble fraction (Figure 2).
Figure 2.
Immunoblots showing HA-Spo14 detergent solubility in meiotic ARF1 arf2 and arf1-myc arf2 diploids. Nonidet P-40–insoluble (I) and –soluble (S) cell fractions were prepared from ARF1 arf2 and arf1-myc arf2 diploids in mitosis (mt) and 15 h after induction for meiosis (me) (see MATERIALS AND METHODS). To accommodate for different efficiencies of lysis, fractions were prepared from lysates containing equivalent amounts of total protein. Equal volumes of Nonidet P-40–insoluble and –soluble fractions were prepared for immunoblot analysis, as described in MATERIALS AND METHODS. The positions of the prestained molecular mass markers are indicted by arrows and are expressed in kilodaltons.
Spo14 becomes modified by phosphorylation during meiosis; phosphorylation does not alter the enzymatic activity of the protein but does correlate with protein movement, suggesting that this modification triggers relocalization (Rudge et al., 1998). Phosphorylation of HA-Spo14 results in a slower-migrating species as detected by SDS-PAGE (Rudge et al., 1998). As seen in Figure 2, HA-Spo14 migrates with a slower mobility in the detergent-soluble meiotic pool in both the ARF1 arf2 and arf1-myc arf2 strains, compared with the detergent-insoluble mitotic pool, indicating that Spo14 is posttranslationally modified correctly in the arf1-myc arf2 mutant.
Prospore Membrane and Spore Wall Formation Is Attenuated in arf1-myc arf2 Diploids
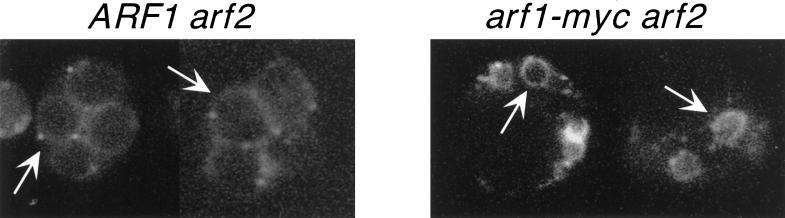
Visualization of a GFP-Spo14 fusion was used to observe the localization of Spo14 in ARF1 arf2 and arf1-myc arf2 diploids induced to undergo meiosis and to monitor spore membrane formation. In both ARF1 arf2 and arf1-myc arf2 mutants, GFP-Spo14 was initially distributed throughout the cell and became concentrated at discrete foci at meiosis I. At meiosis II, GFP-Spo14 localized to the site of the new prospore membrane that first surrounds the nuclei and then expands and fuses with itself to encapsulate each of the haploid nuclei separately (Rudge et al., 1998). Visualization of GFP-Spo14 revealed that the spore compartments formed by the prospore membrane in arf1-myc arf2 diploids were smaller than those formed in wild type (Figure 3).
Figure 3.
GFP-Spo14 staining in living ARF1 arf2 and arf1-myc arf2 cells. The panel on the left shows GFP-Spo14 staining in ARF1 arf2 cells at 20 h after transfer to sporulation medium. The cells have completed sporulation, as evidenced by the uniform circles of GFP-Spo14 staining (arrow). The panel on the left shows GFP-Spo14 staining in arf1-myc arf2 cells 20 h after transfer to sporulation medium. The arrows indicate the small circles of GFP-Spo14 staining; these never progress past this size.
