Abstract
The roles of two kinesin-related proteins, Kip2p and Kip3p, in microtubule function and nuclear migration were investigated. Deletion of either gene resulted in nuclear migration defects similar to those described for dynein and kar9 mutants. By indirect immunofluorescence, the cytoplasmic microtubules in kip2Δwere consistently short or absent throughout the cell cycle. In contrast, in kip3Δ strains, the cytoplasmic microtubules were significantly longer than wild type at telophase. Furthermore, in the kip3Δ cells with nuclear positioning defects, the cytoplasmic microtubules were misoriented and failed to extend into the bud. Localization studies found Kip2p exclusively on cytoplasmic microtubules throughout the cell cycle, whereas GFP-Kip3p localized to both spindle and cytoplasmic microtubules. Genetic analysis demonstrated that the kip2Δ kar9Δ double mutants were synthetically lethal, whereas kip3Δ kar9Δ double mutants were viable. Conversely, kip3Δ dhc1Δ double mutants were synthetically lethal, whereas kip2Δ dhc1Δ double mutants were viable. We suggest that the kinesin-related proteins, Kip2p and Kip3p, function in nuclear migration and that they do so by different mechanisms. We propose that Kip2p stabilizes microtubules and is required as part of the dynein-mediated pathway in nuclear migration. Furthermore, we propose that Kip3p functions, in part, by depolymerizing microtubules and is required for the Kar9p-dependent orientation of the cytoplasmic microtubules.
INTRODUCTION
The migration and orientation of the nucleus and spindle are critical events in mitosis in eukaryotic cells. In many organisms the position of the spindle determines the position of the cleavage furrow at cytokinesis. For example, in Caenorhabditis elegans, the reorientation of the spindle in response to a cortical signal is essential for establishing the orientation of the second cell division (Hyman, 1989). In the budding yeast, Saccharomyces cerevisiae, cytokinesis is not determined by the orientation of the spindle. Instead, the spindle must become oriented with respect to the growth axis of the cell to allow elongation of the spindle into the growing bud.
In S. cerevisiae, the nucleus does not break down during cell division and the microtubule organizing centers (or spindle pole bodies, SPBs) are embedded in the nuclear envelope. Spindle microtubules extend from the SPB into the nucleus, and cytoplasmic microtubules extend outward toward the cell cortex (Byers and Goetsch, 1975). Early in G1/S the nucleus rotates to orient the SPB toward the bud (Snyder et al., 1991). The SPB then duplicates to initiate formation of the bipolar spindle. By the end of G2, before the onset of anaphase, the nucleus migrates to the neck of the mother–bud junction. At this point, one SPB is oriented toward the bud neck with the cytoplasmic microtubules extending into the bud (Byers and Goetsch, 1975). With the onset of anaphase the nucleus both elongates and translocates through the bud neck (Kahana et al., 1995; Yeh et al., 1995). After the separation of the nuclear material at late anaphase and telophase, cytokinesis occurs.
Failure in nuclear orientation and migration can cause severe defects in the partitioning of the nuclear material during the subsequent mitosis. Both the cytoplasmic microtubules and the actin cytoskeleton are required for proper nuclear positioning. Disruption of either results in aberrant spindle orientation before anaphase and a failure of nuclei to migrate into the bud neck. Initially, such defects result in the accumulation of binucleate mother cells (Jacobs et al., 1988; Palmer et al., 1992; Sullivan and Huffaker, 1992). The complete absence of cytoplasmic microtubules leads to the formation of multinucleate mother cells with multiple anucleate buds (Sullivan and Huffaker, 1992).
Several microtubule-associated proteins have been shown to be required for efficient nuclear migration. For example, the minus end-directed microtubule motor protein dynein is thought to act by generating the forces required for translocation of the nucleus (Eshel et al., 1993; Li et al., 1993). Deletion of the dynein gene (DHC1/DYN1) causes an abnormal placement of the spindle relative to the bud neck, resulting in the occurrence of anaphase entirely within the mother cell. Consequently, as many as 10–15% of the mother cells in the dhc1Δ deletion strain are binucleate. One model of nuclear migration places dynein at a cortical site such that its minus-end motor activity would pull on the cytoplasmic microtubules to “tow” the nucleus through the bud neck (Eshel et al., 1993; Li et al., 1993). Consistent with this hypothesis, observations in live cells demonstrated that the cytoplasmic microtubules spend a significant period of time in contact with the bud cortex, and the cortical association is coordinated with the budward movement of the spindle (Carminati and Stearns, 1997). Furthermore, in dynein-deficient cells, microtubule-associated movement and microtubule dynamics were significantly altered (Carminati and Stearns, 1997). However, recent experiments with dynein-green fluorescent protein (GFP)1 hybrids suggest that dynein may be localized along the length of the cytoplasmic microtubules (Shaw et al., 1997). If so, the dynein-dependent force may result from interactions with cytoskeletal elements other than the cortex and act along the length of the microtubules.
Two genes, ACT5 and JNM1, are thought to act with dynein as part of the dynactin complex. ACT5 is the yeast homologue of ARP1 that encodes a component of the vertebrate dynactin complex (Schafer et al., 1994). JNM1 has been proposed to correspond to the vertebrate p50 dynactin component (Geiser et al., 1997). Mutations in either gene result in spindle orientation and nuclear migration defects similar to those observed in dynein heavy chain mutants (Eshel et al., 1993; Clark and Meyer, 1994; McMillan and Tatchell, 1994; Muhua et al., 1994). Double mutants with either of these genes and a dynein deletion are no worse than single mutants alone, indicating that ACT5 and JNM1 function in the same pathway as dynein for nuclear migration (Geiser et al. 1997; Muhua et al. 1994; Tatchell, personal communication).
The cortical protein Kar9p functions in nuclear migration in a pathway separate from, yet partially redundant with, dynein. The kar9Δ mutants exhibit mitotic defects similar to dynein and are synthetically lethal in combination with dhc1, jnm1, and act5 deletion mutants (Miller and Rose, 1998). Unlike the dhc1 mutant, kar9 mutants have misoriented cytoplasmic microtubules in both mitosis and mating. In both cases it is likely that microtubule misorientation results in disrupted nuclear positioning (Miller and Rose, 1998). GFP-tagged Kar9p localizes to a single cortical spot at the tip of the bud and at the tip of mating projections. Because GFP-Kar9p localization at the cell cortex is independent of microtubules, yet required for their orientation, Kar9p may function as a cortical target for the capture of the cytoplasmic microtubules. Capture and stabilization of the cytoplasmic microtubules would then provide a mechanism for the orientation of the microtubules and the mitotic spindle (Miller and Rose, 1998).
It was initially surprising that dynein’s role in positioning the nucleus during nuclear migration is not essential for life. Several general models can be advanced to explain this observation. First, random motion of the nucleus, together with spindle elongation, would allow the nucleus to enter the bud during anaphase. While the proper orientation might occur infrequently, elongation into the bud would be irreversible. Second, other microtubule-dependent motors might provide the force for nuclear movement. Third, the intrinsic dynamic properties of microtubules might provide sufficient force for movement. Support for the latter two hypotheses comes from the observation that the tub2–401 mutation selectively destabilizes the cytoplasmic microtubules and leads to a much more severe nuclear migration defect than the dynein deletion. Candidate motor proteins that might provide compensatory or redundant forces in the absence of dynein include the kinesin-related motor proteins. Of the six kinesin-related genes in S. cerevisiae, three (KAR3, KIP1 and CIN8) have been shown previously to play a role in spindle function (Meluh and Rose, 1990; Hoyt et al., 1992; Roof et al., 1992; Saunders et al., 1997).
The kinesin-related gene KIP2 was previously identified in a screen for kinesin-related genes using degenerate PCR primers to conserved regions of kinesin-related motor domains (Roof et al., 1991, 1992). Unlike conventional kinesin, the Kip2p motor domain is positioned in the center of the molecule. Preliminary work reported that kip2 mutants exhibited a defect in nuclear migration and were not synthetically lethal with dynein mutants (Miller and Rose, 1995). Subsequent work confirmed the role of Kip2p in nuclear migration (Cottingham and Hoyt, 1997). The kinesin-related gene KIP3 was identified during the completion of the yeast genome-sequencing project and encodes an 805-amino acid protein with an N-terminal motor domain. This and concurrent work (Cottingham and Hoyt 1997; DeZwaan et al., 1997) demonstrate that kip3Δ mutants are also defective for nuclear migration during mitosis. DeZwaan et al. (1997) showed that nuclear positioning is random and that the mitotic spindle is misoriented in preanaphase kip3Δ cells. Based upon genetic and morphological data, DeZwaan et al. (1997) proposed that Kip3p and dynein act at different temporal steps to complete anaphase. In addition, Cottingham and Hoyt (1997) provided genetic evidence that suggests that Kip2p and Kip3p act antagonistically to position the mitotic spindle.
The genetic and morphological studies presented in this paper confirm that both Kip2p and Kip3p affect nuclear migration and revealed that they do so via different mechanisms. Our conclusions are based primarily upon the differences observed for kip2Δ and kip3Δ with respect to genetic interaction profiles, microtubule morphology, and cellular localization. The kip3Δ mutation was synthetically lethal with a deletion mutation in dynein (also shown by Cottingham and Hoyt, 1997; DeZwaan et al., 1997) and with deletions in components of the dynactin complex. In contrast, kip2Δ mutations were not synthetically lethal in combination with deletions in dynein (also shown by Cottingham and Hoyt, 1997) or with deletions in components of the dynactin complex. Conversely, the kip2Δ mutations were found to be synthetically lethal with kar9Δ, whereas kip3Δ kar9Δ double mutants were viable. Furthermore, Kip2p was localized to the cytoplasmic microtubules throughout the cell cycle. In contrast, Kip3p was found on both the spindle and cytoplasmic microtubules (also shown by DeZwaan et al., 1997). These results support the view that there are at least two partially redundant pathways for nuclear migration, one of which requires Kar9p and the second of which requires dynein. Overall, our findings add to our expanding knowledge of the components involved in positioning the nucleus during the cell cycle.
