Abstract
The MPS2 (monopolar spindle two) gene is one of several genes required for the proper execution of spindle pole body (SPB) duplication in the budding yeast Saccharomyces cerevisiae (Winey et al., 1991). We report here that the MPS2 gene encodes an essential 44-kDa protein with two putative coiled-coil regions and a hydrophobic sequence. Although MPS2 is required for normal mitotic growth, some null strains can survive; these survivors exhibit slow growth and abnormal ploidy. The MPS2 protein was tagged with nine copies of the myc epitope, and biochemical fractionation experiments show that it is an integral membrane protein. Visualization of a green fluorescent protein (GFP) Mps2p fusion protein in living cells and indirect immunofluorescence microscopy of 9xmyc-Mps2p revealed a perinuclear localization with one or two brighter foci of staining corresponding to the SPB. Additionally, immunoelectron microscopy shows that GFP-Mps2p localizes to the SPB. Our analysis suggests that Mps2p is required as a component of the SPB for insertion of the nascent SPB into the nuclear envelope.
INTRODUCTION
In the budding yeast, Saccharomyces cerevisiae, the spindle pole body (SPB) functions as the sole microtubule organizing center (Byers et al., 1978; Hyams and Borisy, 1978). The SPB is embedded in the nuclear envelope (NE), which remains intact throughout the cell cycle. From its position in the NE, the SPB nucleates both cytoplasmic and nuclear microtubules from its cytoplasmic and nuclear faces, respectively. To set up a proper bipolar mitotic spindle, the SPB must be precisely duplicated during G1 of the cell cycle (Byers and Goetsch, 1975). This process of SPB duplication is thought to occur through a conservative mechanism (Vallen et al., 1992).
Several genes required for SPB duplication have been identified (Winey and Byers, 1993). Among them, the MPS2 (monopolar spindle two) gene is required for a late step in SPB duplication. At the restrictive temperature, cells mutant for MPS2 (mps2–1) contain duplicated SPBs, but the nascent SPB is not inserted into the NE (Winey et al., 1991). The defective SPB lies on the cytoplasmic face of the NE, unable to nucleate nuclear microtubules. As a consequence, the cells arrest in G2 of the cell cycle with large buds and unsegregated DNA, which is associated with the functional SPB. After continued incubation at the restrictive temperature, mps2–1 cells overcome the mitotic arrest and proceed through the cell cycle; however, the nuclear DNA is asymmetrically divided: one cell receives all of the nuclear DNA and the other (daughter) cell receives no DNA. Consequently, cells that contain a mutant MPS2 gene display an increase in ploidy. The NDC1 gene also functions at this late step in SPB duplication: ndc1–1 mutant strains exhibit phenotypes that are indistinguishable from mps2–1 strains at the nonpermissive temperature (Thomas and Botstein, 1986; Winey et al., 1993).
The SPB of S. cerevisiae is a disk-like structure composed of six major layers, with the central layer, or plaque, in the same plane as the NE (Bullitt et al., 1997). The defective SPB in mps2–1 and ndc1–1 mutant cells appears to lack the inner (nuclear) plaque. It was originally proposed that Mps2p and Ndc1p function to insert the nascent SPB into the NE, allowing the inner plaque to form and nucleate microtubules on the nuclear face (Winey and Byers, 1993). Recently, Ndc1p was shown to be a shared component of SPBs and nuclear pore complexes (NPCs) (Chial et al., 1998), consistent with a direct role for Ndc1p in the insertion event.
In addition to the structural details of how the SPB is duplicated, it is also important to understand the regulation of a such a crucial aspect of the cell cycle. A major consequence of unregulated SPB duplication is asymmetric chromosome division. Mutants that fail in SPB duplication lead to aneuploidy (Schild et al., 1981; Rose and Fink, 1987; Winey et al., 1991), which is associated with some human cancer cells (Lengauer et al., 1997). The regulation of cell cycle events by posttranslational modifications such as protein phosphorylation and proteolysis is well documented (for reviews see King et al., 1996; Lew and Kornbluth, 1996). Indeed, phosphorylation and ubiquitin-mediated proteolysis have been implicated in the regulation of the SPB duplication cycle. The Mps1p kinase is essential for proper SPB duplication (Winey et al., 1991; Lauze et al., 1995). Interestingly, two components of the SPB, Spc110p (Friedman et al., 1996) and Spc98p (Pereira et al., 1998), are phosphorylated in a cell cycle-dependent manner, and there is evidence that Mps1p may be involved in the phosphorylation of Spc98p (Pereira et al., 1998).
