Abstract
We have identified a mutant allele of the DAM1 gene in a screen for mutations that are lethal in combination with the mps1-1 mutation. MPS1 encodes an essential protein kinase that is required for duplication of the spindle pole body and for the spindle assembly checkpoint. Mutations in six different genes were found to be lethal in combination with mps1-1, of which only DAM1 was novel. The remaining genes encode a checkpoint protein, Bub1p, and four chaperone proteins, Sti1p, Hsc82p, Cdc37p, and Ydj1p. DAM1 is an essential gene that encodes a protein recently described as a member of a microtubule binding complex. We report here that cells harboring the dam1-1 mutation fail to maintain spindle integrity during anaphase at the restrictive temperature. Consistent with this phenotype, DAM1 displays genetic interactions with STU1, CIN8, and KAR3, genes encoding proteins involved in spindle function. We have observed that a Dam1p-Myc fusion protein expressed at endogenous levels and localized by immunofluorescence microscopy, appears to be evenly distributed along short mitotic spindles but is found at the spindle poles at later times in mitosis.
INTRODUCTION
The mitotic spindle serves to segregate replicated chromosomes to progeny cells during cell division. This sophisticated microtubule-based structure is organized from two spindle poles on opposing sides of the nucleus that contain centrosomes or equivalent organelles (Kellogg et al., 1994). In the budding yeast Saccharomyces cerevisiae, the centrosome-equivalent organelle is the spindle pole body (SPB) (Botstein et al., 1997; Winsor and Schiebel, 1997). The SPB is a multilayered organelle, which remains embedded in the nuclear envelope throughout the yeast cell cycle (Byers and Goetsch, 1975). The SPB contains γ-tubulin (the product of the TUB4 gene) and the associated Spc97p and Spc98p, all of which are required for microtubule nucleation (Knop et al., 1997; Knop and Schiebel, 1997, 1998; Sundberg and Davis, 1997), a key function of the SPB. Centrosome duplication is a critical cell cycle–regulated event required for the formation of a bipolar spindle. In budding yeast, the SPB is duplicated late in the G1 phase of the cell cycle (Byers and Goetsch, 1974, 1975). The duplication pathway has been described morphologically, and several genes required for the process have been identified by mutation (Winey and Byers, 1993; Botstein et al., 1997). Among the genes required for SPB duplication is MPS1 (for monopolar spindle), which encodes an essential protein kinase (Winey et al., 1991; Lauze et al., 1995). Analysis of strains containing different mutant alleles of MPS1 reveals two distinct SPB duplication defects, suggesting that Mps1p has at least two distinct functions in SPB duplication (Schutz and Winey, 1998).
After SPB duplication, the SPBs separate as the mitotic spindle is assembled. Later in the cell cycle, the spindle segregates chromosomes by both anaphase A and anaphase B movements (Winey et al., 1995; Straight et al., 1997). A number of proteins are required for these processes, including several that are localized to the spindle and spindle pole (for review, see (Hoyt and Geiser, 1996; Botstein et al., 1997). These include a variety of microtubule-based molecular motors, both kinesin-related proteins (Cin8p, Kar3p, and Kip1p) and dynein (Dhc1p). There are also nonmotor spindle components such as Stu1p, Ase1p, and Duo1p that are required for spindle function. Finally, components of the kinetochore, which connect the chromosomes to the spindle, contribute to spindle formation and integrity (e.g., Mif2p; Brown et al., 1993). Extensive genetic interactions have been detected between alleles of the various genes required for spindle assembly and function.
The assembly of the mitotic spindle and the execution of mitosis are both under strict cell cycle control. Both SPB duplication and the separation of the SPBs to form the spindle require CDC28/CLN activity (Winey and Byers, 1993; Winsor and Schiebel, 1997). The onset of anaphase requires the degradation of the Esp1p inhibitor Pds1p by the anaphase-promoting complex (Cohen-Fix et al., 1996), and exit from mitosis requires complete inactivation of CDC28/CLBs as well as the degradation of the spindle component Ase1p (Juang et al., 1997). These controls are executed during each round of cell division and serve to coordinate activities of the mitotic spindle with other cell cycle events. A distinct set of cell cycle controls, termed checkpoint pathways, are inducible and function to block mitotic progression when the DNA or the spindle is damaged or when a prerequisite event has not occurred (Elledge, 1996). For example, the spindle assembly checkpoint pathway delays the onset of anaphase when spindle function is compromised by microtubule-depolymerizing agents or by mutations that affect assembly or integrity of the mitotic spindle (Rudner and Murray, 1996; Wells and Murray, 1996). In addition to its role in SPB duplication, Mps1p kinase is also required for activation of the spindle assembly checkpoint, most likely through phosphorylation of Mad1p, another constituent of this signaling pathway (Hardwick et al., 1996; Weiss and Winey, 1996). Thus far, all conditional mutations in MPS1 lie in the kinase domain, and all affect both the SPB duplication and the checkpoint functions (Schutz and Winey, 1998).
We have endeavored to understand how Mps1p functions in its dual roles by identifying factors with which it interacts, either genetically or physically. We previously reported that MPS1 and the molecular chaperone gene CDC37 show a variety of genetic interactions and that Mps1p requires the Cdc37p for kinase activity (Schutz et al., 1997). MPS1 also shows genetic interactions with CIN8, a spindle motor gene, suggesting a potential role for Mps1p at the spindle that may be distinct from its functions in SPB duplication and spindle checkpoint activation (Geiser et al., 1997). Finally, Mps1p has been found by two-hybrid and coimmunoprecipitation experiments to bind Mob1p (Luca and Winey, 1998). Mob1p is required for the completion of mitosis and the maintenance of ploidy; the latter function requires Mps1p activity and may be related to the role of Mps1p in SPB duplication.
Here we report that six genes were identified by mutations that caused inviability in combination with the mps1-1 mutation. One of these genes encodes Bub1p, a kinase that, like Mps1p, is required for the spindle assembly checkpoint (Hoyt et al., 1991; Roberts et al., 1994). Several of the other genes encode molecular chaperones, suggesting a single kinase may require several different chaperones for activity. Last, we identified a novel gene that encodes an essential coiled-coil protein. This gene, DAM1, was independently identified in a screen for proteins that interact with a spindle-associated protein, Duo1p (Hofmann et al., 1998; also see Figure 6), and Dam1p has been shown to bind microtubules in vitro. Our findings indicate that Dam1p is required for spindle integrity during anaphase B elongation, and that the protein is localized to short spindles and spindle poles during the cell cycle. The fact that a mutation in MPS1 exhibits a genetic interaction with an allele of DAM1 reinforces the idea that Mps1p may have essential roles in both SPB and spindle function.
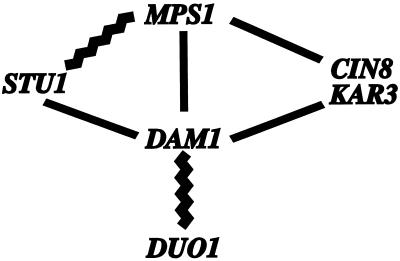
Figure 6.
Summary of genetic and two-hybrid interactions involving DAM1. Jagged lines denote two-hybrid interactions, and straight lines denote synthetic lethal interactions. The CIN8-MPS1 data are from Geiser et al. (1997). The STU1-MPS1 data are from Luca and Winey (personal communication). The DAM1-DUO1 data are from Hofman et al. (1998).
MATERIALS AND METHODS
Yeast and Escherichia coli Culture and Genetic Techniques
The yeast strains used in this study are listed in Table 1. Yeast growth conditions (including media) and yeast genetic techniques were as described (Guthrie and Fink, 1991). Most strains derive from the S288c background. The cell synchronization experiments were performed by treating cultures (0.4 OD units) with 10 μM α-factor (US Biological, Swampscott, MA) for ∼2 h, washing two times, and then resuspending in warmed media. Percentage of large budded cells was measured by counting those cells with a bud size greater than half the size of the mother cell. Depolymerizaton of microtubules was performed as described (Hardwick et al., 1996). Samples were prepared after 3 h in media containing benomyl (DuPont, Wilmington, DE) and nocodazole (US Biological) for flow cytometry (60% large budded; our unpublished results) and immunofluorescence as described below.