MATERIALS AND METHODS
Yeast Strains, Microbial Techniques, and Growth Assays
Yeast strains used in this study are listed in Table 1. Plasmids and bacterial strains are listed in Table 2. Yeast media and standard genetic techniques were essentially as previously described (Rose et al., 1990).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| MS17 | MATα trp1-Δ1 ura3-52 ade2-101 | |
| MS52 | MATα leu2-3 leu2-112 trp1-Δ1 ura3-52 | |
| MS660 | MATα kar3-101::LEU2 leu2-3 leu2-112 ura3-52 his4-539 trp1-Δ1 | |
| MS1554 | MATa ura3-52 leu2-3 leu2-112 ade2-101 his3-Δ200 | |
| MS2305 | MATα kip1-Δ::URA3 ura3-52 leu2-3 leu2-112 trp1-Δ1 | |
| MS2309 | MATa kip2-Δ1::URA3 ura3-52 leu2-3 leu2-112 ade2-101 his3-Δ200 | |
| MS2310 | MATα kip2-Δ1::URA3 ura3-52 leu2-3 leu2-112 trp1-Δ1 | |
| MS2354 | MATα kip2-Δ::TRP1 ura3-52 leu2-3 leu2-112 trp1-Δ1 | |
| MS2442 | MATα kip2::TRP1 trp1-Δ1 leu2-3 leu2-112 his3-Δ200 ura3-52 ade2-101 | |
| MS4062 | MATα kar9-Δ1::LEU2 leu2-3 leu2-112 trp1-Δ1 ura3-52 | |
| MS4262 | MATα dhc1-Δ::URA3 leu2-3 leu2-112 trp1-Δ1 ura3-52 | |
| MS4263 | MATa kar9-Δ1::LEU2 leu2-3 leu2-112 ade2-101 his3-Δ200 ura3-52 | |
| MS4304 | MATa dhc1::URA3 ura3-52 leu2-3 leu2-112 trp1-Δ1 his3-Δ200 | |
| MS4308 | MATa kip2-Δ::URA3 trp1-Δ1 ade2-101 his3-Δ200 ura3-52 leu2-3 leu2-113 | |
| MS4316 | MATα kar9-Δ1::LEU2 leu2-3 leu2-112 ura3-52 ade2-101 his3-Δ200 | |
| MS4321 | MATa jnm1-Δ::LEU2 ura3-52 leu2-3 leu2-112 ade2-101 his3-Δ200 | |
| MS4516 | MATa kip3-Δ::HIS3 his3-Δ200 ade2-101 leu2-3 leu2-112 ura3-52 | |
| MS4586 | MATα act5-Δ::HIS3 trp1-Δ63 lys2-801 his3-Δ200 ade2-101 ura3-52 leu2 | |
| MS4600 | MATα dhc1-Δ::URA3 ura3-52 ade2-101 trp1-Δ1 his4-539 | |
| MS4648 | MATα kip3-Δ::HIS3 kar9-Δ::LEU2 his3-Δ200 ade2-101 leu2-3 leu2-112 ura3-52 | |
| MS4654 | MATα kip3-Δ::HIS3 kar9-Δ::LEU2 his3-Δ200 ade2-101 leu2-3 leu2-112 ura3-52 | |
| MS4691 | MATa kip3-Δ::HIS3 ura3-52 his3-Δ200 leu2-3 leu2-112 trp1 | |
| MS4692 | MATα jnm1-Δ::LEU2 leu2-3 leu2-112 his3-Δ200 ura3-52 trp1-Δ1 | |
| MS4701 | MATα act5-Δ::TRP1 trp1-Δ1 his3-Δ200 ade2-101 ura3-52 | |
| MS4719 | MATa/MATα kip3-Δ::HIS3/kip3-Δ::HIS his3-Δ200/his3-Δ200 ade2-101/ADE2 leu2-3/leu2-3 leu2-112/leu2-112 TRP1/trp1-Δ1 ura3-52/ura3-52 | |
| MS4720 | MATa/MATα ura3-52/ura3-52 leu2-3/leu2-3 leu2-112/leu2-112 ade2-101/ADE2 his3-Δ200/HIS3 trp1-Δ1/TRP1 | |
| MS4734 | MATα bik1-Δ::TRP1 leu2-3 leu2-113 trp1-Δ1 ura3-52 | |
| MS4903 | MATa dhc1::LEU2 ura3-52 leu2-3 leu2-112 ade2-101 his3-Δ200 | |
| MS4915 | MATα bik1::TRP1 leu2-3 leu2-112 trp1-Δ1 ura3-52 his3-Δ200 ade2-101 | |
| MS5001 | MATa dhc1-Δ::URA3 ura3-52 leu2-3 leu2-112 ade2-101 his3-Δ200 | |
| MS5210 | MATa/MATα kar9-Δ1::HIS3/KAR9 kip2-Δ::TRP1/KIP2 leu2-3/leu2-3 leu2-112/leu2-112 ura3-52/ura3-52 ade2-101/ADE2 his3-Δ200/his3-Δ200 trp1-1/trp1-Δ1 | |
| MS5214 | MATa kip2-Δ::TRP1 dhc1-Δ::URA3 ura3-52 leu2-3 leu2-112 trp1-Δ1 his3-Δ200 | |
| BY397 | MATα act5-Δ1-366 trp1-Δ1 ade2-101 ura3-52 | Clark |
| APY4ΔD5 | MATa smy1Δ::URA3 ura3-52 his6 leu2-3 leu2-112 trp1 ade2 | Brown |
| MAY2058 | MATα cin8Δ::LEU2 leu2-3 leu2-112 his3-Δ200 ura3-52 lys2-801 | Hoyt |
| MAY2059 | MATa cin8Δ::LEU2 his3-Δ200 leu2-3 leu2-112 ura3-52 ade2-101 | Hoyt |
Unless indicated otherwise, all strains are from this laboratory.
Table 2.
Plasmids used in this study
| Strain | Genotype | Source |
|---|---|---|
| pMR3144 | KIP2 CEN URA3 | |
| pMR3465 | PGAL-GFP-KAR9 LEU2 CEN4 ARS1 | |
| pMR3769 | GFP#2 | Leibler |
| pMR3777 | KIP2::HA CEN URA3 | |
| pMR3778 | KIP2 2μ URA3 | |
| pMR3779 | KIP2::HA 2μ URA3 | |
| pB1893 | PGAL1-GFP CEN4 ARS1 LEU2 | Broach |
| pMR3635 | PGAL1-GFP-KIP3 CEN4 ARS1 LEU2 | |
| pMR3889 | PGAL1-GFP-KIP2 CEN URA3 | |
| pRS403 | Yip-HIS3 | Hieter |
| pRS426 | 2μ URA3 | Hieter |
| pTRP17-20 | act5Δ1-366::TRP1 disruption plasmid | Clark |
| pJM1432 | jnm1::LEU2 disruption plasmid | Tatchell |
| pBR2-1U | dhc1::URA3 disruption plasmid | Bloom |
| pVB17 | bik1::TRP1 disruption plasmid | Fink |
| pGTEP1 | Triple HA epitope | Futcher |
Unless indicated otherwise, all plasmids are from this laboratory.
Double-deletion mutants were created by standard genetic techniques. The growth phenotypes of spores were scored on the second and third day of growth after tetrad dissection. Colonies were classified as showing normal growth, microcolonies, or no growth. To examine the temperature sensitivity of the germinating spores, 10 tetrads from each cross were dissected onto YPD plates and incubated at 37°, 35°, 23°, 16°, and 14°C.
The growth rates of the kip3Δ::HIS3 strain (MS4516) and the wild-type strain (MS1554) were evaluated as follows. The strains were grown in YPD medium at 30°C until the cultures were in the exponential phase of growth. The cultures were then diluted 10-fold into YPD that had been preincubated at 37°, 30°, 23°, or 14°C. Optical density readings were taken at various time points to estimate cell numbers. Tetrads from a heterozygous kip3Δ/KIP3 diploid were dissected on YPD medium, and identical plates containing 10 tetrads each were incubated at 14°, 16°, 23°, 30°, and 37°C. At all temperatures the kip3Δ strain grew identically to wild type.
DNA Manipulations
For PCR analysis, genomic DNA was prepared from strains MS4516 and MS1554 by a modification of the CsCl method (Rose et al., 1990). DNA preparations for transformations and plasmid construction were made as described previously (Rose et al., 1990). Plasmid transformations into yeast were carried out by the lithium acetate method (Ito et al., 1983). Transformations of bacteria were performed by standard electroporation techniques. Gene disruptions were done as described previously (Rothstein, 1991).
Strain Construction
KIP2 was previously identified using a PCR-based homology screen (Roof et al., 1992). KIP3 was identified by the yeast genome-sequencing project and corresponds to ORF YGL216W. A strain lacking the entire KIP3- coding region was created by the one-step gene replacement method (Rothstein, 1991). The disruption fragment was generated by PCR using the following two oligonucleotides (Syn/Seq Facility, Princeton University, Princeton, NJ). The primer sequences were (KIP3 sequence is in italics, HIS3 is in roman font): 5′-TAC TTG AGT TTT CTT TCC AGC TGT ATA CTA TTG ACA CTA ACA TGC CGT TTT AAG AGC TTG GTG-3′; and 5′- GAA AGA AGT TAT ATT CGA TAG TTT ACG TAG GAT ATG TAT GGT CGA GTT CAA GAG AAA AAA-3′. Plasmid pRS403 (Sikorski and Hieter, 1989) was used as the template for the HIS3 portion of the PCR construct. Transformation of a haploid strain (MS1554) produced a kip3Δ::HIS3 strain (MS4516). The replacement of the entire KIP3 coding region from the ATG start site through the last codon of KIP3 by HIS3 was confirmed at both ends by PCR analysis. All kip3Δ strains described in this study were derived from backcrosses of MS4516.
MS4262, a dhc1Δ::URA3 disruption in a S288C isogenic strain was made as described elsewhere (Miller and Rose, 1998) using pBR2–1U (Li et al., 1993). MS4262 was backcrossed once to create MS4304, a strain with the required auxotrophic markers. MS4321, the jnm1Δ::LEU2 disruption in a S288C isogenic strain, was made as described elsewhere (Miller and Rose, 1998) using pJM1432 (McMillan and Tatchell, 1994). MS4321 was backcrossed once to obtain strain MS4692 used in the analysis. An act5Δ1–366::TRP1 disruption strain BY397 (from Sean Clark, Princeton University) was generated from MS17 using the plasmid pTRP17–20 (Clark and Meyer, 1994) digested with SacI and XhoI. This strain was backcrossed once to obtain MS4701. MS4734, a bik1Δ strain, was made as described previously (Miller and Rose, 1998) using plasmid pVB17 (Fink Laboratory, Whitehead Institute, Cambridge, MA). This strain was backcrossed once to obtain MS4915.