There is also evidence that SPB duplication is regulated by the ubiquitin–proteasome pathway (McDonald and Byers, 1997). In this pathway, specific proteolysis is mediated by the covalent attachment of ubiquitin to substrate proteins, followed by targeting to the proteasome where they are degraded (Hershko and Ciechanover, 1992). The first in vivo evidence supporting a role for the proteasome in a major cell cycle transition came from the finding that some 26S proteasome mutants arrest at the G2/M stage in the cell cycle (Ghislain et al., 1993; Gordon et al., 1993). Recently, a proteasome subunit encoded by the PCS1 gene was shown to be required for SPB duplication (Russell et al., 1996; McDonald and Byers, 1997). A temperature-sensitive pcs1 strain arrests in G2 of the cell cycle with large buds and unsegregated DNA, and electron microscopy revealed that the arrested cells had a single, unduplicated SPB. On the basis of this finding, it was postulated that Pcs1p-containing proteasomes may play a role in the degradation of a particular protein(s) that is specifically required for SPB duplication.
We report here the isolation of the MPS2 gene. Interestingly, we identified MPS2 in a genetic screen with CIM5, a gene that encodes a proteasome subunit required for the G2/M transition (Ghislain et al., 1993), thus linking MPS2 to the ubiquitin–proteasome pathway. In addition, we show that MPS2 encodes an integral membrane protein localized to SPBs and the NE. We propose that Mps2p is directly required for insertion of the nascent SPB into the NE.
MATERIALS AND METHODS
Yeast Strains and General Methods
The yeast strains used in this study are listed in Table 1. Yeast media, growth conditions, and genetic and molecular techniques were as described previously (Sambrook et al., 1989; Guthrie and Fink, 1991). Yeast shuttle vectors and yeast strains used in this study are congenic with S288C (Sikorski and Hieter, 1989). ILM2 was made by transforming CMY826 with the ClaI–SacI fragment from pOC52 containing PDS1-HA (Cohen-Fix et al., 1996). MCL120 and MCL175 strains were made by integration at the URA3 locus of the BsmI-linearized plasmid p20 or p30, respectively, in a mps2Δ::HIS3 strain, followed by loss of the complementing plasmid in nonselective medium. MCL123 was made by transforming strain ILM2 with the EcoRI-linearized plasmid p23 to direct integration at the MPS2 locus. SMY22–5a was made by integrating 9xmycC-MPS2 at the URA3 locus in the strain mps2Δ::KanMX/MPS2, ura3–52/ura3–52, sporulating the diploid, and isolating KanMX+, URA3+ meiotic products. SMY28–2b is the product of a cross between HC32–1C (spc42Δ::LEU2, TRP1:SPC42-GFP, original spc42Δ::LEU2, TRP1:SPC42-GFP strain provided by Ian Adams and John Kilmartin (Medical Research Council, Cambridge, UK) and SMY22–5a (mps2Δ::KanMX,URA3:9xmycC-MPS2). SMY1892 was derived from SMY11–13c (original strain, SWY809, provided by Susan Wente, Washington University, St. Louis, MO), and SMY22–5a. Yeast strains harboring URA3-containing plasmids were counterselected by growth on media containing 5-FOA (United States Biological, Swampscott, MA) as described previously (Boeke et al., 1987). Yeast strains harboring the KanMX4 gene were selected by growth on plates containing 25 μg/ml geneticin (G418) (Wach et al., 1994).
Table 1.