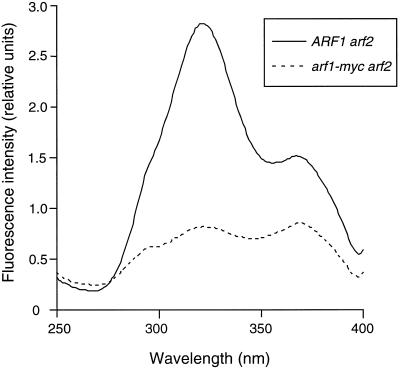
Late in meiosis, components of the spore wall are deposited in the luminal space of the prospore membrane (Byers, 1981). The outermost layer of the spore wall consists of dityrosine (Briza et al., 1986). Because yeast cells synthesize dityrosine only during ascus maturation and dityrosine fluoresces (Briza et al., 1986), spore wall formation can be monitored by fluorescence under UV light. Cell wall components were extracted from sporulating cultures of ARF1 arf2 and arf1-myc arf2 strains, and the fluorescence excitation spectrum of dityrosine was monitored as described in MATERIALS AND METHODS. The excitation spectrum obtained from sporulating ARF1 arf2 diploids was characteristic of dityrosine with a strong peak of fluorescence at 315 nm (Briza et al., 1986; Figure 4). In contrast, the intensity at 315 nm in the arf1-myc arf2 diploid was substantially reduced (Figure 4). This result indicates that spore wall maturation is perturbed in the arf1-myc arf2 mutant.
Figure 4.
Fluorescence excitation spectra of cell wall extracts from ARF1 arf2 and arf1-myc arf2 sporulating cultures. Extracts were prepared and fluorescence was measured as described in MATERIALS AND METHODS. Intensity of emission was measured at 420 nm. The peak at 315 nm corresponds to dityrosine (Briza et al., 1986). Solid line, ARF1 arf2; dotted line, arf1-myc arf2.
Arf1 and Arf2 Do Not Activate Spo14 Activity In Vitro
The ability of Arf1 to directly activate Spo14 was tested by immunoprecipitating HA-Spo14 from spheroplasts prepared from cells 15 h after induction of meiosis and measuring PC-PLD activity in assay buffer known to support Arf protein activation of hPLD1 (Brown et al., 1993; Hammond et al., 1997). As seen in Table 4, PLD activity was unaltered by the addition of recombinant yeast Arf1, with or without the presence of GTPγS. To demonstrate that the yeast Arf1 was capable of activating an Arf-regulated PLD, we immunoprecipitated hPLD1 from undifferentiated HL-60 cells and assayed PLD activity with the same preparation of yeast Arf1. Consistent with previous reports (Brown et al., 1995), hPLD1 was activated two- and fivefold by recombinant yeast Arf1 and human ARF1 protein respectively, in the presence of GTPγS (Table 4). Furthermore, preactivated (GTPγS-bound) yeast Arf2 also failed to activate Spo14, despite being capable of activating hPLD1.
Table 4.
Effects of recombinant yeast and human Arf on HA-Spo14 and hPLD1 activities
| PLD activity (pmol/min/immunoprecipitated PLD)
|
||||||
|---|---|---|---|---|---|---|
| No additions | + 100 μM GTPγS | + 6 μM yeast Arf1 | + 6 μM human ARF1 | + 6 μM yeast Arf1 + 100 μM GTPγS | + 6 μM human ARF1 + 100 μM GTPγS | |
| HA-Spo14 | 0.16 ± 0 | 0.16 ± 0.02 | 0.17 ± 0.02 | 0.16 ± 0.01 | 0.14 ± 0.01 | 0.15 ± 0.02 |
| hPLD1 | 0.41 ± 0.06 | 0.36 ± 0.04 | 0.26 ± 0.05 | 0.31 ± 0.04 | 0.71 ± 0.06 | 2.24 ± 0.27 |
HA-Spo14 and hPLD1 were immunoprecipitated from yeast cells 15 h after induction for meiosis and from undifferentiated HL-60 cells, respectively, as described in MATERIALS AND METHODS. PLD assays were performed in triplicate in identical assay buffer at 30°C, using the same preparations of preactivated GTP-binding proteins. SDs were calculated with PLD values obtained from individual experiments.