Nuclear and Microtubule Morphology in Mitotic and Mating Cells
To assay for defects in mitosis, strains were grown to exponential phase at 30°, 14°, and 12°C. After 4 h at 30°C (24 h at 14° and 12°C), cells were centrifuged and fixed in methanol:acetic acid (3:1) for 30 min on ice. After washing in PBS, cells were stained with the fluorescent DNA dye DAPI at 1 μg/ml for 30 min. The cells were then analyzed microscopically using differential interference contrast optics to assess the cellular morphology and UV fluorescence to evaluate the nuclear morphology.
To assay karyogamy, selected strains of opposite mating types were grown to early exponential phase at 23°C, mixed in a 1:1 ratio, and collected by filtration on 0.45-μm filters (Millipore, Bedford, MA). The mating mixtures on the filters were incubated on YPD media for 2.5 h at 30°C. Cells were then resuspended in PBS, fixed in methanol-acetic acid, and stained with DAPI to visualize nuclear material as described above.
To visualize microtubules, indirect immunofluorescence was carried out as described previously (Rose et al., 1990). Selected strains were grown to exponential phase at 30°C and shifted to 14°C for 16–24 h. Microtubules were visualized with the rat anti-tubulin antibody YOL1/34 (Accurate Chemical and Scientific, Westbury, NY) undiluted, and FITC-conjugated goat anti-rat secondary antibody (Amersham, Arlington Heights, IL) at a 1:100 dilution.
Localization of Kip2p
To localize Kip2, in some experiments a hemagglutinin (HA) epitope-tagged form of KIP2 was used. A fragment coding for a triple HA tag with HindIII ends was synthesized by PCR using the GTEPI plasmid (from Bruce Futcher, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), as template. The following primers were used: 5′-GCC CAA GCT TAT ACC CAT ACG AT-3′, and 5′-GCC TTA AGC TTG CAG CGT AAT CTG G-3′. The KIP2-containing CEN plasmid pMR3144 was cut with HindIII, and the HA-containing insert was ligated into it to produce the plasmid pMR3777. To create a 2μ version of this construct, pMR3777 was cut with KpnI and SacI, and the 2.6-kilobase (kb) fragment containing KIP2::HA was gel purified and ligated to pRS426 (Sikorski and Hieter, 1989), to produce pMR3779. To construct a nonepitope-tagged control plasmid, the KpnI/SacI fragment containing KIP2 was excised from pMR3144 and ligated to pRS426 (Sikorski and Hieter, 1989), creating the 2μ KIP2 URA3 plasmid, pMR3778.
To localize Kip2p-HA, a kip2Δ strain (MS2354) containing pMR3779 was fixed using a modified procedure described elsewhere (Roberts et al., 1991). Five-milliliter cultures of cells grown to midexponential phase were fixed in 4% paraformaldehyde for 4 h at 23°C. No preceding formaldehyde fixation was used. Cells were then incubated in TEB (200 mM Tris-HCl, pH 8.0, 20 mM EDTA, 1% 2-mercaptoethanol, prepared fresh) for 10 min at 23°C. Cells were resuspended in SPM (1.2 M sorbitol, 50 mM potassium phosphate, pH 7.3, 1 mM MgCl2) and stored at 4°C for 16 h. The cell walls were digested for 1 h at 30°C with 50 μl Glusulase (DuPont, Wilmington, DE) and 15 μl Zymolyase 100T (10 mg/ml) (ICN Immunobiologicals, Lisle, IL). Cells were extracted with 1% SDS/1.2 M sorbitol/PBS for 10 min at 23°C. The extracted cells were then diluted fivefold with 1.2 M sorbitol/PBS and washed three times with 1.2 M sorbitol/PBS. The cells were then attached to poly-l-lysine coated coverslips and blocked with BSA/PBS (5 mg/ml) for 10 min. Monoclonal HA antibody (12CA5) was used at 1:300 dilution and goat anti-mouse FITC-conjugated secondary was used at 1:25 dilution. Both antibodies were preabsorbed using a kip2Δ strain.
In other experiments a GFP-Kip2p hybrid was used to localize Kip2p. For this purpose, plasmid pMR3769 (M. Elowitz and S. Leibler, Princeton University) containing Aquoreus victoria GFP mutant 2 (Cormack, 1996) was used as template in a PCR reaction to generate a GFP fragment flanked by HindIII recognition sites. The following oligonucleotides were used as primers: 5′-CGG CGC CCA AGC TTG ATG AGT AAA GGA GAA G-3′, and 5′-CCC AAG CTT TTG TAT AGT TCA TCC ATG-3′. The resulting PCR product was digested with HindIII and ligated into the HindIII site of KIP2 on pMR3144, creating pMR3889. This construct complemented the growth defect of a kip2Δ kar9Δ double mutant, produced by transforming and sporulating the kip2Δ/KIP2 kar9Δ/KAR9 heterozygous diploid, MS5210.
To visualize GFP-Kip2p, strains containing pMR3889 were grown at 23°C to early exponential phase in synthetic complete media minus uracil. To assess nuclear material, cells were fixed in 3.7% formaldehyde for 5 min, washed twice in PBS, stained with DAPI for 5 min, and washed in PBS twice as was described above. Cells were viewed using a Ziess Axiophot microscope equipped with a High Q FITC filter set (no. 41001 from Chroma Technology, Brattleboro, VT), a 100× plan-neufluoar objective (1.3 N.A.) (Carl Zeiss, Thornwood, NY), and a cooled charge-couple device camera (Princeton Instruments, Princeton, NJ) connected to a Hamamatsu Video Camera 3200 and a Hamamatsu Image Processor C2400 (Hamamatsu, Hamamatsu City, Japan).
Visualization of GFP-Kip3p and GFP-Kar9p
The KIP3 gene was amplified with PCR using the following flanking primers: 5′-CCG CCG TCG ACT ATT GAC ACT AAC ATG-3′ and 5′-CGG GAT CCG CTG GCG GAA AGA AGT TA-3′. The PCR product was digested with SalI and BamHI and ligated into a PGAL-GFP#1 vector pB1893 (Corey Davis and James Broach, Princeton University) cut with the same enzymes to create the PGAL-GFP-KIP3 construct, pMR3635.
A wild-type strain (MS1554) containing the PGAL-GFP-KIP3 construct was grown at 30°C to early exponential phase in synthetic complete medium minus leucine containing 2% raffinose. GFP-Kip3p expression was induced by the addition of 2% galactose for 3–4 h. The localization was determined using the microscopy conditions described for GFP-Kip2p.
The localization of GFP-Kar9p was performed and scored as described previously (Miller and Rose, 1998) except that glucose-modulated galactose inductions for Kar9p-GFP expression were not used; instead, the expression was fully induced using 2% galactose for 2–3 h.
RESULTS
kip2Δ and kip3Δ Mutants Display Defects in Mitosis but Not in Karyogamy
To assess the functions of the two kinesin-related genes, KIP2 and KIP3, strains containing deletions of each were constructed. In both cases, the deletion strains were viable and exhibited wild-type growth rates at a wide range of temperatures. We next examined them for more subtle defects in cell cycle progression. During mitosis in wild-type cells, the nucleus migrates to the neck of the mother–bud junction and anaphase occurs through the space of the neck (Kahana et al., 1995; Yeh et al., 1995). In both the kip2Δ and kip3Δ strains, a variety of nuclear migration defects could be observed. These included 1) large-budded cells with a single nucleus that had failed to migrate to the bud neck; 2) large-budded cells with anaphase occurring exclusively within the mother cell; and 3) large-budded cells with the mother cell containing two nuclei (Figure 1 and Table 3A). Such aberrant cells occurred infrequently in the isogenic wild-type strain (∼2%). In contrast, at 30°C, ∼7% of the kip2Δ cells exhibited nuclear migration defects (Table 3A). At 12°C, the frequency of aberrant cells increased to ∼20%, while in the wild-type strain the frequency remained at 2%. Most of the increase occurred in the class of large-budded cells with two nuclei in the mother cell (e.g., for the kip2Δ::TRP1 strain, the number of binucleate mother cells increased from 2 to 16%).
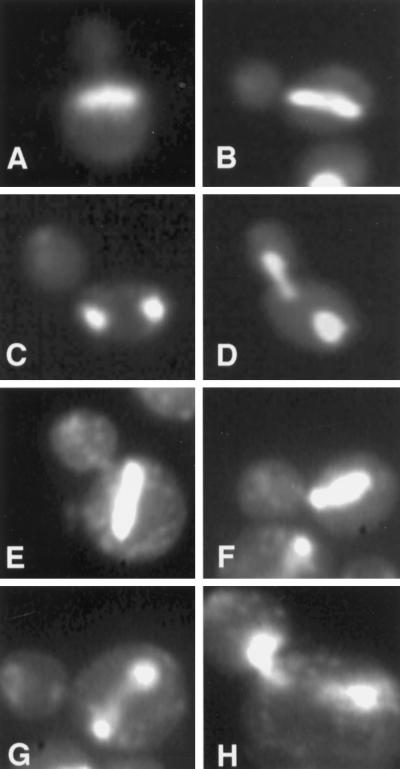
Figure 1.
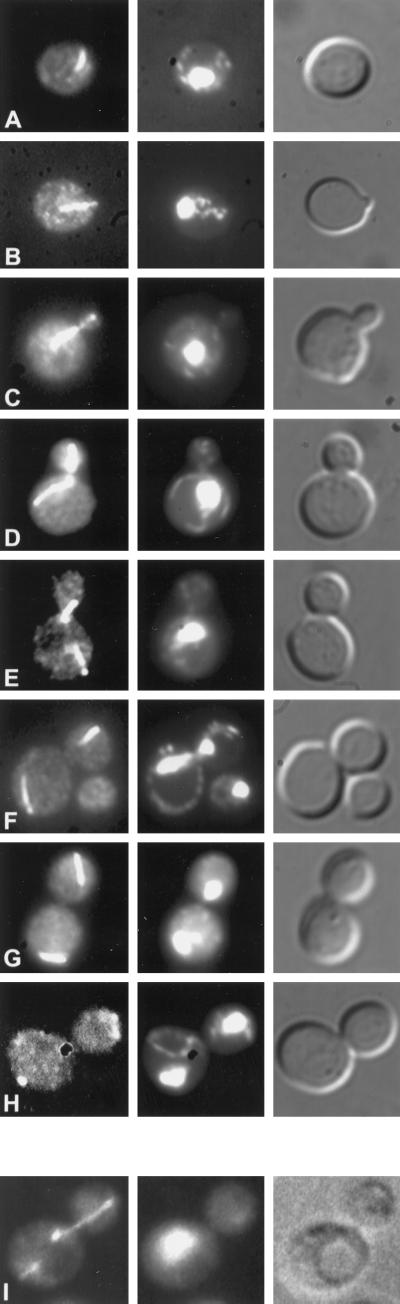
DAPI staining of abnormal nuclear morphologies in kip2Δ and kip3Δ mutants. The kip2Δ strain, MS2309 (A–D), and the kip3Δ strain, MS4516 (E–H), were grown to early exponential phase in YPD medium at either 14°C (MS2309) or 12°C (MS4516) and stained with DAPI to visualize the nuclear material.