Yeast strains used in this study
| Yeast strains | Genotypes |
|---|---|
| CMY763 | MATα ura3-52 leu2Δ1 cim3-1 |
| CMY765 | MATα ura3-52 leu2Δ1 his3Δ200 cim5-1 |
| CMY826 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 bar1::HIS3 |
| ILM2 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 bar1::HIS3 PDS1::PDS1-HA-URA3 |
| MCL29 | MATa/α ura3-52/ura3-52, leu2Δ1/+, lys2-801/lys2-801, his3Δ200/his3Δ200, trp1Δ1/trp1Δ1, ade2-101/ade2-101 MPS2/mps2Δ::HIS3 |
| MCL44 | MATα ura3-52 trp1Δ1 his3Δ200 leu2Δ1 lys2-801 ade2-10 mps2Δ::HIS3 plus pRS316(URA3)-pGAL1-MPS2 |
| MCL94 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 bar1::HIS3 plus pCM190(URA3)-pTet-9mycN-MPS2 |
| MCL120 | MATa ura3-52 trp1Δ1 his3Δ200 LEU2 lys2-801 ade2-101 mps2Δ::HIS3 plus pRS306(URA3)-9mycN-MPS2 |
| MCL123 | MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 bar1::HIS3 PDS1::URA3-PDS1-HA plus pRS304(TRP1)-9mycN-MPS2 |
| MCL175 | MATa ura3-52 trp1Δ1 his3Δ200 LEU2 lys2-801 ade2-101 mps2Δ::HIS3 plus pRS306(URA3)-GFP-3mycN-MPS2 |
| SC55 | MATa/α ura3-52/ura3-52, leu2Δ1/+ lys2-801/lys2-801 his3Δ200/his3Δ200 trp1Δ1/trp1Δ1 ade2-101/ade2-101 |
| AM610 | MATa/α MPS2/mps2Δ(116-387)::HIS3 ura3-52/ura3-52 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 |
| D8BX5Ca | MATa/α ura3-52/ura3-52 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 |
| WX178-1a | MATα mps2-1 trp1Δ1 ura3-52 |
| SMY28-2b | MATa/a mps2Δ::KanMX/mps2Δ::KanMX URA3::9xmycC-MPS2/URA3::9xmycC-MPS2 spc42Δ::LEU2/spc42Δ::LEU2 TRP1::SPC42-GFP(3x)/TRP1::SPC42-GFP(3x) trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 ura3-52/ura3-52 leu2-3,112/leu2-3,112 |
| SMY22-5a | MATa mps2Δ::KanMX URA3::9xmycC-MPS2 trp1Δ1 his3Δ200 ura3-52 leu2-3,112 |
| HC32-1C | MATα spc42Δ::LEU2 TRP1::SPC42-GFP(3x) trp1Δ1 his3Δ200 ura3-52 leu2-3,112 ade2 |
| SMY1892 | MATa/α mps2Δ::KanMX/mps2Δ::KanMX URA3::9xmycC-MPS2/URA3::9xmycC-MPS2 nup49-1::URA3/nup49-1::URA3 nup49ΔGLFG::GFP-S65T-TRP1/nup49Δ::GFP-S65T-TRP1 ura3-52/ura3-52 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 |
| SMY11-13c | MATα nup49-1::URA3 nup49Δ::GFP-S65T-TRP1 ura3-52 his3Δ200 leu2-3,112 |
MPS2 Isolation
The genomic plasmid bank used to clone MPS2 was a URA3-marked genomic library (Rose and Fink, 1987). Two clones with overlapping inserts, designated E27 and E28, were able to rescue the temperature-sensitive phenotype of mps2–1 cells. Plasmid E13 was created by excising a 6-kb insert from E27 with SalI and XhoI, then ligating it into the SalI site of pRS313. The E13 plasmid was cut with BamHI, excising a 2.6-kb BamHI fragment and was then religated to create E14. Plasmid E150 was constructed by digesting E14 with XhoI and XbaI and ligating this fragment into the same sites of pRS316. To define the boundaries of the complementing ORF, the E150 insert was digested from either side to generate a nested set of partial deletions.
The MPS2 gene was sequenced with the Sequenase Version 2.0 DNA Sequencing Kit (Amersham International, Cleveland, OH) according to the manufacturer. Our sequence is identical to that for YGL075c (Saccharomyces Genome Database). For sequencing of the mps2–1 mutant lesion, genomic DNA was prepared from strain Wx178–1a as described previously (Hoffman and Winston, 1987). Primers to MPS2 were used to generate a series of PCR fragments, which were then used as templates for sequencing using the Sequenase PCR Product Sequencing Kit (Amersham International).