Arf Proteins Do Not Activate PC-PLD Activity in Mitotically Dividing Cells
Arf activation of hPLD1 is synergistic with other regulators, including the regulatory domain of PKC-α and activated Rho proteins (Singer et al., 1996; Hammond et al., 1997). To test the ability of Arf proteins to regulate the PLD activity of Spo14 in total yeast cell lysates, likely to contain other regulatory components in an Arf-PLD signaling pathway, we assayed total cellular PLD activity in wild type, arf1 and arf2 deletion strains, and in wild-type strains overexpressing dominant activating ([Q71L]Arf1) or inactivating ([N126I]Arf1) proteins (Kahn et al., 1995). No significant differences were observed in PLD activity assayed in total cell lysates (Table 5), indicating that Arf proteins do not activate Spo14 in vivo.
Table 5.
Total cell lysate PC-PLD activity
| Genotype | PC-PLD activity (pmol/min/mg protein) |
|---|---|
| ARF1 ARF2 ADE2a | 22 ± 10 |
| arf1::HIS3 ARF2 ADE2b | 23 ± 2.5 |
| ARF1 ARF2 ade2-101c | 24 ± 2.6 |
| ARF1 arf2::LEU2 ade2-101d | 24 ± 2.9 |
| ARF1 ARF2 ADE2 + GAL-ARF1 2μ URA3e | 22 ± 0.3 |
| ARF1 ARF2 ADE2 + GAL-arf1Q71L 2μ URA3f | 23 ± 0.6 |
| ARF1 ARF2 ADE2 + GAL-arf1N126I 2μ URA3g | 25 ± 1.4 |
Total cell lysates were prepared from mitotically dividing yeast in triplicate and assayed for Spo14 PLD activity as described in MATERIALS AND METHODS. SDs were calculated with PLD values obtained from individual experiments.
Strain PSY315.
Strain TT104.
Strain PSY316.
Strain TT139.
Strain RT143.
Strain RT149.
Strain RT150.
Arf Proteins Do Not Activate ScPLD2
S. cerevisiae contain a second PLD activity, termed ScPld2, which is easily assayed in spo14 deletion strains (Mayr et al., 1996; Waksman et al., 1997). The function of this second activity is unknown; however, it is clear that this activity cannot substitute for Spo14 in meiosis. Furthermore, the biochemical properties of this second activity are distinct from Spo14; ScPld2 preferentially hydrolyses phosphatidylserine and phosphatidylethanolamine (Mayr et al., 1996), requires calcium for catalytic activity, but not PIP2, and does not catalyze transphosphatidylation in the presence of alcohols (Mayr et al., 1996; Waksman et al., 1997). To determine whether Arf1 or Arf2 activates ScPld2, we assayed total cell lysates for PE-PLD activity in spo14 deletion, spo14 arf1, spo14 arf2, and spo14 deletion strains overexpressing dominant activating ([Q71L]Arf1) or inactivating ([N126I]Arf1) proteins (Kahn et al., 1995). As seen in Table 6, PE-PLD activity was equivalent in all total cell lysates assayed, indicating that Arf proteins are not activators of ScPld2. The differences in activity observed do not correlate with the ARF genotype and suggests that strain backgrounds influence the ability to assay PE-PLD activity.
Table 6.
Total cell lysate PE-PLD activity
| Genotype | PE-PLD activity (pmol/min/mg protein) |
|---|---|
| ARF1 ARF2 spo14::URA3 ADE2a | 180 ± 15 |
| arf1::HIS3 ARF2 spo14::URA3 ADE2b | 182 ± 22 |
| ARF1 ARF2 spo14::URA3 ade2-101c | 292 ± 70 |
| ARF1 arf2::LEU2 spo14::URA3 ade2-101d | 295 ± 86 |
| ARF1 ARF2 spo14::LEU2 ADE2 + GAL-ARF1 2μ URA3e | 174 ± 11 |
| ARF1 ARF2 spo14::LEU2 ADE2 + GAL-arf1Q71L 2μ uRA3f | 168 ± 69 |
| ARF1 ARF2 spo14::LEU2 ADE2 + GAL-arf1N126I 2μ URA3g | 156 ± 18 |
Total cell lysates were prepared from mitotically dividing yeast in triplicate and assayed for ScPld2 activity as described in MATERIALS AND METHODS. SDs were calculated with PLD values obtained from individual experiments.