Table 1.
Yeast strains
| Strain | Genotype |
|---|---|
| 7d | MATa, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 12a | MATα, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b | MATα, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 12a30-69 | MATα, moe1, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 12a30-70 | MATα, moe2, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b20-11 | MATα, moe2, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-5 | MATα, moe7, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801(MPS1-URA3,ADE3,CEN) |
| 15b45-68 | MATα, moe13, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-188 | MATα, moe21, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-279 | MATα, moe27, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-341 | MATα, moe34, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-69 | MATα, moe14(1), mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-248 | MATα, moe25, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-298 | MATα, moe29, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-69 | MATα, moe14(2), mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b30-4 | MATα, moe4, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-59 | MATα, moe12, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-156 | MATα, moe15, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-240 | MATα, moe24, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-262 | MATα, moe26, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-295 | MATα, moe28, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-335 | MATα, moe33, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 15b45-350 | MATα, moe35, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801 (MPS1-URA3,ADE3,CEN) |
| 7d45-1 | MATa, moe37, mps1-1, ade2, ade3, his3Δ200, leu2-3,112, ura3-52, lys2-801, (MPS1-URA3,ADE3,CEN) |
| WX241-20c(295) | MATa, mps1-1, ura3-52, his3Δ200 |
| DB8WX257-15ca | MATa, mps1Δ::HIS3, ura3-52, leu2-3,112, trp1Δ1, his3Δ200 (MPS1-URA3,CEN) |
| WX257-17c(371) | MATa, ura3-52, leu2-3,112, trp1Δ1, his3Δ200 |
| D8bX5cA(375)a | MATa/MATα, ura3-52/ura3-52, leu2-3,112/leu2-3,112, trp1Δ1/trp1Δ1, his3Δ200/his3Δ200 |
| MJYM14-9b(889) | MATa, dam1-1, ura3-52, leu2-3,112, trp1Δ1, his3Δ200 |
| moe1b/1d (908) | MATa/MATα, dam1-1/dam1-1, ura3-52/ura3-52, leu2-3,112/leu2-3,112, TRP/trp1Δ1, HIS3/his3Δ200 |
| MHJ19-5b(1325) | MATa, dam1-1, mad1Δ2::URA3, ura3-52, leu2-3,112, his3Δ200, ade2-1 |
| MHJ39-8c(1491) | MATa, dam1-1, mad2Δ::URA3, ura3-52, leu2-3,112, trp1Δ1 |
| MHJ39-8c(1495) | MATa, dam1-1, mad3Δ::URA3, ura3-52, leu2-3,112, trp1Δ1, his3Δ200, ade2-1, can1-100 |
| MHJ22-1b(1341) | MATa, dam1-1, cin8Δ::HIS3, ura3-52, leu2-3,112, trp1Δ1, his3Δ200, lys2-801, cyh2R, can1-100 (DAM1-URA3-CEN) |
| MHJ38-2a(1485) | MATα, dam1-1, kar3Δ::LEU2, ura3-52, leu2-3,112, his3Δ200 (KAR3-URA3-2μ) |
| MHJ37-8b(1547) | MATa, DAM1-MYC::HIS3, SPC42-GFP::TRP, SPC42Δ::LEU2, ura3-52, leu2-3,112, his3Δ200, trp1-1 |
| 8bX15a-1(1549) | MATa/MATα, DAM1-MYC::HIS3/DAM1-MYC::HIS3, SPC42-GFP::TRP/SPC42-GFP::TRP, SPC42Δ::LEU2/SPC42Δ::LEU2, ura3-52/ura3-52, leu2-3,112/leu2-3,112, his3Δ200/his3Δ200, trp1-1/trp1-1 |
| MHJMM (1345) | MATa/MATα, DAM1-MYC::HIS3/DAM1-MYC::HIS3, ura3-52/ura3-52, leu2-3,112/leu2-3,112, his3Δ200/his3Δ200, TRP/trp1Δ1 |
| MHJΔDAM1 (950) | MATa/MATα, dam1Δ::HIS3/DAM1, ura3-52/ura3-52, leu2-3,112/leu2-3,112, his3Δ200/his3Δ200, trp1Δ1/trp1Δ1, |
| WX257-14c(372) | MATa, ura3-52, leu2-3,112, trp1Δ1, his3Δ200 |
All strains from this study except aLauze et al. (1995).
Synthetic Lethal Screen and Gene Cloning
Mutant strains that require MPS1 at the normally permissive (ambient, ∼23°C) mps1-1 temperature (mps1-1 enhancer [moe]) were derived from three strains, 12a, 15b, and 7d (Table 1). These strains contain the mps1-1 ts allele and a mutant alleles of ade2, ade3, and ura3. They harbor a plasmid with the yeast markers ADE3 and URA3 and a wild-type copy of MPS1. The ADE3 and URA3 yeast markers allowed us to assess the requirement of these strains for the wild-type copy of MPS1 using the adenine red–white sectoring assay (Bender and Pringle, 1991) and resistance to 5-fluoroorotic acid (5-FOA, a suicide substrate for strains with the URA3 gene product; Boeke et al., 1987). Strains 12a, 15b, and 7d were mutagenized using methane sulfonic acid ethyl ester (Sigma, St. Louis, MO) to between 40 and 50% viability (Guthrie and Fink, 1991). Mutagenized cultures were plated on rich (YPD) media, and 93,000 colonies were screened for a solid red colony phenotype that would indicate the strain was unable to lose the wild-type copy of MPS1. Seven hundred eighty-seven (0.8%) nonsectoring colonies were further analyzed for their requirement for wild-type MPS1 by checking them for 5-FOA sensitivity. Twenty-two of these strains were consistently nonsectoring and 5-FOA resistant. To demonstrate the above phenotypes were due to the requirement of these strains for MSP1, a LYS2-based plasmid containing MPS1 was transformed into the strains, and all 22 were subsequently shown to sector and be 5-FOA resistant.
To distinguish between a bona fide moe mutation and the creation of an mps1 chromosomal null within these strains, they were crossed to an mps1Δ strain (DB8WX257-5C; Table 1) supported by a URA3-based plasmid harboring a wild-type copy of MPS1. The resulting diploids were struck to 5-FOA-containing medium at an mps1-1 permissive temperature to determine whether they required the wild-type copy of MPS1. Nine of the diploids from these crosses were 5-FOA sensitive, indicating that an additional mutation within mps1-1 had generated a chromosomal null; these mps1Δ strains behave phenotypically as moe strains by being nonsectoring and 5-FOA sensitive.
The nature of the moe mutations was assessed by crossing the strains to a mps1-1 strain (WX241-20c; Table 1). All 13 of the mps1-1 homozygous, moe heterozygous diploids were 5-FOA resistant, suggesting that the moe mutations were recessive. To determine whether the synthetic lethal phenotype in the original moe strains was due to a mutation in a single gene, these diploids were sporulated, and resulting tetrads were dissected. Spores were analyzed for viability on 5-FOA medium. For all but two of the 13 moe strains, the tetrads segregated 2:2 for synthetic lethality, suggesting that the moe phenotype was due to a mutation in a single gene.
Complementation analyses were carried out to determine the number of genes represented by the 13 remaining moe strains. Because many of the moe strains identified were of the same mating type, the strains were crossed to a mps1-1 strain (WX241-20c; Table 1) to obtain the mutation in the opposite mating type. Spores resulting from this cross that were 5-FOA sensitive, and hence contained both the mps1-1 and moe mutations, were analyzed for their mating type. Both MATa and MATα moe mutant strains were intercrossed, and resulting diploids were replica plated to 5-FOA medium to determine whether the moe strains could complement one another. This analysis revealed seven genes, six of which are known (Table 2).