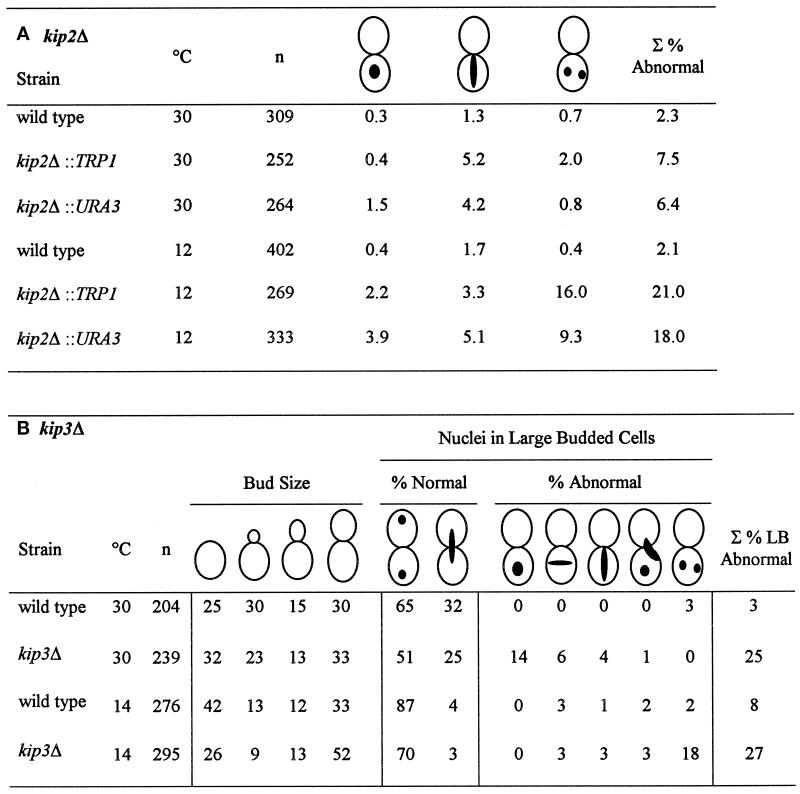
Table 3.
Nuclear migration defects of kip2Δ and kip3Δ mutants
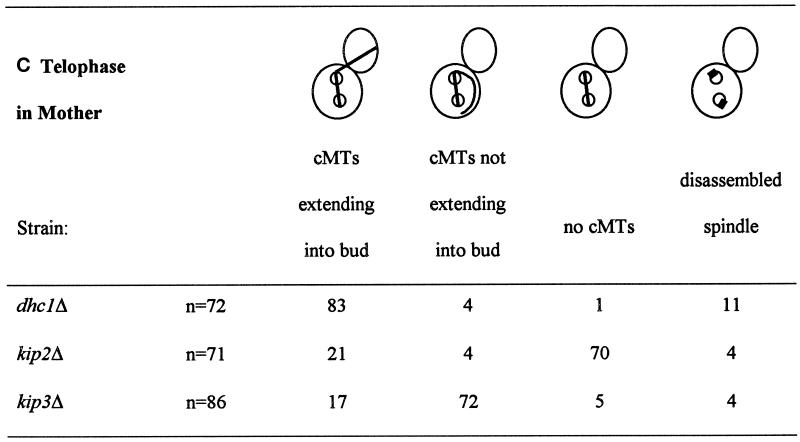
For both panels A and B, a representative experiment is shown. In repeat experiments, percentages vary by about 5%. (A) Percentage of nuclear positioning defects in kip2Δ mutant cultures. Cultures of kip2Δ::TRP1 (MS2354), kip2Δ::URA3 (MS2310), and wild-type, WT (MS52) strains were grown to early exponential phase at 12° and 30°C. The position of nuclear DNA was visualized by DAPI. The percentages of abnormal nuclear position phenotypes are shown relative to the total cell culture. The defects scored included: 1) large-budded cells with a single nucleus in the mother cell, 2) mitosis occurring within the mother cells, and 3) large-budded cells with two nuclei per mother cell and are depicted schematically above the columns. ∑ represents the percentage of the total culture with abnormal mitotic phenotypes. (B) The percentage of nuclear positioning defects in kip3Δ mutant cultures. A wild-type (WT) strain (MS1554) and a kip3Δ strain (MS4516) were grown to early exponential phase in YPD medium at 14° and 30°C and then stained with DAPI to visualize nuclear material. Each strain was scored for bud size as unbudded, small, medium, or large budded. Large-budded cells were also examined for their nuclear phenotype and scored as either normal or abnormal (nuclear morphology corresponding to the phenotypes schematically depicted). The percentages of bud size are from the total cells counted (n > 200). The percentages for the nuclear phenotypes are derived from the 30 to 52% of large-budded cells. The percentage of total defects in large-budded cells is shown in the ∑% LB Abnormal column.
For the kip3Δ strain at 30°C, 9% of the culture (25% of the large- budded cells) displayed defects in nuclear positioning (Table 3B) compared with 3% for wild type. At 14°C, the frequency of aberrant cells increased to 15% (27% of large-budded cells). While the total increase in the frequency of aberrant cells was not as large as the kip2Δ strain, the specific spectrum of defects for the kip3Δ strain shifted dramatically. At 30°C, in most of the aberrant cells the nuclei failed to migrate to the bud neck. In contrast, at 14°C most of the aberrant cells were binucleate mother cells (increased from 0 to 18%). In addition, the percentage of cells that were unbudded binucleates increased from 0 to 5% at 14°C (our unpublished observations). Growth in the cold also caused a shift in the cell cycle distribution as indicated by bud size. At 30°C, 33% of kip3Δ mutants were large budded; at 14°C, 52% were large budded (Table 3B). Despite these mitotic defects, the kip3Δ mutants did not show any growth defects at a variety of temperatures compared with wild type (see MATERIALS AND METHODS).
Clearly, both the kip2 and kip3 mutant strains displayed nuclear positioning defects in mitosis. For this reason, we wanted to determine whether these genes also played a role in karyogamy, another cellular function involving nuclear migration. During mating, cells arrest in G1 in response to mating pheromones and initiate polarized growth toward each other. The intervening cell wall breaks down and the two cells fuse. The zygotic nuclei then migrate toward each other and fuse, forming a diploid zygote. This movement is microtubule dependent and requires the Kar3p motor protein (Meluh and Rose, 1990; Kurihara et al., 1994; Marsh and Rose, 1997). As a direct assay for nuclear fusion, microscopic examination of zygotes was performed. In wild-type zygotes, more than 98% have a single fused nucleus before the appearance of the first bud. In matings of kip3Δ crossed to wild type and matings of MATa kip3Δ crossed to MATα kip3Δ, greater than 95% of the unbudded zygotes contained a single fused nucleus. Similar microscopic analyses of kip2Δ matings revealed no defect in nuclear fusion, confirming previously reported results using a quantitative mating assay (Roof et al., 1992). Thus, kip2Δ and kip3Δ mutants do not have karyogamy defects. These results confirm that Kar3p is the sole kinesin responsible for nuclear migration during mating (Meluh and Rose, 1990).
Distinct Microtubule Morphologies in kip2Δ and kip3Δ
Since nuclear migration and spindle orientation have been shown to depend on the cytoplasmic microtubules (Sullivan and Huffaker, 1992), we next examined whether deletions in either KIP2 or KIP3 caused altered microtubule morphology. In wild-type cells at telophase, the mitotic spindle aligns along the mother–bud axis, extending directly from the nuclear material in the mother cell through the nuclear material in the bud (Figure 2A). The cytoplasmic microtubules at this stage appeared as short extensions at one or both ends of the spindle in 79% of the wild-type cells (Figure 2A and Table 4A). One striking feature of the kip2Δ cultures grown at 14°C was the absence of the cytoplasmic microtubules in 53% of the telophase cells (Figure 2D and Table 4A). In contrast, in wild-type cultures only 14% of the telophase cells displayed no discernible cytoplasmic microtubules. These results suggest that Kip2p may play a role in stabilizing the cytoplasmic microtubules at this stage of the cell cycle. Interestingly, in the dhc1Δ strain, 22% of the cells had no obvious cytoplasmic microtubules, indicating that dynein may also play a minor role in stabilizing the cytoplasmic microtubules. In contrast to kip2Δ and dhc1Δ, in the kip3Δ culture only 5% of the telophase cells lacked cytoplasmic microtubules (Figure 2, M–U, and Table 4A). Indeed, under these conditions 35% of the kip3Δ cells had abnormally long cytoplasmic microtubules (Figure 2 M and Table 4A). This aberrant morphology was not observed in the kip2Δ and dhc1Δ mutants or the wild-type strain. The long microtubules most often extended from the bud tip back into the mother cell (Figure 2M). Regardless of their abnormal microtubule phenotypes, more than 80% of the kip3Δ cells had segregated their nuclei normally. The effect on microtubules was temperature dependent because the kip3Δ cultures grown at 30°C appeared to have normal microtubules.
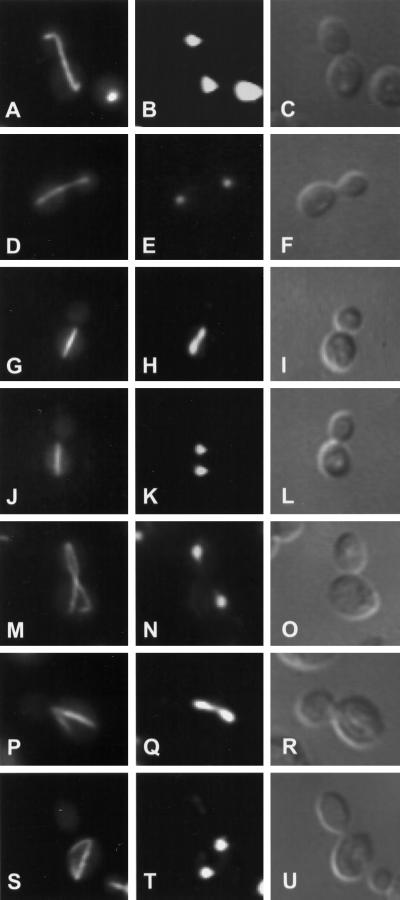
Figure 2.