Strain Y1125.
Strain Y1127.
Strain Y1126.
Strain Y1128.
Strain Y1129.
Strain Y1130.
Strain Y1131.
DISCUSSION
The results presented in this study provide evidence for a specific role of Arf protein(s) in the sporulation process. Analyses of mutants of ARF1, particularly arf1-myc, uncovered a required role for Arf protein(s) to form spores. This role for Arf in sporulation can be clearly resolved from its role in mitotic cell growth, because the arf1-myc allele completely restores growth to wild-type rates, at all temperatures. That Arfs are direct activators of mammalian PLD1 offered one potential explanation for the roles of both Arf and Spo14 in sporulation. However, we were unable to obtain any evidence of a change in Spo14 activity resulting from deletion or expression of dominant (activating or inhibitory) alleles of ARF1 or when PLD was assayed in immunoprecipitates of Spo14 with purified recombinant Arf proteins. Furthermore, there was no change in the residual PLD activity of spo14 deletion yeast extracts when assayed in the presence of activated yeast Arfs. The lack of any detectable Arf-sensitive PLD activity in yeast leads us to conclude that other cellular effectors must mediate the regulation of both the sporulation and secretory processes by Arfs. Thus, there may exist fundamentally different molecular mechanisms for Arf regulation of cell functions between yeast and mammalian cells. This would be surprising given the very high degree of conservation of structure and function of Arf proteins throughout eukaryotic evolution. Alternatively, these results may be interpreted as providing uncertainty to the proposed role of PLD activity as the mediator of Arf’s effects on protein traffic in mammalian and yeast cells.
Diploids homozygous for arf1-myc and arf2 entered meiosis and successfully completed both meiotic divisions but were unable to form spores. This obligate role for Arf in sporulation was evident from studies with the arf1-myc allele (Figure 1 and Table 2). However, other mutations in ARF1 (e.g., arf1–3) also are defective in sporulation. It is likely that other essential functions of Arf proteins, specifically roles in mitotic growth, growth on nonfermentable carbon sources, and membrane traffic, can obscure the importance of Arf in sporulation. Because the Arf1-myc protein contains the full Arf1 protein it must be the additional residues at the C terminus that interfere with Arf function in meiosis. Deletion of up to six residues from the C terminus of Arf1 resulted in an Arf protein that was fully wild type by all assays mentioned above (Cavenagh and Kahn, unpublished observation). Thus, it seems likely that steric hindrance with effector binding is responsible for the sporulation defect, rather than a change in the structure of the very C terminus of Arf1.
Arf-activated PLD1 and the consequent production of PA have been shown to be necessary for the formation of in vitro coatomer-coated vesicles from the trans-Golgi complex (Ktistakis et al., 1996). Because the formation of the prospore membrane is thought to require both the fusion of vesicles derived from the trans-Golgi complex (Neiman, 1998) and the PLD activity of Spo14 (Rudge et al., 1998), the failure of arf1-myc arf2 mutants to sporulate may have been explained by the inability of Arf1-myc to directly activate Spo14. However, the PLD activity of Spo14 in meiotic ARF1 arf2 and arf1-myc arf2 total cell lysates was indistinguishable (Table 3). We had previously reported a fivefold increase in PLD activity during meiosis (Rose et al., 1995). The induction of PLD activity was more modest (approximately twofold) in the strains used in this study. This difference may be a consequence of the differences in strain backgrounds and/or in the different lipid substrates used to assay PLD activity (32P-labeled PC; Rose et al., 1995; vs. BODIPY-PC; this study). Nonetheless, no differences in PLD activity were seen between cells carrying ARF1 arf2 and arf1-myc arf2 alleles.
Ktistakis et al. (1996) showed that the requirement for Arf in the formation of Golgi-derived coated vesicles could be overcome by the addition of an activated PLD. In a related manner, Bi et al. (1997) found that added PA could overcome primary alcohol-mediated inhibition of glycoprotein transport between the ER and Golgi compartments. In contrast to these effects on in vitro transport, we found that increasing the PLD activity 10-fold, by overexpressing Spo14, did not rescue the sporulation defect of the arf1-myc arf2 strain.