Table 2.
moe mutations
| Complementation group | Members | Previously identified genesa |
|---|---|---|
| I | dam1-1 | YGR113W/DAM1 |
| II | moe2 | YDJ1 |
| III | moe3, moe7, moe13 | CDC37 |
| IV | moe21, moe27, moe34 | STI1 |
| V | moe14(1), moe25 | HSC82 |
| VI | moe29 | BUB1 |
| VII | moe14(2) | Not yet identified |
| VIII | moe4, moe12, moe15, moe24, moe26, moe28, moe33, moe35, moe37 | mps1 null alleles |
See text for literature citations.
Complementation groups I, II, and III (Table 2) were cloned by complementation of the ts phenotype of the respective mutation upon transformation of these strains with a LEU-based centromeric yeast genomic library (a gift from C. Connelly and P. Heiter, University of British Columbia, Vancouver, Canada) and found to be the known genes, DAM1, YDJ1, and CDC37. To ensure that we had cloned the gene responsible for the mps1-1 synthetic lethal phenotype, we determined that the corresponding null strain (see below) did not complement the original ts strain. Genes representing complementation groups IV, V, and VI were cloned by restoration of their sectoring and 5-FOA resistance phenotypes upon transformation with the LEU-based centromeric yeast genomic library. The gene corresponding to moe142 is being isolated (VII; Table 2).
DAM1 Plasmids, DAM1 Gene Disruption, and myc-DAM1
Multicopy plasmids containing the DAM1 gene were constructed by inserting a 5-kb XbaI–SalI fragment containing the DAM1 gene into plasmids pRS424 and pRS426 (Sikorski and Hieter, 1989) cut with Spe1–SalI. A precise deletion of the DAM1 gene and replacement with the HIS3 gene was done by one-step gene replacement (Baudin et al., 1993) in wild-type diploid strain 375 (Table 1) with a PCR product containing 42 bp of DAM1 gene flanking sequences on either side of the HIS3 gene. Histidine prototrophs were selected, and correct integration was confirmed using PCR amplification (Luca and Winey, 1998). A haploid strain containing the DAM1 null allele was obtained by sporulation and dissection. Oligonucleotide primers were purchased from Life Technologies (Gaithersburg, MD). The strain containing the myc-DAM1 fusion gene was constructed by transformation of wild-type strain with a PCR product containing the following sequence: from −80 bp to the stop codon of the DAM1 gene, a triple repeat of the myc epitope (Kolodziej and Young, 1991), the HIS3 gene, and the 3′ untranslated region of DAM1 from +76 to +148. Histidine prototrophs were selected and analyzed by PCR amplification to confirm correct integration and by Western analysis to confirm protein expression as described (Luca and Winey, 1998), using a mouse anti-myc 1° antibody (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA) and sheep anti-mouse 2° antibody conjugated to HRP (1:20,000; Amersham Pharmacia Biotech, Piscataway, NJ).
Cytological Techniques
Flow cytometric analysis of cells was performed as described using the DNA stain propidium iodide (Sigma) (Hutter and Eipel, 1979). Samples were analyzed on a Becton Dickinson (Mountain View, CA) FACScan flow cytometer using CELL QUEST software to obtain and analyze data (BDIS, San Jose, CA).
Fixation techniques and immunofluorescence of microtubules using 1° monoclonal antibody YOL1/34 and 2° anti-rat conjugated to FITC or Texas Red (Accurate Chemical and Scientific, Westbury, NY) and visualization of DNA using DAPI stain were performed as described (Pringle et al., 1991). The techniques used to visualize Dam1p-myc involved either the technique described for microtubules or a formaldehyde–methanol–acetone technique as described (Chial et al., 1998), in the second case, using a mouse anti-myc 1° antibody (1:300; Santa Cruz Biotechnology) and a sheep anti-mouse 2° antibody conjugated to Texas Red or FITC (1:800; Jackson ImmunoResearch, West Grove, PA). Standard fluorescence microscopy was carried out using either a Zeiss (Thornwood, NY) fluorescence microscope with an Empix charge-coupled device camera and Metamorph Software (Universal Imaging, Westchester, PA) or a Leica DMRXA/RF4/V automated microscope with a Cooke SensiCam digital camera and Slidebook software (Intelligent Imaging Innovations, Denver, CO). Yeast cells were prepared for thin sectioning as described by (Byers and Goetsch, 1991). Serial sections were viewed on a Philips CM10 electron microscope (Philips Electronic Instruments, Mahwah, NJ).
RESULTS
Synthetic Lethal Screen
A screen for mutations that are lethal in combination with the mps1-1 mutation was conducted to identify genes whose products interact with the Mps1p kinase (moe = mps one enhancer). At restrictive temperatures the mps1-1 allele is defective for SPB duplication (Winey et al., 1991) and, subsequently, in the spindle assembly checkpoint (Weiss and Winey, 1996). The mutation in this allele lies in the kinase domain and greatly decreases protein kinase activity in vitro (Schutz and Winey, 1998). Mutations lethal in combination with mps1-1 at its permissive temperature were initially identified in a plasmid-sectoring assay based on the adenine biosynthetic pathway (Bender and Pringle, 1991). After mutagenisis (see MATERIALS AND METHODS) nonsectoring solid red colonies were selected as candidates because they appeared to require the MPS1-containing plasmid. The inability of the solid red colonies to lose the wild-type copy of MPS1 was confirmed by their failure to grow on 5-FOA medium that selects against cells harboring the plasmid-borne URA3 gene (Boeke et al., 1987). Finally, to demonstrate that the nonsectoring and 5-FOA sensitivity phenotypes were due to the strains’ requirement for the plasmid-borne wild-type MPS1, a second MPS1-containing plasmid was transformed into candidate strains, and the transformed strains were shown to sector and grow on 5-FOA medium. Twenty-two independently isolated strains were identified in which colonies both fail to sector and fail to grow on 5-FOA medium, but in which both phenotypes were restored when another MPS1 plasmid was introduced into the strain.
Genetic analysis of the 22 original isolates involved eliminating chromosomal null alleles of MPS1 (nine were recovered; see MATERIALS AND METHODS), screening for additional phenotypes, and segregation and complementation analysis. For all but 2 of the 13 remaining strains, the synthetic lethality segregated as a single mutation. One of these two was not studied further, because the synthetic lethal phenotype was due to mutations in more than one gene. However, for the second aberrantly segregating strain two separable mutations were identified, each of which was lethal in combination with mps1-1. Three of the 13 strains were found to have temperature-sensitive growth defects. The ts phenotype cosegregated with the synthetic lethality in all three strains. Complementation analysis (Table 2) revealed that each of the synthetic lethal mutations that give temperature-sensitive growth defects defines separate complementation groups and that two of these groups each contain other nonconditional synthetic lethal alleles. The remaining nonconditional synthetic lethal mutations identified four complementation groups, two with multiple members (Table 2).