Microtubule morphologies of kip2Δ and kip3Δ mutants. The wild-type strain, MS1554 (A–C), the kip2Δ strain, MS2309 (D–L), and the kip3Δ strain, MS4516 (M–U), were grown to early exponential phase at 14°C. Cells were fixed in formaldehyde and stained with anti-tubulin YOL1/34 antibody (A, D, G, J, M, P, and S). Cells were also stained with DAPI to visualize nuclear material (B, E, H, K, N, Q, and T). Visualization of cells by Nomarski optics is seen in panels C, F, I, L, O, R, and U.
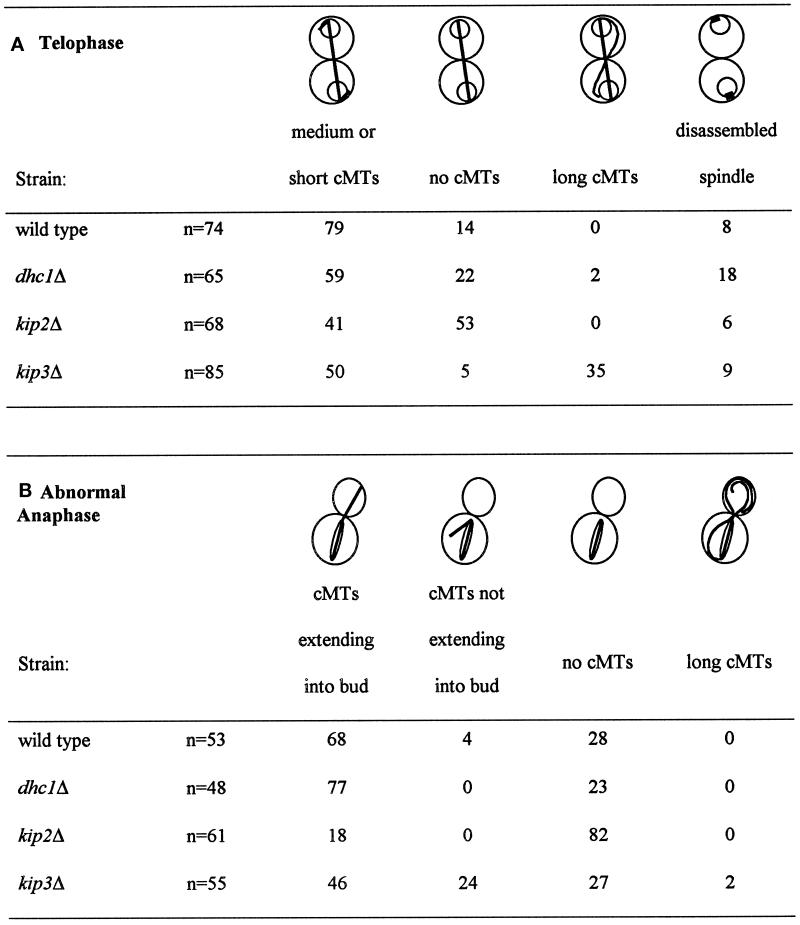
Table 4.
Quantification of microtubule morphologies
The wild-type strain (MS1554), the kip2Δ strain (MS2309), the kip3Δ strain (MS4516), and the dhc1Δ strain (MS4903) were grown to early exponential phase at 14°C and fixed for indirect immunofluorescence with anti-tubulin (see MATERIALS AND METHODS). The data for the wild-type and dhc1Δ controls were also reported by Miller and Rose (1998).
In addition to the cells showing a normal telophase, we specifically examined the microtubules in cells showing abnormal nuclear morphologies (Table 4, B and C, and Figure 2, G, J, P, and S). In the wild-type strain at 14°C, 28% of the cells in which mitosis was occurring within the mother cell had no detectable cytoplasmic microtubules (Table 4B). However, in the kip2Δ strain, 70–82% of the cells had no detectable cytoplasmic microtubules, regardless of whether nuclear division had been completed (Figure 2, G and J, and Table 4, B and C).
The morphology of the cytoplasmic microtubules in the kip3Δ mutants was strikingly different from that of the kip2Δ mutants. In anaphase, the percent of kip3Δ cells without cytoplasmic microtubules was virtually identical to wild type. However, many of the kip3Δ mutants had cytoplasmic microtubules that were misoriented and failed to extend into the bud (Figure 2, P–U, and Table 4, B and C). In the kip3Δ cells in which anaphase was occurring within the mother cell, 24% exhibited misoriented cytoplasmic microtubules. This frequency was similar to the 30% misoriented cytoplasmic microtubules found in the kar9Δ mutant (Miller and Rose, 1998). When two nuclei were present within the mother cell, 72% of kip3Δ cells had misoriented cytoplasmic microtubules (Figure 2S and Table 4C). This frequency was similar to the 70% misoriented microtubules observed in the equivalent class of kar9Δ cells (Miller and Rose, 1998). While the kip3Δ microtubule orientation defect is similar to that observed for the kar9Δ mutant, abnormally long cytoplasmic microtubules were not observed in kar9Δ strains (Kurihara et al., 1994; Miller and Rose, 1998). In contrast to kip3Δ, the dhc1Δ mutants nearly always contained a cytoplasmic bundle that extended into the bud, even in aberrant cells exhibiting nuclear positioning defects (Table 4, B and C) in agreement with previous reports (Li et al., 1993).
To summarize the microtubule morphology data, both kip2Δ and kip3Δ influence the length of the cytoplasmic microtubules but appear to have opposite effects. By inference, Kip2p seems to be required to stabilize or assemble the cytoplasmic microtubules, whereas Kip3p appears to destabilize them. In addition, Kip3p may have a second function that is important for the orientation of cytoplasmic microtubules.
Genetic Analysis of kip2Δ Mutants Places Kip2p in the Dynein/Dynactin Pathway of Nuclear Positioning
Our previous work showed that DHC1 and KAR9 function in partially redundant pathways for nuclear migration (Miller and Rose, 1998). Because the mitotic defects observed in the kip2Δ and kip3Δ mutants were similar to those seen in dhc1Δ and kar9Δ mutants, we wanted to determine whether Kip2p and Kip3p might function in either of these two pathways. To carry out the analysis, we performed crosses with strains bearing deletions in each of the kinesin-related genes along with several other relevant genes required for spindle function and/or nuclear positioning. The viability and growth characteristics of the resulting double-mutant strains were then used as an assay for possible genetic interactions. Table 5 summarizes the results from the crosses, and Figure 3 depicts a typical cross in which a synthetic growth defect was apparent.
Table 5.
Viability of double mutants in combination with kip2Δ or kip3Δ
| Mutant combination | Tetrads analyzed | No. of predicted mutants | % of mutants dead or forming microcolonies | Viability of double or triple mutant |
|---|---|---|---|---|
| 1. kip2Δ kar9Δ | 33 | 33 | 100 | SL |
| 2. kip2Δ dhc1Δ | 33 | 31 | 6 | viable |
| 3. kip2Δ jnm1Δ | 11 | 10 | 0 | viable |
| 4. kip2Δ act5Δ | 12 | 13 | 0 | viable |
| 5. kip2Δ bik1Δ | 10 | 11 | 0 | viablea |
| 6. kip2Δ cin8Δ | 21 | 18 | 100b | SL |
| 7. kip2Δ kip1Δ | — | — | — | viablec |
| 8. kip2Δ kar3Δ | — | — | — | viablec |
| 9. kip2Δ smy1Δ | 8 | 7 | 0 | viable |
| 10. kip3Δ kar9Δ | 29 | 23 | 12 | viable |
| 11. kip3Δ dhc1Δ | 16 | 15 | 100 | SL |
| 12. kip3Δ jnm1Δ | 29 | 29 | 100 | SL |
| 13. kip3Δ act5Δ | 13 | 9 | 100 | SL |
| 14. kip3Δ bik1Δ | 13 | 12 | 100 | SL |
| 15. kip3Δ cin8Δ | 18 | 24 | 75d | SL |
| 16. kip3Δ kip1Δ | 9 | 9 | 0 | viable |
| 17. kip3Δ kar3Δ | 17 | 13 | 100 | SL |
| 18. kip3Δ kip2Δ | 17 | 14 | 0 | viable |
| 19. kip2Δ dhc1Δ kar9Δ | 18 | 8 | 100 | SL |
| 20. kip3Δ dhc1Δ kar9Δ | 12 | 5 | 100 | SL |
| 21. kip2Δ kip3Δ kar9Δ | 54 | 19 | 100 | SL |
Standard meiotic crosses were used to construct the indicated double mutants. The size of colonies was scored 2-3 d after germination. SL indicates that synthetic lethality was observed. The following crosses were carried out to create the indicated double and triple mutants: 1. kip2Δ kar9Δ, MS2310 × MS4263; 2. kip2Δ dhc1Δ, MS2354 × MS4304; 3. kip2Δ jnm1Δ, MS2310 × MS4321; 4. kip2Δ act5Δ, MS4308 × MS4586; 5. kip2Δ bik1Δ, MS4308 × MS4734; 6. kip2Δ cin8Δ, MS2310 × MAY2059; 9. kip2Δ smy1Δ, APY4ΔD5 × MS2354; 10. kip3Δ kar9Δ, MS4516 × MS4316; 11. kip3Δ dhc1Δ, MS4516 × MS4600; 12. kip3Δ jnm1Δ, MS4692 × MS4516; 13. kip3Δ act5Δ, MS4691 × MS4701; 14. kip3Δ bik1Δ, MS4691 × MS4915; 15. kip3Δ cin8Δ, MS4516 × MAY2058; 16. kip3Δ kip1Δ, MS4516 × MS2305; 17. kip3Δ kar3Δ, MS4516 × MS660; 18. kip3Δ kip2Δ, MS2442 × MS4691; 19. kip2Δ dhc1Δ kar9Δ, MS5214 × MS4062; 20. kip3Δ dhc1Δ kar9Δ, MS4654 × MS4304; and 21. kip2Δ kip3Δ kar9Δ, MS4308 × MS4648.
Identical results were observed when kip2Δ was crossed to a bik1Δ in a nonisogenic strain (our unpublished observations).
The viability of cin8Δ single mutants in this cross was 52%. This double mutant was also examined by plasmid loss assay on 5-FOA using a KIP2 URA3 CEN plasmid, which demonstrated that the cin8Δ kip2Δ double mutant lost a KIP2 CEN plasmid much less frequently than either wild-type or either single mutant.