The regulation of Spo14 activity includes changes in activity, phosphorylation status, solubility in detergent extracts, and cellular localization (Rudge et al., 1998); none of which are affected by changes in Arf activity. During meiosis, Spo14 is modified by phosphorylation and becomes readily solubilized by nonionic detergent in whole-cell extracts. Neither of these changes was affected by the arf1-myc allele. The change in phosphorylation correlates temporally with a change in the localization of Spo14 to the new prospore membrane during meiosis. This relocalization was monitored with the expression of GFP-tagged Spo14 in live cells, and we found that Spo14 localized to the prospore membrane in arf1-myc arf2 cells. However, in contrast to wild-type (ARF1 arf2) cells, the development of the prospore membrane was attenuated in arf1-myc arf2 mutants. The prospore membrane had fused with itself to capture each of the haploid nuclei separately but had failed to elongate. Consequently, the individual spore compartments in arf1- myc mutants were smaller that those formed in wild-type cells. Taken together, the data presented here suggest that the failure of arf1-myc arf2 diploids to sporulate is not a consequence of Spo14 mislocalizing or from a reduction in PLD activity. Furthermore, in diploids homozygous for spo14, few cells complete meiosis II, no prospore membrane is formed, and no spores are made (Honigberg et al., 1992; Rose et al., 1995; Rudge et al., 1998). The ability of diploids homozygous for arf1-myc arf2 to proceed through meiosis II and to form an abnormal prospore membrane indicates that the defect resulting from mutation of ARF1 is downstream of Spo14 action and confirms that the action of each gene product is essential for sporulation but independent of the others. That Spo14 is not the sole effector for Arf functions in yeast was already evident from the fact that ARF genes together form an essential pair (Stearns et al., 1990a), whereas spo14 null mutants are viable (Honigberg et al., 1992; Rose et al., 1995; Ella et al., 1996; Waksman et al., 1996).
Although the critical function of Spo14 is in meiosis, Spo14 is expressed and active in mitotically dividing cells (Rose et al., 1995), and recent evidence suggests that it plays a role in Golgi function (Patton-Vogt et al., 1997). However, spo14 mutants do not display the phenotypes associated with arf1 strains, i.e., slow growth, cold sensitivity, and supersensitivity to fluoride ions (Rudge and Engebrecht, unpublished observations). Furthermore, PC-PLD activity assayed from mitotically dividing cells was found to be independent of Arf activity, even in the presence of dominant activating or inactivating alleles, which ultimately will prove lethal to yeast cells (Kahn et al., 1995). These results further support our conclusion that SPO14 and ARF genes function in different signaling pathways.
This clear difference in Spo14 and Arf1 action, and the failure to detect any changes in PLD activity assayed in total cell lysates, prompted us to investigate whether any yeast PLD activity could be altered by activated Arf proteins. In contrast to human, porcine, or rat PLD1 (Brown et al., 1993; Cockcroft et al., 1994; Hammond et al., 1997), we found (Table 4) that yeast Arf1 or Arf2 or human Arf1 did not increase the PLD activity of Spo14 either with or without the Arf activator GTPγS. Under the conditions used in our PLD assay, both yeast and human Arf proteins stimulated the activity of the human PLD1 (Table 4) and bound guanine nucleotides normally.