Identification of the Genes
We have previously reported alleles of the yeast CDC37 gene, which encodes a molecular chaperone, that are lethal in combination with mps1-1 (Schutz et al., 1997). We tested to see whether any of the seven complementation groups of synthetic lethal mutations could be complemented by CDC37. One group (III; Table 2) consists of both conditional and nonconditional alleles of CDC37. Five of the six remaining genes were identified by complementation using a yeast genomic library (see MATERIALS AND METHODS). Consistent with our previous findings that the Mps1p kinase requires chaperone function for its activity, three of the genes identified, HSC82, YDJ1, and STI1 (V, II, and IV; Table 2), encode chaperonins. HSC82 encodes one of the two S. cerevisiae Hsp90 proteins that exhibit substrate specificity for kinases (Pratt, 1992; Jakob and Buchner, 1994; Nathan and Lindquist, 1995). STI1 encodes an Hsp70p of the Ssa subclass that has been found in a complex with Hsp90p (Chang and Lindquist, 1994). The third chaperone identified in this screen, Ydj1p, encodes a dnaJ homologue that has been shown to interact with and regulate the Ssa subclass of Hsp70s (Cyr et al., 1992; Cyr, 1995; Cyr and Douglas, 1994). The strain that contained two mutations that were synthetically lethal with mps1-1 had a mutation in HSC82 and another as yet unidentified locus. Interestingly, an allele of the BUB1 gene that encodes another protein kinase involved in the spindle assembly checkpoint was identified in the screen (VI; Table 2; Roberts et al., 1994)). This allele of BUB1, called bub1-656, behaved similarly to previously identified bub1 alleles (Hoyt et al., 1991), in that it exhibited slow growth and benomyl sensitivity, suggesting that the checkpoint activity of bub1-656 was compromised (our unpublished observation).
A conditional allele representing the final complementation group (I; Table 2) was rescued at the nonpermissive temperature by the uncharacterized YGR113W gene on chromosome VII. We constructed a null allele of this gene by exact gene replacement (see MATERIALS AND METHODS) and found that the gene is essential for viability. This gene, named DAM1 (for Duo1p and Mps1p Interactor) has been independently identified in a screen for proteins that interact with a spindle-associated protein, Duo1p. Dam1p has also been shown to localize to the spindle when overexpressed and to bind microtubules in vitro (Hofmann et al., 1998).
dam1-1 Strains Fail in Mitosis
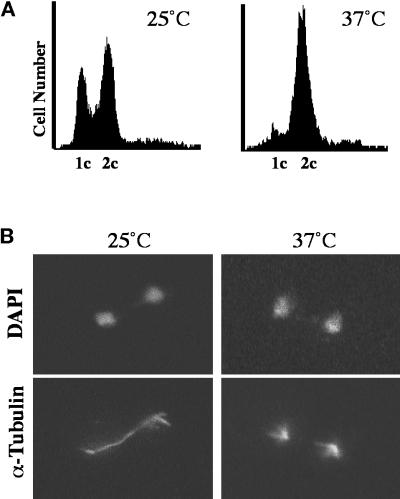
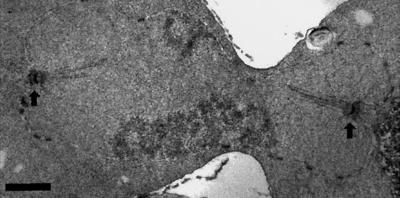
We examined cells containing the ts dam1-1 mutation (908; Table 1) at the nonpermissive temperature to understand more about the role of Dam1p in the cell. After 2.5 h at the nonpermissive temperature (37°C), a significant number of dam1-1 cells arrest with large buds, as determined by budding index (47%; n = 400 cells), and with 2C DNA content as determined by flow cytometry (Figure 1A). The chromatin in these cells, as visualized by DAPI staining, is separated into two masses of approximately equal staining intensity, indicating that the cells have begun anaphase elongation of the spindle (Figure 1B, DAPI). In anaphase, dam1-1 cells at the permissive temperature (Figure 1B, 25°C, α-Tubulin) and wild-type cells (our unpublished observation) contain mitotic spindles with interdigitated microtubules from each SPB that appear as a solid bar of microtubules between and overlapping the DAPI staining region. Strikingly, this structure is absent in dam1-1 strains incubated at the nonpermissive temperature (Figure 1B, 37°C, α-Tubulin); instead, these cells contain two distinct microtubule-staining regions that appear to be the result of loss of integrity in the mitotic spindle. This “broken spindle” phenotype was confirmed by electron microscopic examination of the spindles in these cells. The cells were found to contain two SPBs, each with a distinct array of nuclear mictrotubules, but these microtubules are not interconnected as in a normal spindle (Figure 2 and adjoining serial sections; our unpublished observation; see legend for quantitation). Consistent with these mitotic abnormalities, a significant loss of viability (>66% lethality upon return to the permissive temperature after 2.5 h at the nonpermissive temperature) was observed in dam1-1 cells.
Figure 1.
Characterization of the asynchronously growing diploid dam1-1 strain at the permissive temperature (25°C) and after shift to the nonpermissive (37°C) temperature for 2.5 h. (A) Flow cytometric analysis to measure DNA content. 1C and 2C, DNA content (propidium iodide fluorescence) before and after DNA replication, respectively. Each histogram represents 5000 cells. (B) Immunofluorescence staining of DNA (DAPI) and tubulin (α-Tubulin) in cells grown at 25 and 37°C, as described in MATERIALS AND METHODS.
Figure 2.
Electron micrograph of the asynchronously growing diploid dam1-1 strain after shift to the nonpermissive (37°C) temperature for 2.5 h. Arrows denote spindle pole bodies. Bar, 0.5 μm. Of spindles examined from 13 large-budded cells, 11 were discontinuous as shown here, and 2 appeared normal.
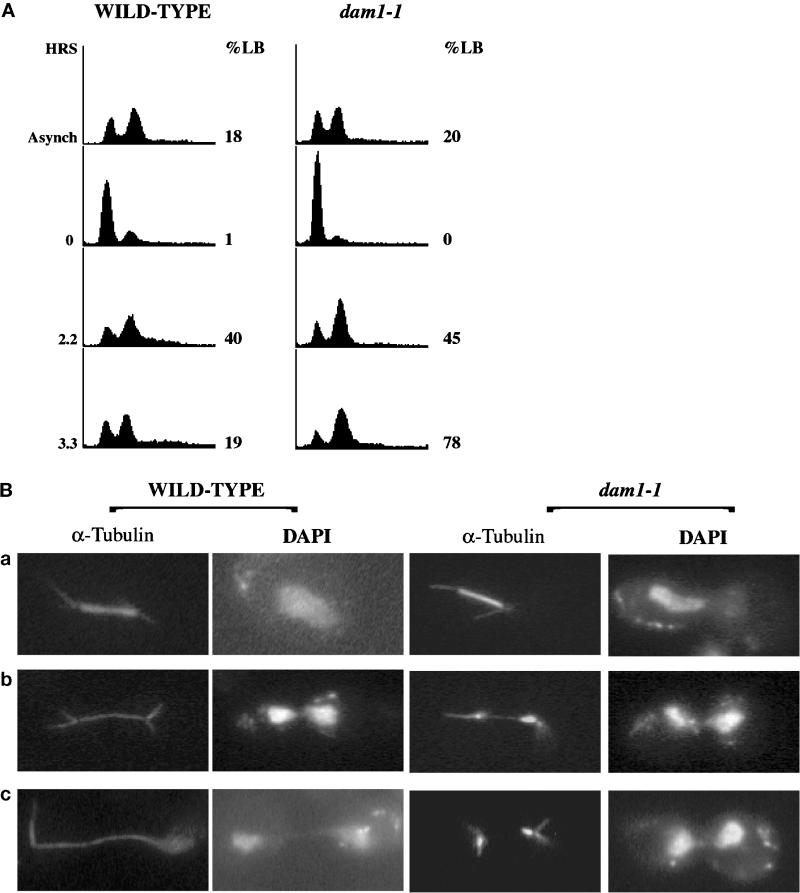
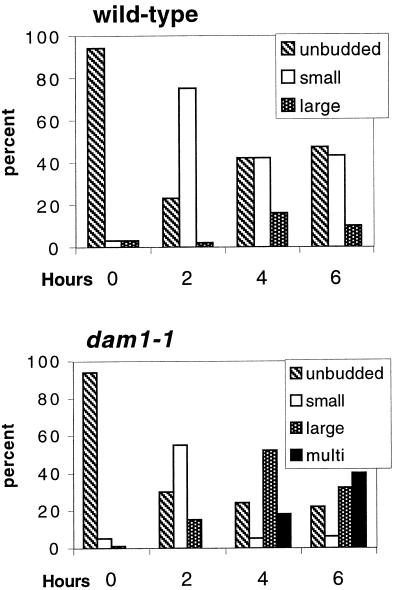
We examined dam1-1 cells proceeding synchronously through the cell cycle at the nonpermissive temperature to determine the timing of development of the defective spindle phenotype. Wild-type and dam1-1 strains (372 and 889; Table 1) were arrested in the G1 phase of the cell cycle at permissive temperature by treatment with α-factor and then released from this G1 arrest at the nonpermissive temperature (see MATERIALS AND METHODS). Cell cycle progression was monitored over time by quantitating the number of large-budded cells (%LB) and quantitating DNA content in the population of cells (Figure 3A). The mitotic spindles in these cells were monitored by immunofluorescence staining of microtubules (Figure 3B, α-Tubulin), and the DNA was detected by DAPI staining (Figure 3B, DAPI). Early mitotic spindles, before visible separation of the chromatin, appear to be similar in wild-type and dam1-1 cells (Figure 3B, a). However, as cells proceed with anaphase elongation of the spindle (Figure 3B, b and c), the spindle remains intact in wild-type cells but breaks in dam1-1 cells, yielding two distinct microtubule-staining arrays and partially separated chromatin. After 3.3 h at the nonpermissive temperature, 78% of dam1-1 cells are large-budded, the majority containing defective spindles (see quantitation in figure legend). Additional experiments were conducted to determine the fate of these cells by monitoring the budding index of wild-type and dam1-1 cells up to 6 h after release into the nonpermissive temperature (Figure 4). Although both strains start with a large percentage of unbudded cells because of treatment with α-factor, wild-type cells show a second increase in unbudded cells after 4 h, presumably because of normal cytokinesis to complete the cell cycle. In contrast, no unbudded cell population reappears in dam1-1 cells over the course of the experiment; instead an increase in large and multibudded cells is observed at 4 h. The population of multibudded cells increases further after 6 hours, consistent with the idea that dam1-1 cells do not undergo cytokinesis. Taken together, these results suggest a direct role for Dam1p during anaphase spindle elongation and another role, possibly indirect, during cytokinesis.