Data from Roof et al. (1992).
The viability of cin8Δ single mutants in this cross was 67%.
Figure 3.
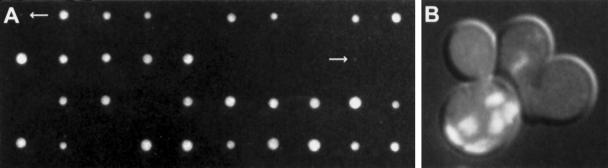
A representative cross exhibiting synthetic lethality. Diploids resulting from the cross between kip2Δ and kar9Δ were sporulated, and the tetrads were dissected on YPD plates. After 2–3 d of growth at 30°C, the colonies were scored for normal growth, microcolonies, or no growth. A dissection plate after 2 d is shown in panel A. The small arrows indicate two kip2Δ kar9Δ microcolonies. Several other microcolonies became apparent after prolonged incubation. Panel B shows a typical multinucleate mother cell with several anucleate buds from a kip2Δ kar9Δ microcolony. The cells were stained with the fluorescent DNA-specific dye, DAPI, to allow visualization of the nuclei, and photographed using fluorescence and differential interference contrast optics simultaneously.
First, the kip2Δ mutant was crossed to the mutants that exhibit defects in nuclear migration (Table 5, top panel). We found that all kip2Δ kar9Δ double mutants were inviable, or synthetically lethal (Figure 3 and Table 5, line 1), suggesting that Kip2p and Kar9p act in separate pathways for nuclear migration. Cells from the kip2Δ kar9Δ microcolonies exhibited a severe defect in nuclear migration; many were multinucleated and had anucleate buds (Figure 3B). In contrast, double mutants with kip2Δ in combination with deletions in genes of the dynein/dynactin complex (dhc1Δ, jnm1Δ and act5Δ) were viable and displayed no visible differences in growth compared with wild type (Table 5, lines 2, 3, and 4). Furthermore, all kip2Δ dhc1Δ kar9Δ triple mutants were indistinguishable in size from the kip2Δ kar9Δ or dhc1Δ kar9Δ double mutants (Table 5, line 19). Taking the single, double, and triple mutant analyses together, the data suggest that Kip2p functions in the dynein/dynactin branch of the nuclear migration pathway and not in a third redundant pathway.
Genetic Interactions Between kip2Δ and Mutations Affecting Microtubule Function
BIK1encodes a microtubule-associated protein that, like Kip2p, is thought to stabilize microtubules and also functions in nuclear positioning in mitosis and mating (Berlin et al., 1990). Given these phenotypes, we wanted to determine whether there are genetic interactions between kip2Δ and bik1Δ. All of the kip2Δ bik1Δ double mutants were viable (Table 5, line 5). These results are consistent with the hypothesis that Kip2p and Bik1p might stabilize microtubules through a common pathway. In support of this, mutations in bik1 show synthetic lethality with mutations in TUB2, the gene for β-tubulin (Berlin et al., 1990), and the kip2Δ mutation was found to be synthetically lethal in combination with mutations in TUB1 (our unpublished observations).
Given the precedence of functional redundancy between kinesin-related proteins (Roof et al., 1992; Saunders and Hoyt, 1992), we next performed crosses between kip2Δ and deletions of the other kinesin-related genes. The kinesin-related gene SMY1 appears to act in concert with a myosin, Myo2p, in the secretory pathway (Lillie and Brown, 1992). In crosses, the kip2Δ smy1Δ double mutants were viable (Table 5, line 9). Genetic interactions with cin8Δ, a kinesin-related gene known to function in mitosis, were also performed. All kip2Δ cin8Δ double mutants were inviable (Table 5, line 6). Previous work showed that kip2Δ is not synthetically lethal with a deletion of KIP1 or with a deletion of KAR3, two other mitotic kinesin-related genes (Roof et al., 1992). Finally, we crossed the kip3Δ mutation and the kip2Δ mutation. All kip2Δ kip3Δ double mutants were viable and exhibited normal growth. In summary, in genetic crosses with all of the known kinesin-related genes, kip2Δ was found to be synthetically lethal only with cin8Δ.
Genetic Analysis of kip3Δ Mutants Places Kip3p in the KAR9 Pathway of Nuclear Positioning
To understand the functions of KIP3 in nuclear migration, crosses to kar9Δ, dhc1Δ, and other mutations affecting nuclear migration were performed. We found that 88% of the kip3Δ kar9Δ double mutants were viable (Table 5, line 10). Thus, kip3Δ, unlike kip2Δ, was not more severe when combined with kar9Δ. Furthermore, kip3Δ resulted in severe growth defects in combination with mutations in genes in the dynein/dynactin complex. In crosses to dhc1Δ, jnm1Δ and act5Δ, all of the predicted double mutants were dead or produced microcolonies (Table 5, lines 11–13). Cells from the kip3Δ dhc1Δ microcolonies exhibited an extreme nuclear migration defect; many were multinucleated, some had fragmented nuclei, and nearly 25% had lysed. These results suggest that the kip3Δ defect interferes with a pathway that is partially redundant with the dynein–dynactin pathway for nuclear migration. Furthermore, we found that all kip3Δ dhc1Δ kar9Δ triple mutants were indistinguishable from the kip3Δ dhc1Δ and dhc1Δ kar9Δ double mutants (Table 5, line 20). Taking the single, double, and triple mutant data together, we conclude that Kip3p functions in the KAR9 branch of the nuclear migration pathway and not in a third redundant pathway.
Finally, we examined the kip2Δ kip3Δ kar9Δ triple mutant and found that it was inviable (Table 5, line 21), unlike the kip2Δ kip3Δ double mutant. This result suggests that, although Kar9p and Kip3p act in the same pathway, Kar9p must still be active in the kip2Δ kip3Δ double mutant.
Genetic Interactions Between kip3 and Mutations Affecting Microtubule Function
The kip3Δ mutation was also tested in combination with mutations in genes known to affect microtubule stability and/or spindle function. All kip3Δ kip1Δ double mutants were viable (Table 5, line 16). In contrast, all of the kip3Δ bik1Δ (Table 5, line 14), kip3Δ kar3Δ (Table 5, line 17), and most of the kip3Δ cin8Δ (Table 5, line 15) double mutants were inviable. Therefore, kip3Δ mutants exhibited severe growth defects in combination with kar3Δ, cin8Δ, and bik1Δ mutants, but not kip1Δ mutants. Thus, kip2Δ and kip3Δ differed with respect to their interactions with bik1Δ and kar3Δ, but were similar with respect to their interactions with kip1Δ and cin8Δ.
It is striking that the two mutations that primarily affect spindle elongation, cin8Δ and kip1Δ, do not produce distinct genetic interactions with kip2Δ and kip3Δ. One interpretation of this observation is that any strong defects in spindle elongation will be synthetically lethal with any mutation that causes strong defects in spindle orientation, regardless of the mechanism. In contrast, Bik1p and Kar3p both have significant effects on the cytoplasmic microtubules and show differential interactions with kip2Δ and kip3Δ. It seems likely that the specificity of the interaction reflects their quite different roles in cytoplasmic microtubule function.
kip2 Mutants Show Altered Cellular Localization of Kar9p
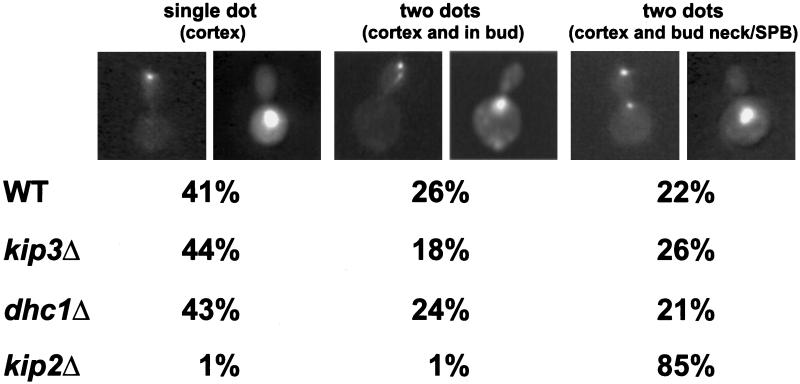
In support of the hypothesis that Kip2p and Kip3p function differently in nuclear migration, we found that localization of GFP-Kar9p was altered in a kip2Δ, but not in kip3Δ or dhc1Δ mutants (Figure 4). As was reported previously (Miller and Rose, 1998), in a majority of wild-type preanaphase cells, GFP-Kar9p localized exclusively to one or two dots (41 and 26%, respectively) at the cortex of the bud (Figure 4). In a minority of wild-type cells (22%), in addition to the cortical localization, a second spot was observed on the edge of the nucleus close to the bud neck bud, presumably associated with the SPB. For both kip3Δ and dhc1Δ, the pattern of GFP-Kar9p localization was indistinguishable from wild type. In contrast, in the kip2Δ, GFP-Kar9p was exclusively localized to the bud tip cortex in less than 2% of the cells. In most kip2Δ cells (85%), GFP-Kar9p localized both to the tip of the bud and to edge of the nucleus very close to the bud neck (Figure 4).
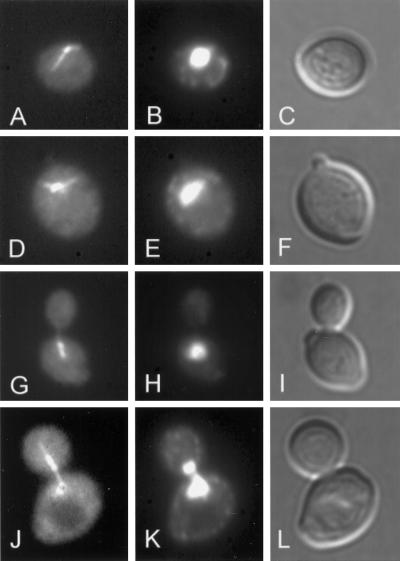
Figure 4.
GFP-Kar9p localization in nuclear migration mutants. Representative cells show the pattern of fluorescence of GFP-Kar9p (right) and the corresponding nuclear and cellular morphologies (left). The two left-most panels are a cell with GFP-Kar9p at the tip of the bud (single-dot cortex). The middle panels are of a cell with GFP-Kar9p at the tip of the bud and elsewhere in the bud (two dots, cortex and in bud). The right-most panels show a cell with GFP-Kar9p at the tip of the bud in and at the nuclear periphery (two dots, cortex and bud neck/SPB). The wild-type (WT), (MS1554), kip3Δ (MS4516), dhc1Δ (MS5001), and kip2Δ (MS2309) strains were transformed with a GFP-KAR9 (pMR3465) construct and scored for GFP-Kar9p localization in preanaphase cells (medium-budded cells with a single nucleus). Between 242 and 508 cells were counted for each strain. For the various strains, percentages of total cells with the localization patterns are shown under the representative cell. Between 3 and 6% of the cells showed no fluorescence, and between 6 and 10% showed patterns that did not fall into the three classes represented in the figure.