Yeast also contain a second PLD activity, termed ScPld2 (Mayr et al., 1996; Waksman et al., 1997). In contrast to Spo14 (Rose et al., 1995; Waksman et al., 1996), PLD1 (Hammond et al., 1995, 1997), and PLD2 (Colley et al., 1997; Kodaki and Yamashita, 1997), ScPld2 does not require PIP2 for catalysis and does not perform transphosphatidylation (Mayr et al., 1996; Waksman et al., 1997). Moreover, ScPld2 requires calcium for activity and preferentially hydrolyses phosphatidylserine and phosphatidylethanolamine (Mayr et al., 1996; Waksman et al., 1997). These biochemical properties of ScPld2 indicate that it is unrelated to Spo14. Indeed, sequencing of the S. cerevisiae genome has shown that there are no other sequences that display similarity to the SPO14/PLD gene family (Morris et al., 1996). However, in common with Spo14, we could find no evidence that ScPld2 is regulated by yeast Arf proteins (Table 6). This observation is consistent with earlier work performed by Mayr et al. (1996); they reported that the catalytic activity of ScPld2 was not activated by the addition of GTPγS and cytosolic factors.
Although in vivo PLD activity in mammalian cells is routinely measured by transphosphatidylation in the presence of a primary alcohol, Spo14 performs this reaction inefficiently (Rose et al., 1995) and ScPLD2 not at all (Mayr et al., 1996; Waksman et al., 1997). Consequently, we and others (Mayr et al. 1996) have failed to detect PLD-dependent transphosphatidylation in vivo. Thus, we examined the effect of Arf proteins on PLD activity using an in vitro assay. The failure to detect changes in PLD activity using this assay assumes that the conditions of cell lysis do not affect enzymatic activity. Consistent with this assumption, PLD activity of Spo14 immunoprecipitates derived from meiotic cells lysed under different conditions is also not stimulated by Arf proteins.
Our data support the conclusions that, in S. cerevisiae, the essential mitotic and meiotic functions of Arf proteins are not mediated by the activation of a PLD. Although we cannot eliminate the possibility that yeast contain an unidentified Arf-activated PLD, we conclude that in yeast there must be as yet undiscovered mediators of Arf action(s). In support of this conclusion, recent work has shown that mammalian ARF1 can promote the binding of AP-1 and coatomer onto the trans-Golgi network independently of PLD and has led to the suggestion of the existence of PLD– and non-PLD–mediated pathways of ARF function (West et al., 1997).
Finally, the failure to find any Arf-sensitive PLD activity in yeast raises the intriguing possibility that the effects of Arf on membrane traffic are also not mediated by PLD activation in mammalian cells. To date, the only PLDs shown to be directly activated by Arf proteins are those requiring PIP2 and using PC as the preferred substrate. Differences in cellular lipids between yeast and mammals may have necessitated the evolution of differences in PLD activation mechanisms. Regulation of human PLD1 activity is proving to be a very complicated process, involving multiple protein and lipid coactivators. Given this complexity and the “negative” nature of our results, failing to link Arf and PLD activities, such an extrapolation to mammalian cells from data in yeast is risky. However, it may be worth noting that the data supporting a role for Arf-activated PLD activity in membrane transport are also indirect and largely rely on the use of nonspecific (e.g., neomycin and ethanol) or partially specific (e.g., brefeldin A) reagents. It is likely that more work and better reagents will be required before faithful descriptions of the actions of these important signaling molecules in cells can be formulated.
ACKNOWLEDGMENTS
We thank J. Trimmer for helpful discussions and comments on the manuscript and K. Kachel and E. London for help in measuring dityrosine fluorescence. This work was supported by National Institutes of Health grants GM4863903 (to J.E.), GM50388 and GM54641 (to A.J.M.), and GM55148 and GM55823 (to R.K.). S.A.R. had an Affiliate Fellowship from New York State Heart Association (grant 950209).