Figure 3.
Characterization of synchronized wild-type and dam1-1 haploid strains after shift to the nonpermissive (37°C) temperature. (A) Flow cytometric analysis to measure DNA content, as in Figure 1. Asynch, asynchronously growing cells at 25°C; 0, 2.2, and 3.3, hours at 37°C after removal of α-factor, %LB, percentage of cells with bud size more than half the size of the mother cell; 200–400 cells were counted. (B) Immunofluorescence staining of DNA (DAPI) and tubulin (α-Tubulin) in wild-type and dam1-1 cells grown at 37°C for 2.2 h (a and b) or 3.3 h (c). Of spindles from large-budded dam1-1 cells with two separated DAPI-staining regions at 2.2 h, 90% are discontinuous or abnormally thin, as in b and c, 6% are otherwise abnormal (e.g., not associated with DNA), and 4% appear normal (n = 200).
Figure 4.
Budding indices of wild-type and dam1-1 cells after release from α-factor arrest to the nonpermissive temperature represented as a bar graph. Each sample represents 200–400 cells examined.
dam1-1 Strains Do Not Constitutively Require the Spindle Assembly Checkpoint but Do Require Mad1p to Accumulate Mitotic Cells at the Nonpermissive Temperature
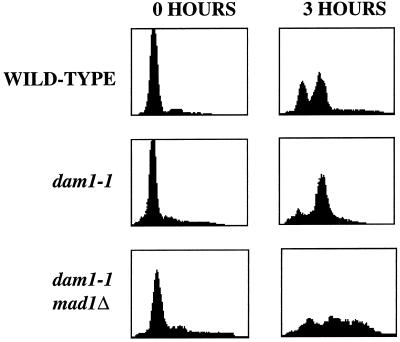
As described, mps1-1 strains show documented defects both in SPB duplication and in the spindle assembly checkpoint pathway. The failure of the mitotic spindle in dam1-1 strains suggested that the genetic interaction between MPS1 and DAM1 might reflect Mps1p’s role in the checkpoint, i.e., that the normally nonessential checkpoint role of Mps1p is essential in a dam1-1 strain. A constitutive checkpoint requirement has been postulated for the spindle defect caused by a deletion of the CIN8 gene, because mutation in all but one of the seven checkpoint genes causes lethality in this mutant strain (a lesser effect was seen in the MAD3Δ strain; Geiser et al., 1997). Although mps1-1 strains do not show a checkpoint defect at the permissive temperature (Weiss and Winey, 1996), it seems possible that even a slight checkpoint defect could be lethal in the presence of the dam1-1 mutation. We tested this model by making double mutants between the dam1-1 mutation and the mad1, mad2, and mad3 null mutations (1325, 1491, and 1495; Table 1), each of which are defective in the spindle assembly checkpoint (Li and Murray, 1991). We found that these double mutant strains were viable with normal growth characteristics, indicating that dam1-1 strains do not require the spindle assembly checkpoint for viability at the permissive temperature. Because dam1-1 cells at the nonpermissive temperature do not display a short mitotic spindle with duplicated, unseparated DNA typical of cells arrested through the spindle assembly checkpoint pathway, it is possible that the mitotic bias observed in dam1-1 cells is independent of this pathway. We tested this idea by comparing synchronized dam1-1 and dam1-1, mad1 null double mutant strains 3 h after release into the nonpermissive temperature (Figure 5). Although dam1-1 cells show the previously described mitotic bias (Figure 5, dam1-1), this peak is missing in the cells lacking Mad1p (Figure 5, dam1-1, mad1Δ), suggesting that the defect caused by the dam1-1 mutation at the nonpermissive temperature activates the spindle assembly checkpoint.
Figure 5.
DNA content of wild-type, dam1-1, and dam1-1mad1null cells. Left panels (0 h), flow cytometric data of cells at α-factor arrest; right panels (3 h), flow cytometric data (performed as in Figures 1 and 3) of cells 3 h after release from α-factor to the nonpermissive temperature. Thirty-four percent (n = 400 cells) of dam1-1, mad1Δ cells were multibudded at this time point compared with 0% for both wild-type and dam1-1 cells.
The dam1-1 Mutation Exhibits Genetic Interactions with Mutations in Other Spindle Proteins
The requirement for Dam1p in mitotic spindle function led us to determine whether the dam1-1 mutation exhibits genetic interactions with mutations in other genes encoding proteins involved in mitosis, in addition to its genetic interaction with MPS1. No interactions were detected between the dam1-1 mutation and alleles of genes encoding integral SPB components (tub4-1, spc98-2, and cmd1-1), kinetochore components (ndc10-42 and mif2-3), or several proteins that act late in mitosis (ase1Δ and mob1-77) (our unpublished observations, mutations in genes encoding the mitotic spindle apparatus; for review, see Botstein et al., 1997). Given the common occurrence of allele-specific interactions, these results clearly do not rule out the possibility of genetic interactions among different alleles of these genes. However, the dam1-1 mutation was found to be lethal in combination with a deletion of either of the kinesin-like motor proteins encoded by CIN8 or KAR3 (but not with a deletion of DYN1) and to exacerbate stu1-5, a mutation in a microtubule-binding protein (Table 3). We examined dosage suppression of dam1-1 with STU1, KAR3, and CIN8 multicopy plasmids and of stu1-5 and cin8Δ with a DAM1 multicopy plasmid, but none was detected (our unpublished observation). Finally, we have observed that dam1-1 strains are hypersensitive to growth on benomyl, a microtubule-destabilizing agent (our unpublished results). Genetic interactions between DAM1 and genes that encode spindle components, together with the dam1-1 phenotype and the in vitro microtubule-binding activity of Dam1p (Hofmann et al., 1998), are consistent with a role of Dam1p in the function of the mitotic spindle. In addition, Stu1p has been shown to interact with Mps1p in 2-hybrid experiments (Luca and Winey, personal communication), and CIN8 shows genetic interactions with MPS1 (Geiser et al., 1997), providing a further connection between DAM1 and MPS1. The genetic interactions described here together with genetic and two hybrid interactions from the work of others (Hofmann et al., 1998; Luca and Winey, personal communication) are summarized in Figure 6.