Kip2p and Kip3p Show Distinct Cellular Localization Patterns
To further define the roles of these kinesin-related proteins in mitosis, we next determined their intracellular location. For visualization of Kip2p, we generated two different tagged protein constructs, GFP-Kip2p and Kip2p-HA. Expression of either construct suppressed the growth defects of kip2Δ kar9Δ double mutants, demonstrating that the proteins were functional. Visualization of GFP-Kip2p using fluorescence microscopy (Figure 5, A–H) and Kip2p-HA by indirect immunofluorescence microscopy (Figure 5I) showed that Kip2p localized exclusively on cytoplasmic microtubules. At all stages of the cell cycle, GFP-Kip2p fluorescence corresponded to the cytoplasmic microtubule pattern (Figure 5). Furthermore, Kip2p localization was observed on the cytoplasmic microtubules in both the mother and the bud. Localization to the nuclear microtubules was not detected. The localization pattern was identical in kip2Δ and KIP2 strains.
Figure 5.
Kip2p localization. To localize GFP-Kip2p, a kip2Δ strain (MS2354) containing the GFP-Kip2 plasmid (pMR3889) was grown to early exponential phase, formaldehyde fixed, and stained with DAPI. Left panels are fluorescence, middle panels are DAPI staining, and right panels are Nomarski images of the same cell. Representative cells at different stages of the cell cycle are shown in the figure. (A) Unbudded or G1 cell, n = 89; (B and C) small budded, n = 97; (D and E) medium budded, n = 107; (F–H) large budded, n = 93. The bottom panels (I) show indirect immunofluorescence of HA on the left, DAPI staining in the middle, and Nomarski image on the right. To localize Kip2p using an HA epitope, a kip2Δ strain (MS2354) containing pMR3779 (a Kip2p::HA 2μ construct) was grown to early exponential phase and paraformaldehyde fixed (see MATERIALS AND METHODS).
Kip3p localization was determined using a PGAL1-GFP-KIP3 construct transformed into a wild-type diploid (MS4720). Early in the cell cycle in unbudded and small-budded cells, GFP-Kip3p localized to the SPB and the associated cytoplasmic microtubules (Figure 6A). After spindle formation, GFP-Kip3p fluorescence was associated with the SPBs, the spindle, and the cytoplasmic microtubules (Figure 6, D–J). Less than 5% of cells appeared to have no GFP-Kip3p signal, but this was not specific to any particular cell-cycle stage. Identical localization patterns were found using GFP-KIP3 in the homozygous kip3Δ omit strain.
Figure 6.
GFP-Kip3p localization. To localize GFP-Kip3p, a wild-type strain (MS4720) containing the GFP-Kip3p plasmid (pMR3635) was grown at 30°C, formaldehyde fixed, and DAPI stained. GFP-Kip3p localization is shown in panels A, D, G, and J. DAPI staining is shown in panels B, E, H, and K. Nomarski images are shown in C, F, I, and L.
Many of the cells overexpressing GFP-KIP3 showed abnormal morphologies. Some cells were arrested as large-budded cells with a single nucleus that had not elongated or migrated to the bud neck. This phenotype was noted in 41% of large-budded cells when expressed in the wild-type strain, and in 50% when expressed in the kip3Δ strain MS4719. Because of the abnormal overexpression phenotypes, it was not possible to determine whether the GFP-KIP3 expressed functional Kip3p.
In summary, although both proteins show localization patterns consistent with microtubule structures, Kip2p appears to localize exclusively to cytoplasmic microtubules, whereas Kip3p is found on both the spindle and cytoplasmic microtubules. The results from the localization further confirm that the two proteins play distinct roles in mitosis and nuclear positioning.
DISCUSSION
In this study, we confirm that the kinesin-related genes, KIP2 and KIP3, are involved in yeast nuclear migration during mitosis (Miller and Rose, 1995; Cottingham and Hoyt, 1997; DeZwaan et al., 1997). Consistent with defects in nuclear migration, the cytoplasmic microtubules were aberrant in kip2Δ and kip3Δ strains. As was found by others (Cottingham and Hoyt, 1997), in kip2Δ strains the cytoplasmic microtubules were absent or dramatically shorter than wild type. In contrast, we found that in kip3Δ strains the cytoplasmic microtubules were present but misoriented and often failed to enter the bud. Moreover, we confirmed that the cytoplasmic microtubules were much longer than in wild type (Cottingham and Hoyt, 1997; DeZwaan et al., 1997). In our study this was most pronounced at telophase. Genetic analysis showed that Kip2p and Kip3p function very differently in nuclear migration. Specifically, mutations in the two genes had complementary patterns of synthetic lethality when combined with mutations in genes that affect cytoplasmic microtubule function. Extending earlier observations, kip2Δ was synthetically lethal in combination with kar9Δ, but was viable in combination with dhc1Δ, jnm1Δ, and act5Δ. In contrast, kip3Δ was viable in combination with kar9Δ mutations but was synthetically lethal when combined with dhc1Δ, jnm1Δ, and act5Δ. These genetic results suggest that Kip2p functions as part of the dynein pathway for nuclear migration, whereas Kip3p functions as part of the Kar9p pathway. Localization studies also showed that Kip2p and Kip3p function differently in mitosis. Kip2p localized exclusively to the cytoplasmic microtubules, whereas we and others (DeZwaan et al., 1997) found that Kip3p was present on both spindle and cytoplasmic microtubules. Taken together, these data support a model of nuclear migration in which Kip2p-dependent microtubule stability is required for dynein-dependent nuclear movement, whereas Kip3p is required for the cytoplasmic microtubule orientation function of Kar9p.
kip2Δ and kip3Δ Mutants Display Distinct Cytoplasmic Microtubule Morphologies
The morphology of the microtubules in the kip2Δ strain suggest that wild-type Kip2p plays a role in stabilizing the cytoplasmic microtubules. Consistent with this, we observed that overexpression of Kip2p on a 2μ plasmid resulted in 10% of the cells exhibiting hyperelongated microtubules that looped around the inside of the cells. In addition, increased overexpression using a galactose-inducible promoter was toxic to cells (our unpublished observations).
In contrast to kip2Δ, the kip3Δ mutants displayed normal cytoplasmic microtubules during the early stages of mitosis. However, at telophase the cytoplasmic microtubules were often significantly longer than normal. By inference, these observations suggest that Kip3p destabilizes the cytoplasmic microtubules during at least one period in mitosis. In support of the suggestion that Kip3p shortens the microtubules, overexpression of Kip3p-GFP appeared to diminish the cytoplasmic microtubules during anaphase (Figure 6, G–L).
How might proteins that are thought to function in microtubule-dependent movements affect the stability/assembly of the microtubules in opposite ways? First, it is possible that a kinesin-related protein may not act as a motor, but may instead bind in a stable manner to microtubules. As such, a kinesin-related protein could help stabilize a microtubule against depolymerization. For example, mutant forms of Kar3p (kar3–1 and Kar3p-β-galactosidase) that cannot hydrolyze ATP do stabilize the cytoplasmic microtubules during mating (Meluh and Rose, 1990; Vallen et al., 1992). Second, some kinesins, such as Kar3p and XKCM1, have the intrinsic ability to depolymerize microtubules (Endow et al., 1994; Walczak et al., 1996). Finally, in the absence of ATP, kinesin and kinesin-related proteins can “track” along the ends of microtubules as they depolymerize (Coue et al., 1991; Lombillo et al., 1995). Indeed, the sliding attachment can couple depolymerization to the movement of attached organelles, effectively converting disassembly into work. It seems reasonable to propose that the association of kinesin-related proteins with the depolymerizing ends of microtubules would have significant effects on the stability of microtubules, with variable and distinct effects depending on the properties of the specific protein.
Kip2p and Kip3p Have Different Roles in Nuclear Migration
Broadly speaking, dynein and Kar9p define two partially redundant pathways for efficient nuclear migration; the former largely providing the force for movement and the latter largely providing the orientation. It is striking that the genetic analysis indicated that kip2Δ and kip3Δ each specifically affect the dynein and Kar9p pathways, respectively. The complementary nature of the genetic interactions provides important clues as to their roles in nuclear movement.
One interpretation of our finding that kip2Δ affects the dynein pathway is that Kip2p interacts directly with dynein or the proteins in the dynactin complex. Although native cytoplasmic dynein has not yet been localized in yeast, a functional GFP-dynein hybrid localizes along the length of the cytoplasmic microtubules (Shaw et al., 1997). The similar localization of Kip2p-GFP and Kip2p-HA along the length of the cytoplasmic microtubules is consistent with the suggestion that the two proteins interact. However, there is no biochemical evidence to support a direct interaction. One possible role for Kip2p would be as a transporter to move dynein back toward the plus ends of the microtubules. One explicit prediction of this hypothesis is that the localization of Kip2p and dynein along the cytoplasmic microtubules would be interdependent.
However, the fact that dynein does not appear to be restricted to a cortical site raises the possibility that dynein acts to produce force over the entire length of the cytoplasmic microtubules. Presumably, dynein would also interact with other cytoskeletal elements to provide the counterbalance for force production. Assuming that dynein does act over the entire length of the microtubules, the total force exerted by dynein would be directly related to the total length of the cytoplasmic microtubules. Accordingly, any mutation that leads to destabilization or failure to assemble the cytoplasmic microtubules would also compromise dynein-dependent force production. Thus, Kip2p would genetically appear to be a member of the dynein pathway because of the reduced length of the cytoplasmic microtubules and not because of any specific association between dynein and Kip2p. In support of this view, we observed that mutations in BIK1, which also destabilize the cytoplasmic microtubules, were synthetically lethal with kip3Δ and kar9Δ (Miller and Rose, 1998) but not with kip2Δ. The viability of the bik1Δ kip2Δ double mutant suggests that the microtubules may not be significantly more destabilized than in either single mutant.