REFERENCES
- Bi K, Roth MG, Ktistakis NT. Phosphatidic acid formation by phospholipase D is required for transport from the endoplasmic reticulum to the Golgi complex. Curr Biol. 1997;7:301–307. doi: 10.1016/s0960-9822(06)00153-9. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;76:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Briza P, Winkler G, Kalchhauser H, Breitenbach M. Dityrosine is a prominent component of the yeast ascospore wall. A proof of its structure. J Biol Chem, 1986;261:4288–4288. [PubMed] [Google Scholar]
- Broach JR, Volkert FC. Circular DNA plasmids of yeasts. In: Broach J, Pringle RJR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1991. pp. 297–331. [Google Scholar]
- Brown HA, Gutowski S, Kahn RA, Sternweis PC. Partial purification and characterization of Arf-sensitive phospholipase D from porcine brain. J Biol Chem. 1995;270:14935–14943. doi: 10.1074/jbc.270.25.14935. [DOI] [PubMed] [Google Scholar]
- Brown HA, Gutowski S, Moomaw CR, Slaughter C, Sternweis PC. ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern E, Jones W, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1981. pp. 59–96. [Google Scholar]
- Chen Y-G, Siddhanta A, Austin CD, Hammond SM, Sung T-C, Frohman MA, Morris AJ, Shields D. Phospholipase D stimulates release of nascent secretory vesicles from the trans-Golgi network. J Cell Biol, 1997;138:495–504. doi: 10.1083/jcb.138.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft S, Thomas GM, Fensome A, Geny B, Cunningham E, Gout I, Hiles I, Totty NF, Truong O, Hsuan JJ. Phospholipase D: a downstream effector of ARF in granulocytes. Science. 1994;263:523–526. doi: 10.1126/science.8290961. [DOI] [PubMed] [Google Scholar]
- Colley WC, Sung T-C, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provoke cytoskeletal reorganization. Curr Biol. 1997;7:191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- Ella KM, Dolan JW, Qi C, Meier KE. Characterization of Saccharomyces cerevisiae deficient in expression of phospholipase D. Biochem J. 1996;314:15–19. doi: 10.1042/bj3140015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J, Masse S, Davis L, Rose K, Kessel T. Yeast meiotic mutants proficient for the induction of ectopic recombination. Genetics. 1998;148:581–598. doi: 10.1093/genetics/148.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faundez V, Horng J-T, Kelly RB. ADP ribosylation factor is required for synaptic vesicle budding in PC12 cells. J Cell Biol. 1997;138:505–515. doi: 10.1083/jcb.138.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen H, Lunz R, Doyle S, Segall J. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 1994;8:2162–2175. doi: 10.1101/gad.8.18.2162. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Altshuller YM, Sung TC, Rudge SA, Rose K, Engebrecht J, Morris AJ, Frohman MA. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J Biol Chem. 1995;270:29640–29643. doi: 10.1074/jbc.270.50.29640. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Jenco JM, Nakashima S, Cadwallader K, Gu Q, Cook S, Nozawa Y, Preswich GD, Frohman MA, Morris AJ. Characterization of two alternatively spliced forms of phospholipase D1. J Biol Chem. 1997;272:3860–3868. doi: 10.1074/jbc.272.6.3860. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg SM, Conicella C, Esposito RE. Commitment to meiosis in Saccharomyces cerevisiae: involvement of the SPO14 gene. Genetics. 1992;130:703–716. doi: 10.1093/genetics/130.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Clark J, Rulka C, Stearns T, Zhang C-J, Randazzo PA, Terui T, Cavenagh M. Mutational analysis of Saccharomyces cerevisiae ARF1. J Biol Chem. 1995;270:143–150. doi: 10.1074/jbc.270.1.143. [DOI] [PubMed] [Google Scholar]
- Kahn RA, Gilman AG. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. J Biol Chem. 1986;261:7906–7911. [PubMed] [Google Scholar]
- Kodaki T, Yamashita S. Cloning, expression, and characterization of a novel phospholipase D complementary DNA from rat brain. J Biol Chem. 1997;272:11408–11413. doi: 10.1074/jbc.272.17.11408. [DOI] [PubMed] [Google Scholar]