Table 3.
Summary of synthetic interactions with dam1-1
| Yeast strain | Relative growth
|
|||
|---|---|---|---|---|
| 23°C | 30°C | 34°C | 37°C | |
| dam1-1 | + | + | + | − |
| dam1-1, mps1-1 | − | ND | ND | ND |
| dam1-1, cin8Δ | − | ND | ND | ND |
| dam1-1, kar3Δ | − | ND | ND | ND |
| stu1-5 | + | + | + | ± |
| dam1-1, stu1-5 | + | ± | − | − |
+, size of colonies similar to a wild-type control; ±, size of colonies <20% of a wild-type control; −, no growth; ND, not determined.
Dam1p Is Localized to the Spindle and Peripheral to the Spindle Pole
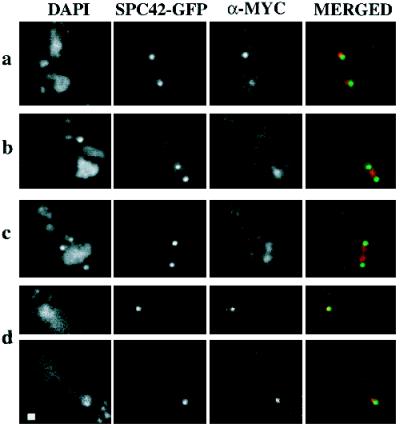
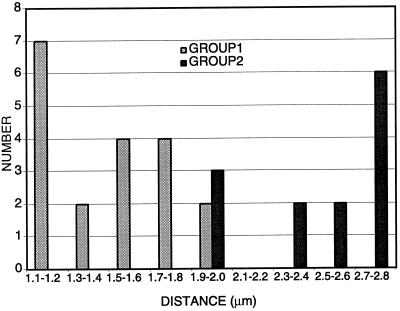
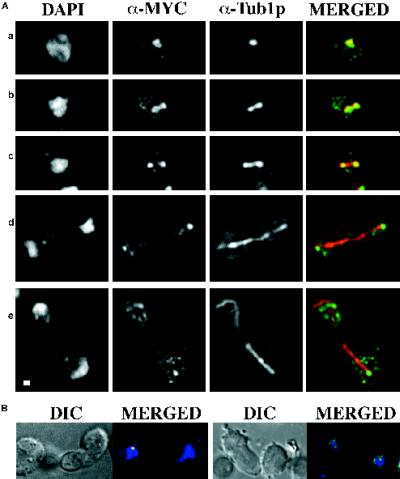
We investigated subcellular localization of the Dam1p protein at endogenous levels to determine whether the spindle defect observed in dam1-1 mutant strains might be due to a direct role of Dam1p at the spindle. The chromosomal DAM1 gene in a haploid strain was epitope tagged by integration of a triple c-myc epitope at its 3′ end (see MATERIALS AND METHODS). C-terminally myc-tagged Dam1p was detectable on Western blots (our unpublished observation), and the myc-DAM1 strain (1345, Table 1) grows at a rate indistinguishable from wild-type strains at all temperatures checked (15–37°C), suggesting that the tagged DAM1 allele is fully functional. Initial immunofluorescence experiments using an anti-myc antibody indicated that myc-Dam1p localizes with structures suggestive of spindle poles and short mitotic spindles (our unpublished observation). To verify this localization, the myc-DAM1 allele was introduced into a strain containing an SPC42p–green fluorescent protein (GFP) fusion (Schutz and Winey, 1998) to visualize SPBs (1549; Table 1), or the myc-DAM1 strain was doubly stained with both anti-myc and anti-tubulin (Tub1p) antibodies to visualize mitotic spindles. Using deconvolution microscopy on the myc-DAM1, SPC42-GFP strain (Figure 7), we observed myc-Dam1p localization immediately adjacent to SPBs when a single SPB was present (Figure 7a) and at a late stage in mitosis (Figure 7d). When the duplicated SPBs are still close together, myc-Dam1p clearly localizes between them (Figure 7b), presumably on the mitotic spindle, and starts to separate into two discrete foci as the SPBs move apart (Figure 7c). To quantitate this transition, we measured spindle lengths in a synchronized population of myc-DAM1, SPC42-GFP cells (1547; Table 1). Myc-Dam1p is present in a continuous bar of length 1.1–2.0 μm between SPBs (Figure 8, GROUP 1) and begins to separate into two discrete foci when the distance is ≥1.9 μm (Figure 8, GROUP 2). The spindle localization was confirmed in the myc-DAM1 strain (1345; Table 1) by immunofluorescence experiments using α-myc and α-Tub1p antibodies (Figure 9). Through the short spindle stage (Figure 9A, a–c), myc-Dam1p colocalizes with tubulin and generally reflects microtubule density. At later stages in mitosis, myc-Dam1p staining appears more strongly at the spindle poles (Figure 9A, d and e) and does not appear to correlate with microtubule density. In this experiment, we also were able to detect punctate nuclear myc-Dam1p staining throughout the cell cycle by using a more sensitive FITC-conjugated secondary antibody to the α-myc antibody (not possible with the SPC42-GFP strain). Treatment with the microtubule-destabilizing drugs nocodozole and benomyl abolishes the majority of microtubule and myc-Dam1p staining (Figure 9B, left panel, right cell), although we observed the punctate nuclear staining in the absence of microtubules (Figure 9B, right panel). In cells with residual microtubule staining, there remains proportionally reduced myc-Dam1p staining as well (Figure 9B, left panel, left cell).
Figure 7.
Localization of myc-Dam1p in wild-type yeast cells using immunofluorescence microscopy. DAPI, DNA staining; SPC42-GFP, autofluorescence of a GFP-tagged version of Spc42p; α-MYC, anti-myc 1° antibody plus Texas Red anti-mouse 2° antibody; MERGED, merging of fluorescence of SPC42-GFP (green) and α-MYC (red). (a) Two unbudded cells. (b and c) Budded cells with short spindles before anaphase B elongation. (d) Two focal planes of a large budded cell in late anaphase. Bar, 1 μm.
Figure 8.
Transition of myc-Dam1p localization from one to two foci. Bar graph of spindle lengths in cells with the following patterns of myc-Dam1p staining. GROUP 1, myc-Dam1p localization as an uninterrupted bar between the SPBs (n = 19; average = 1.5 μm; SD = 0.3); GROUP 2, myc-Dam1p localization as two staining regions adjacent to the SPBs (n = 13; average = 2.4 μm; SD = 0.3). Spindles were measured using Slidebook software with an error of ±0.1 μm.
Figure 9.
Colocalization of myc-Dam1p with microtubules using immunofluorescence microscopy. (A) DAPI, DNA staining; α-MYC, anti-myc 1° antibody plus FITC anti-mouse 2° antibody; α-Tub1-GFP, anti-tubulin 1° antibody plus Texas Red anti-rat 2° antibody. The granular staining using Texas Red is present in controls lacking primary antibody and is considered background. MERGED, merging of α-MYC (green) and α-Tub1p fluorescence (red). (a) Unbudded cell. (b and c) Budded cells with short spindles before anaphase B elongation. (d and e) Large budded cells in late anaphase. (B) myc-Dam1p localization after treatment of cells with microtubule-depolymerizing agents. DIC (differential interference contrast) shows cell outline. MERGED, merging of DAPI (blue), α-MYC (green), and α-Tub1p (red) fluorescence. The left panel shows residual colocalization (yellow) of myc-Dam1p (green) and tubulin (red), and the right panel shows punctate nuclear myc-Dam1p staining (green) in the absence of microtubules (red). Bar, 1 μm.