The synthetic lethality of kip2Δ with kar9Δ demonstrates that Kar9p is still at least partially active in the kip2Δ mutant. Unlike dynein, GFP-Kar9p localized to a cortical site at the ends of the cytoplasmic microtubules (Miller and Rose, 1998). Accordingly, Kar9p’s function should be much less dependent upon microtubule length. Microtubules undergoing dynamic growth would become long enough to interact with Kar9p at the cortex, and Kar9p would then be able to stabilize and orient them. It is important to note that indirect immunofluorescence of cytoplasmic microtubules in fixed cells underestimates both their length and number (Carminati and Stearns, 1997). Thus, the cytoplasmic microtubules in the kip2Δ mutant may be longer and more abundant in live cells.
It is striking that the Kar9-GFP showed a significantly different localization in kip2Δ compared with wild type. While localization of Kar9p to the cortex does not require microtubules (Miller and Rose, 1998), the effect of kip2Δ implies that microtubules can nevertheless influence its localization. The secondary localization of Kar9p to the nuclear periphery/bud neck region in kip2Δ mutants might allow the short microtubules to aid in nuclear migration by interacting with Kar9p near the bud neck.
The model that dynein and Kar9p function on different sites of the cytoplasmic microtubules predicts that more effective microtubule-destabilizing mutants would have a much more severe defect in nuclear migration than kar9Δ or dhc1Δ. The absence of cytoplasmic microtubules would necessarily inactivate both pathways for nuclear migration. This is precisely what was observed for tub2–401 and other tub2 mutations under conditions that specifically depolymerize the cytoplasmic microtubules (Sullivan and Huffaker, 1992).
In contrast to KIP2, our analysis of KIP3 mutations suggested that Kip3p acts in the Kar9p pathway. Given GFP-Kip3p’s localization along the length of both the nuclear and cytoplasmic microtubules, Kar9p’s localization at the ends of the cytoplasmic microtubules (Miller and Rose, 1998) would seem to preclude a simple model of Kar9p–Kip3p interaction. One explanation for the genetic data would be that Kar9p does not interact directly with microtubules, but instead interacts with Kip3p on the microtubules. According to this view, Kip3p would appear to have more than one function in the cell. One function for Kip3p would be to destabilize or depolymerize microtubules via interactions at various sites within the cell. A second function would be for Kip3p to serve as a microtubule “adapter” for Kar9p. This model predicts that Kar9p’s association with microtubules would be completely dependent on Kip3p and that kip3Δ mutants would be effectively Kar9−.
While the “adapter” model is superficially attractive, the two mutations act differently in two important respects. First, whereas kip2Δ is synthetically lethal with kar9Δ, kip2Δ is not synthetically lethal with kip3Δ. Second, although kip2Δ mutations suppress the synthetic lethality of kip3Δ dhc1Δ double mutants (Cottingham and Hoyt, 1997), kip2Δ does not suppress the synthetic lethality of kar9Δ dhc1Δ double mutants. Therefore, although both kar9Δ and kip3Δ have similar defects with respect to microtubule orientation, they cannot be said to be equivalent. By inference, kip3Δ mutants must be effectively Kar9+. In support of this, we found that the kip2Δ kip3Δ kar9Δ triple mutant was inviable. Furthermore, we found that Kar9p localization in the kip3Δ was identical to wild type. An alternative model for the kip3Δ defect in nuclear migration would unify the two apparent functions for Kip3p, microtubule destabilization and orientation. Theoretical analysis of microtubule dynamics has demonstrated that the time required for microtubules to find their targets in space is critically dependent upon dynamic instability (Holy and Leibler, 1994). Computer simulations demonstrated that nondynamic microtubules take several orders of magnitude longer time to find distal targets. According to this model, the kip3Δ would cause a defect in microtubule orientation as a consequence of the increased length and presumably increased stability of the cytoplasmic microtubules. In the kip3Δ mutant, Kar9p would still be functional but unable to interact with the persistently misoriented microtubules. Accordingly, kip3Δ would mimic the kar9Δ with respect to the microtubule orientation defect and show lethality with mutations affecting dynein and dynactin subunits.
A complication for the model for nuclear migration described above concerns the viability of the kip2Δ kip3Δ double mutant. The simple expectation from their patterns of genetic interactions is that they should also exhibit synthetic lethality. However, we and others found that the two deletion mutations have opposite effects on microtubule length (Cottingham and Hoyt, 1997; DeZwaan et al., 1997). Furthermore the two mutations show several compensatory effects in mitosis. In particular, kip3Δ suppresses the benomyl sensitivity of kip2Δ mutants (Cottingham and Hoyt, 1997). Therefore, it is likely that the viability of the double mutant occurs because of the compensatory effects of the two mutations on microtubule stability. In one scenario, kip2Δ would shorten the hyperelongated microtubules observed in kip3Δ, allowing them to interact with Kar9p. From this argument, the inviability of the bik1Δ kip3Δ double mutant becomes somewhat less clear. However, bik1Δ may have a more severe defect for cytoplasmic microtubule stability than kip2Δ, or bik1Δ may affect the nuclear microtubules as well as the cytoplasmic microtubules (Berlin et al., 1990).
Genetic Interactions with Spindle-Elongation Mutations
In our study, both kip2Δ and kip3Δ mutations were found to be synthetically lethal with cin8Δ but not with kip1Δ. Cin8p and Kip1p play partially redundant functions in spindle assembly and maintenance (Hoyt et al., 1992; Roof et al., 1992). However, Cin8p appears to play a larger role for spindle function because mutations in CIN8 lead to a conditional growth defect, whereas mutations in KIP1 do not. Previous reports demonstrating that dynein deletions are synthetically lethal with CIN8 deletions have been interpreted to mean that dynein can provide part of the force for spindle elongation, presumably by acting on the cytoplasmic microtubules (Saunders et al., 1995). Our finding of lethality for the cin8Δ kip2Δ double mutant (also observed by Cottingham and Hoyt, 1997) is consistent with this hypothesis. By the same argument, the lack of synthetic lethality between the CIN8 deletion and the KAR9 deletion (Miller and Rose, 1998) implies that the orientation function of Kar9p is not required for the redundant force that powers spindle elongation in the cin8Δ. Consequently, our finding of lethality for the kip3Δ cin8Δ double mutant was initially surprising. However, Kip3p clearly has functions that Kar9p does not, which can be discerned in the kip3Δ mutant by the hyperelongated microtubules. Furthermore, the fact that Kip3p-GFP localizes to both nuclear and cytoplasmic microtubules suggests that Kip3p participates in spindle function as well as nuclear migration. Thus, it seems more likely that the synthetic lethality of the kip3Δ cin8Δ double mutant arises from defects in spindle function rather than nuclear movement. We note that synthetic lethality of kip3Δ cin8Δ was not observed by DeZwaan et al., (1997). The basis for this difference in our results is unclear at present. Finally, kip2Δ and kip3Δ also differed in their interaction with kar3Δ; kip2Δ kar3Δ was viable whereas kip3Δkar3Δ was inviable (our present results and Roof et al., 1992; Cottingham and Hoyt, 1997; DeZwaan et al., 1997). While the exact function of Kar3p in mitosis remains unclear, it appears both to antagonize the function of Cin8p and Kip1p and promote microtubule disassembly (Saunders and Hoyt, 1992; Endow et al., 1994; Saunders et al., 1997). It is striking that both Kip3p and Kar3p appear to destabilize microtubules; this suggests that their synthetic defect might arise from a combined and catastrophic defect in microtubule disassembly.
Conclusion
Three lines of investigation, microtubule morphology, genetic interactions, and localization of the proteins, support the hypothesis that Kip2p and Kip3p function in nuclear migration by two different pathways. Our data strongly suggest that Kip2p function is required for normal dynein-dependent movement, whereas Kip3p function is required for Kar9p-dependent cytoplasmic microtubule orientation.
ACKNOWLEDGMENTS
We are grateful to the laboratories of G. Fink, B. Futcher, K. Bloom, K. Tatchell, J. Cooper, S. Brown, M.A. Hoyt, and P. Hieter for generously supplying plasmids and strains. We thank Stanislas Leibler for the use of his CCD camera and GFP-filter sets. National Institute of Health grants (GM-37739 and GM-52526) awarded to M. Rose supported this work. A National Institute of Health postdoctoral fellowship supported R. Miller.
Footnotes
GFP, green fluorescent protein; HA, hemagglutinin; SBP, spindle pole body;
REFERENCES
- Berlin V, Styles CA, Fink GR. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae colocalizes with tubulin. J Cell Biol. 1990;111:2573–2586. doi: 10.1083/jcb.111.6.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati J, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SW, Meyer DI. ACT3: a putative centractin homologue in S. cerevisiae is required for proper orientation of the mitotic spindle. J Cell Biol. 1994;127:129–138. doi: 10.1083/jcb.127.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Visser S, Jauniaux J-C, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Schott EJ, Kingsbury TJ, Cole NB, Totis LJ, Bhattacharyya G, He L, Hoyt MA. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol Biol Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holy TE, Leibler S. Dynamic instability of microtubules as an efficient way to search in space. Proc Natl Acad Sci USA. 1994;91:5682–5685. doi: 10.1073/pnas.91.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs CW, Adams AEM, Szaniszlo PJ, Pringle JR. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana JA, Schnapp BJ, Silver PA. Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose MD. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J Cell Biol. 1994;126:911–923. doi: 10.1083/jcb.126.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-Y, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Suppression of a myosin defect by a kinesin-related gene. Nature. 1992;356:358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- Marsh L, Rose MD. In: The pathway of cell and nuclear fusion during mating in Saccharoymces cerevisiae. In: The Molecular and Cellular Biology of the Yeast Saccharomyces, vol. 2: Cell Cycle and Cell Biology. Pringle JR, Broach JR, Jones EW, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. pp. 827–888. [Google Scholar]
- McMillan JN, Tatchell K. The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle. J Cell Biol. 1994;125:143–158. doi: 10.1083/jcb.125.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Miller RK, Rose MD. KAR9 and the kinesin-related gene, KIP2, are required for nuclear migration and microtubule associated processes. Mol Biol Cell. 1995;6:256a. (Abstract) [Google Scholar]
- Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua L, Karpova TS, Cooper JA. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Sullivan SD, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD. Multiple kinesin-related proteins in yeast mitosis. Cold Spring Harbor Symp Quant Biol. 1991;56:693–703. doi: 10.1101/sqb.1991.056.01.078. [DOI] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods of Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle pole bodies to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D, Gill S, Cooper J, Heuser J, Schroer T. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 1994;126:403–412. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubules dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–22. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Gehrung S, Page BD. Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. J Cell Biol. 1991;114:515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Hiller MA, Scherson TY, Rose MD. Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J Cell Biol. 1992;117:1277–1287. doi: 10.1083/jcb.117.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]