- Ktistakis NT, Brown HA, Waters MG, Sternweis PC, Roth M. Evidence that phospholipase D mediates ADP ribosylation factor-dependent formation of Golgi coated vesicles. J Cell Biol. 1996;134:295–306. doi: 10.1083/jcb.134.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M, Byers B, Esposito RE, Mitchell AP. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle JR, Broach JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces Cell Cycle. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mayr JA, Kohlwein SD, Paltauf F. Identification of a novel, Ca2+-dependent phospholipase D with preference for phosphatidylserine and phosphatidylethanolamine in Saccharomyces cerevisiae. FEBS Lett, 1996;393:236–240. doi: 10.1016/0014-5793(96)00893-9. [DOI] [PubMed] [Google Scholar]
- Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AJ, Engebrecht J, Frohman MA. Structure and regulation of phospholipase D. Trends Pharmacol Sci. 1996;17:182–185. doi: 10.1016/0165-6147(96)10016-x. [DOI] [PubMed] [Google Scholar]
- Morris AJ, Rudge SA, Mahlum CE, Jenco JM. Regulation of phosphoinositide-3-kinase by G protein beta-gamma subunits in a rat osteosarcoma cell line. Mol Pharmacol. 1995;48:532–539. [PubMed] [Google Scholar]
- Neiman AM. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol. 1998;140:29–37. doi: 10.1083/jcb.140.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Provost JJ, Bae CD, Ho WT, Exton JH. Cloning and characterization of phospholipase D from rat brain. J Biol Chem. 1997;272:29263–29271. doi: 10.1074/jbc.272.46.29263. [DOI] [PubMed] [Google Scholar]
- Patton-Vogt JL, Griac R, Sreenivas A, Bruno V, Dowd S, Swede JJ, Henry SA. Role of the yeast phosphatidylinositol/phosphatidylcholine transfer protein (Sec14p) in phosphatidylcholine turnover and INO1 regulation. J Biol Chem. 1997;272:20873–20883. doi: 10.1074/jbc.272.33.20873. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Weiss O, Kahn RA. Preparation of recombinant ADP-ribosylation factor. Methods Enzymol. 1992;219:362–369. doi: 10.1016/0076-6879(92)19036-6. [DOI] [PubMed] [Google Scholar]
- Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J. Phospholipase D signaling is essential for meiosis. Proc Natl Acad Sci USA. 1995;92:12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics, A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol, 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rudge SA, Morris AJ, Engebrecht J. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J Cell Biol. 1998;140:81–90. doi: 10.1083/jcb.140.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini T, Orci L, Amherdt M, Brunner M, Kahn RA, Rothman JE. ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell. 1991;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- Singer WD, Brown HA, Bokoch GM, Sternweis PC. Resolved phospholipase D activity is modulated by cytosolic factors other than Arf. J Biol Chem. 1996;270:14944–14950. doi: 10.1074/jbc.270.25.14944. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kahn RA, Botstein D, Hoyt MA. ADP-ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol Cell Biol. 1990a;10:6690–6699. doi: 10.1128/mcb.10.12.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Willingham MC, Botstein D, Kahn RA. ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc Natl Acad Sci USA. 1990b;87:1238–1242. doi: 10.1073/pnas.87.3.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman M, Eli Y, Liscovitch M, Gerst JE. Identification and characterization of a gene encoding phospholipase D activity in yeast. J Biol Chem. 1996;271:2361–2364. doi: 10.1074/jbc.271.5.2361. [DOI] [PubMed] [Google Scholar]
- Waksman M, Tang X, Eli Y, Gerst JE, Liscovitch M. Identification of a novel Ca2+-dependent, phosphatidylethanolamine-hydrolyzing phospholipase D in yeast bearing a disruption in PLD1. J Biol Chem. 1997;272:36–39. doi: 10.1074/jbc.272.1.36. [DOI] [PubMed] [Google Scholar]
- West MA, Bright NA, Robinson MS. The role of ADP-ribosylation factor and phospholipase D in adapter recruitment. J Cell Biol. 1997;138:1239–1254. doi: 10.1083/jcb.138.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C.J., Cavenagh, M.M., and Kahn, R.A. (1998). A family of Arf effectors defined as suppressors of the loss of Arf function in the yeast Saccharomyces cerevisiae. J. Biol. Chem. (in press). [DOI] [PubMed]