DISCUSSION
We have reported the isolation of mutations in three distinct classes of genes that are synthetically lethal with the mps1-1 mutation. The first includes genes that encode the molecular chaperones CDC37, YDJ1, HSC82, and STI1. The second class is defined by BUB1, a gene required for the spindle assembly checkpoint. The final class contains the DAM1 gene that encodes an essential spindle- and spindle pole-localized protein required for mitotic spindle function.
We previously demonstrated that Mps1p activity requires the Cdc37p chaperone (Schutz et al., 1997). In this study, we isolated both conditional and nonconditional alleles of CDC37 that are lethal in combination with the mps1-1 mutation. Cells containing the conditional allele of CDC37 arrest in G1 with unreplicated DNA at the nonpermissive temperature as do the previously characterized alleles of this gene (Schutz et al., 1997; Schutz, Ph.D. thesis). In addition to CDC37, we found that mps1-1 strains require wild-type activity of YDJ1, HSC82, and STI1 for their viability. As mentioned previously, the HSC82-encoded Hsp90p and Sti1p are found in a macromolecular chaperone complex, and Ydj1p interacts with and regulates Sti1p (Cyr et al., 1992; Cyr and Douglas, 1994; Cyr, 1995). Mutations in members of this chaperone complex and in CDC37 have been identified in synthetic lethal screens with other defective kinases, including kin28-ts3 and cdc28-109 (Valay et al., 1995; Zarzov et al., 1997). It is possible that this group of chaperones functions to stabilize the compromised kinases at their normally permissive temperatures. Similar to alleles of CDC37, strains harboring mutant alleles of other chaperones also exhibit SPB defects. Cell harboring either ydj1-10 or hsf1-82, which is defective in the synthesis of Hsp90p, have been found to arrest with large buds and a single focus of microtubules at restrictive temperatures. Electron microscopic analysis of hsf1-82 cells revealed an unduplicated SPB with an enlarged half-bridge, reminiscent of the mps1-1 SPB defect (Zarzov et al., 1997). Our results suggest that this phenotype may be due to instability of Mps1p caused by decreased Hsp90p in the cell. Although growth in sti1 null mutants is compromised at restrictive temperatures, detailed cytology of these mutants is not yet available (Nicolet and Craig, 1989). Based on the different terminal SPB phenotypes in cdc37-1 and ydj1-10 and hsf1-82 mutants, it is likely that these genes affect different steps in Mps1p activation or define more than one requirement for chaperones to activate or maintain Mps1p kinase activity and possibly the activity of other kinases.
In addition to requiring molecular chaperones, the mps1-1 mutation shows synthetic lethality with a checkpoint-defective allele of the gene encoding the Bub1p protein kinase (Roberts et al., 1994). Interestingly, mps1-1 is viable in combination with other mutations in the checkpoint pathway (Hardwick et al. 1996), suggesting that the synthetic lethality is not due to a requirement for other checkpoint proteins in general but instead that a closer link may exist between MPS1 and BUB1 than previously suspected. MPS1 and BUB1 (and BUB3, whose product forms a complex with Bub1p; Roberts et al., 1994) are distinct from other checkpoint genes in that null mutations in them have drastic effects on cell viability indicative of other roles in the cell in addition to their nonessential checkpoint functions (Roberts et al., 1994). Also, mutations in both MPS1 and BUB1 have been found to be synthetically lethal with a deletion allele of the CIN8 kinesin-like motor (Geiser et al., 1997). Regardless of the molecular nature of this interaction, it is interesting that these two kinases appear to interact in some way beyond their role in the spindle assembly checkpoint.
The third class of genes reported here is represented by DAM1. Dam1p shows no homology to other genes in S. cerevisiae or in other organisms, but it does contain coiled coil domains as defined by the COILS program (Lupas et al., 1991; our unpublished observation), often seen in structural proteins. DAM1 was independently identified in a two-hybrid screen with the DUO1 gene, and Dam1p has been shown to bind microtubules in vitro (Hofmann et al., 1998). Furthermore, overexpression of DAM1 (and DUO1) results in spindle defects. Consistent with these data, we report that Dam1p is required for the integrity of the spindle during anaphase B elongation. However, we uncovered DAM1 using a very different approach, in an mps1-1-based screen designed to detail interactions necessary for SPB duplication and the spindle assembly checkpoint functions. We eliminated the possibility that the dam1-1 defect at the permissive temperature leads to a constitutive requirement for the checkpoint pathway by demonstrating viability of double mutants containing dam1-1 and a null allele of one of several other checkpoint-related genes. At the nonpermissive temperature, dam1-1 cells do appear to be defective in maintenance of a checkpoint arrest state; however, activation of the checkpoint, the step in which Mps1p acts, appears to be intact (Figure 5).
Therefore, the interaction between MPS1 and DAM1 is probably through the role of Mps1p in SPB duplication or through an as yet uncharacterized role in spindle dynamics. The latter role has been suggested by a demonstrated physical interaction between Mps1p and Stu1p (Luca and Winey, personal communication); Stu1p is a spindle and spindle pole component that was identified originally as an extragenic suppressor of a mutation in the β-tubulin-encoding gene TUB2 (Pasqualone and Huffaker, 1994). In addition, two nonconditional alleles of MPS1 have been shown to be synthetically lethal with a deletion of CIN8 (Geiser et al., 1997), a gene encoding a kinesin-like motor protein involved in spindle assembly and maintenance (Hoyt et al., 1992; Roof et al., 1992; Saunders et al., 1995), and in the “rapid” phase (Straight et al., 1998) of anaphase B chromosome separation (Saunders et al., 1997). We have shown genetic interactions between the dam1-1 mutation and mutations in the CIN8, KAR3, and STU1 genes (Figure 6) and favor the idea that DAM1 interacts with MPS1 through spindle functions after SPB duplication in the cell cycle. This idea will be tested with the isolation of mps1 mutations specific to one of its several roles. Also, further studies involving localization of Dam1p in the mps1-1 mutant strain may shed light on the molecular basis of the dam1-1, mps1-1 synthetic lethality. We do not observe a physical interaction between Mps1p and Dam1p using two-hybrid or coimmunoprecipitation assays (our unpublished observation), suggesting that if a physical association exists between these two proteins, it is likely to be transient or limited to the context of the spindle and spindle pole. It will also be interesting to test whether Dam1p is phosphorylated, because the genetic interaction may be indicative of a kinase–substrate relationship.
We have shown that an epitope-tagged version of Dam1p at endogenous levels exhibits an interesting pattern of localization during the cell cycle, associating with short spindles initially and moving to spindle poles as spindles elongate during mitosis. The presence of Dam1p at the spindle pole is likely mediated through its microtubule-binding activity (Hofmann et al., 1998); this idea is consistent with the dramatic decrease in staining upon depolymerization of microtubules and genetic interactions observed between DAM1 and genes encoding spindle components, such as Stu1p, Cin8p, and Kar3p. The pattern we observe is a subset of that seen upon overexpression of the protein in which it appears to bind the entire spindle and spindle poles at all times during the cell cycle (Hofmann et al., 1998). One explanation for this difference is that increasing the amount of protein drives its association with the spindle, eventually leading to the observed toxicity for the cell. Interestingly, Duo1p, a binding partner of Dam1p, is found on spindles and spindle poles at endogenous levels throughout the cell cycle (Hofmann et al., 1998).
Mitotic spindles contain two general classes of microtubules that can be defined functionally and morphologically (Winey et al., 1995; Straight et al., 1997). One class of microtubules interdigitate in the spindle midzone to define a core bundle that lengthen during anaphase spindle elongation. Some microtubule-associated proteins in yeast, such as Ase1p, appear to be localized to this group of spindle microtubules (Pellman et al., 1995). The other class of spindle microtubules makes up the kinetochore fibers. In budding yeast, a kinetochore fiber is comprised of a single microtubule joining the SPB to a chromosome (Peterson and Ris, 1976; Winey et al., 1995). There are no reported proteins that bind specifically to kinetochore microtubules. Thus far, it is not clear whether Dam1p is specifically associated with one or both of these types of microtubules. The terminal broken spindle phenotype in dam1-1 strains occurs during anaphase B elongation, consistent with a potential role for Dam1p in stabilizing the region of overlap in the spindle midzone, i.e., in the first class of microtubles. However, the redistribution of the wild-type Dam1p from entire spindle to spindle pole during mitotic progression is more reminiscent of the dynamics of kinetochore movement, and a similar pattern of localization is seen with the kinetochore proteins Ndc10p (Goh and Kilmartin, 1993), Ndc80p (Wigge et al., 1998), and Cse4p (Meluh et al., 1998). The SPB-proximal staining is similar to kinetochore proteins Ctf19p (Hyland et al., 1999) and Mif2p (Meluh and Koshland, 1997). Kinetochore fibers occupy a significant amount of space in short spindles but early in mitosis diminish in length until very short remnants remain at the spindle poles during later stages of mitosis (Winey et al., 1995). In fact, the reduction during anaphase A in kinetochore microtubule number and length as documented by electron microscopy (Winey et al., 1995) occurs in spindles of ∼2 μm in length, showing good correlation with the spindle length in which we observe Dam1p concentrating at the poles. In addition, the unusual discontinuous spindle phenotype observed in the dam1-1 mutant strain resembles, at least superficially, that described for a mutation in MIF2 (Brown et al., 1993). However, we have not detected any genetic interactions between DAM1 and NDC10 or MIF2. Although it is difficult to envision how a kinetochore microtubule-binding protein could affect anaphase elongation, one possibility is that dam1-1 could cause a defect in spindle structure, resulting in a tension imbalance in the spindle that is manifested as a broken spindle phenotype.
A higher-resolution analysis of the Dam1p localization will be necessary to reveal its exact location in mitotic spindles. In addition, further analysis of Dam1p will be necessary to understand the basis of the genetic interaction between the dam1-1 and mps1-1 mutations. This analysis should yield a clearer understanding of the function of this novel microtubule-binding spindle protein.
ACKNOWLEDGMENTS
We thank D. Drubin, G. Barnes, and I. Cheeseman for communicating results before publication, M. Brown, C. Connelly, P. Heiter, T. Davis, M. Hoyt, K. Hardwick, T. Huffaker, J. Kilmartin, F. Luca, P. Meluh, D. Pellman, E. Schiebel, and A. Straight for reagents, L. Pillus, T. Su, C. Troxel, and J. Yucel for critical reading of the manuscript, our reviewers for several helpful insights, and members of the Winey and Drubin–Barnes laboratories for helpful discussion. This work was supported by fellowships from the National Institutes of Health (GM-19566 to A.R.C.) and the Cancer League of Colorado (to J.B.B.) and by grants from the National Institutes of Health (GM-51312) and the National Science Foundation (MCB-09357033). Deconvolution microscopy was made possible, in part, by a gift from Virginia and Mel Clark.
Abbreviations used:
- 5-FOA
5-fluoroorotic acid
- GFP
green fluorescent protein
- SPB
spindle pole body
REFERENCES
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Pringle JR. Use of a screen for synthetic lethal and multicopy suppresser mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Botstein D, Amberg D, Mulholland J, Huffaker T, Adams A, Drubin D, Stearns T. The yeast cytoskeleton. In: Pringle JR, Broach JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 1–90. [Google Scholar]
- Brown MT, Goetsch L, Hartwell LH. MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Preparation of yeast cells for thin-section electron microscopy. Methods Enzymol. 1991;194:602–608. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- Chang HC, Lindquist S. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J Biol Chem. 1994;269:24983–24988. [PubMed] [Google Scholar]
- Chial HJ, Rout MP, Giddings TH, Jr, Winey M. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J Cell Biol. 1998;143:1789–1800. doi: 10.1083/jcb.143.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Cyr DM. Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Douglas MG. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Geiser JR, Schott EJ, Kingsbury TJ, Cole NB, Totis LJ, Bhattacharyya G, He L, Hoyt MA. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol Biol Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology, vol. 194. Methods in Enzymology. San Diego: Academic Press; 1991. [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Hofmann C, Cheeseman IM, Goode BL, McDonald KL, Barnes G, Drubin DG. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J Cell Biol. 1998;143:1029–1040. doi: 10.1083/jcb.143.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Geiser JR. Genetic analysis of the mitotic spindle. Annu Rev Genet. 1996;30:7–33. doi: 10.1146/annurev.genet.30.1.7. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hutter KJ, Eipel HE. Microbial determinations by flow cytometry. J Gen Microbiol. 1979;113:369–375. doi: 10.1099/00221287-113-2-369. [DOI] [PubMed] [Google Scholar]
- Hyland KM, Kingsbury J, Koshland D, Hieter P. Ctf19p: a novel kinetochore protein Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U, Buchner J. Assisting spontaneity: the role of Hsp90 and small Hsps as molecular chaperones. Trends Biochem Sci. 1994;19:205–211. doi: 10.1016/0968-0004(94)90023-x. [DOI] [PubMed] [Google Scholar]
- Juang YL, Huang J, Peters JM, McLaughlin ME, Tai CY, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast gamma-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej PA, Young RA. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- Lauze E, Stoelcker B, Luca FC, Weiss E, Schutz AR, Winey M. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Luca FC, Winey M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol Biol Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes & Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolet CM, Craig EA. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3638–3646. doi: 10.1128/mcb.9.9.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualone D, Huffaker TC. STU1, a suppressor of a beta-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman D, Bagget M, Tu YH, Fink GR, Tu H. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J Cell Biol. 1995;130:1373–1385. doi: 10.1083/jcb.130.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JB, Ris H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J Cell Sci. 1976;22:219–242. doi: 10.1242/jcs.22.2.219. [DOI] [PubMed] [Google Scholar]
- Pratt WB. Control of steroid receptor function and cytoplasmic-nuclear transport by heat shock proteins. Bioessays. 1992;14:841–848. doi: 10.1002/bies.950141209. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Roberts BT, Farr KA, Hoyt MA. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol Cell Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Saunders W, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz AR. MPS1 and CDC37: Analysis of Two Genes Required for Spindle Pole Body Duplication in Budding Yeast. Ph.D. Thesis. Boulder, CO: University of Colorado; 1997. [Google Scholar]
- Schutz AR, Giddings TH, Jr, Steiner E, Winey M. The yeast CDC37 gene interacts with MPS1 and is required for proper execution of spindle pole body duplication. J Cell Biol. 1997;136:969–982. doi: 10.1083/jcb.136.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz AR, Winey M. New alleles of the yeast MPS1 gene reveal multiple requirements in spindle pole body duplication. Mol Biol Cell. 1998;9:759–774. doi: 10.1091/mbc.9.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg HA, Davis TN. A mutational analysis identifies three functional regions of the spindle pole component Spc110p in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:2575–2590. doi: 10.1091/mbc.8.12.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valay JG, Simon M, Dubois MF, Bensaude O, Facca C, Faye C. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells WA, Murray AW. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Byers B. Assembly and functions of the spindle pole body in budding yeast. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor B, Schiebel E. Review: an overview of the Saccharomyces cerevisiae microtubule and microfilament cytoskeleton. Yeast. 1997;13:399–434. doi: 10.1002/(SICI)1097-0061(199704)13:5<399::AID-YEA126>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Zarzov P, Boucherie H, Mann C. A yeast heat shock transcription factor (Hsf1) mutant is defective in both Hsc82/Hsp82 synthesis and spindle pole body duplication. J Cell Sci. 1997;110:1879–1891. doi: 10.1242/jcs.110.16.1879. [DOI] [PubMed] [Google Scholar]