Genetic Screen with cim5
MPS2 was also identified in a screen for genes whose overexpression is toxic in a cim5 proteasome mutant at a semipermissive temperature, but not in the wild-type strain. A cim5–1 temperature-sensitive mutant (strain CMY765) was transformed with a GAL1-regulated S. cerevisiae genomic DNA expression library (Ramer et al., 1992) in the centromeric (URA3) pYES vector, (Elledge et al., 1991). Cells were plated to synthetic medium containing glucose but lacking uracil (−Ura) and grown at the semipermissive temperature of 30°C. Approximately 30,000 colonies were then replica-plated to selective media (−Ura) plates containing either glucose (no library expression) or galactose plus raffinose (gal–raff) (overexpression conditions) at the same temperature. Raffinose was added to facilitate yeast growth on galactose. Colonies able to grow on the glucose plates but showing little or no growth on the gal–raff plates were selected. Only rho+ colonies showing the inhibitory effect on the gal–raff plates in a URA3 plasmid-dependent manner were selected for plasmid recovery in Escherichia coli. CMY765 (cim5–1), CMY763 (cim3–1), and CMY826 (isogenic wild-type) strains were then transformed with the selected plasmids, and growth inhibition on the gal–raff plates at 30°C was retested. Fourteen independent plasmids contained yeast DNA fragments whose overexpression caused a specific inhibitory effect on growth of the cim mutants. One of these DNA fragments corresponded to the MPS2 gene.
Disruption of MPS2
Two different null alleles of MPS2 were constructed using a PCR-based method (Baudin et al., 1993). One allele was made by replacing amino acids 116–387 with the HIS3 gene using the one-step method (Baudin et al., 1993). The other deletion was made by replacing the entire MPS2 ORF with the Kanamycin resistance gene (Wach et al., 1994; Brachmann et al., 1998), KanMX4, using a two-step gene replacement technique (Rothstein, 1991). Both the mps2Δ::KanMX-containing fragment and the mps2Δ116–387::HIS3 PCR product were used to transform a wild-type diploid strain (D8BX5CA) as described previously (Schiestl and Gietz, 1989). HIS3 prototrophs or G418-resistant isolates, respectively, were examined by PCR for the correct recombination event.
Plasmids
p17: pCM190 (2 μ, URA3)-pTet-9mycN-MPS2.
Nine copies of the myc epitope (EQKLISEEDL) were introduced after the initiation codon by “gene SOEing” or overlap extension (Horton, 1995). Oligonucleotides TG1 (5′-CCCAGCTTTGTTTAAACATGCGCGGTGGCGGCCGCTCTAGA-3′) and TG2 (5′-ATCAAACGCACCGTTACTCATCAGCCCGGGGGATCCACTAGT-3′) were used to amplify by PCR a DNA fragment containing the nine-myc epitope sequence from a plasmid kindly provided by Kim Nasmyth (Institute of Molecular Pathology, Vienna, Austria). Oligonucleotide TG1 introduces an initiation codon in-frame with the first myc epitope, and TG2 contains a 3′ 21-bp overlapping region with the 5′ end of MPS2 ORF. The 444-bp PCR-amplified fragment was called PCR-TG-A. The MPS2 ORF was PCR amplified from a MPS2-containing plasmid (p2) using oligonucleotides TG3 (5′-ATGAGTAACGGTGCGTTTGAT-3′) and TG4 (5′-CCAAAACTGCAGGGCCAAGGTTTAAAT-3′), and the 1185-bp PCR fragment was called PCR-TG-B. In a subsequent PCR reaction, the overlap between PCR-TG-A and PCR-TG-B served as a primer for extension using oligonucleotides TG1 and TG4, creating a recombinant molecule containing the 9xmyc tag after the initiation codon in-frame with the MPS2 coding sequence. The PmeI–PstI fragment from the final PCR product was cloned into pCM190 (2 μ, URA3, pTet) (Gari et al., 1997) to obtain p17. The function of the 9xmycN-MPS2 gene was verified by its ability to suppress the mps2 temperature-sensitive mutant. In p17, 9mycN-MPS2 is expressed from a tetracycline-repressible promoter.
p20: pRS306 (integrative, URA3)-9mycN-MPS2 and p23: pRS304 (integrative, TRP1)-9mycN-MPS2.
A recombinant DNA molecule was generated by “gene SOEing” containing the endogenous MPS2 promoter and the 9mycN-MPS2 ORF. Oligonucleotides SOE1 (5′-GGTACCGGGCCCCCCCTCGAGG-3′) and SOE2 (5′-TCTAGAGCGGCCGCCACCGCGCATACTTACGTTGTCAAAGACAG-AATT-3′) were used to amplify by PCR a 671-bp fragment (PCR1) containing the endogenous MPS2 promoter from a MPS2-carrying plasmid. The 9mycN-MPS2 ORF and terminator sequences were PCR amplified from pCM190-pTet-9mycN-MPS2 (p17) using oligonucleotides SOE3 (5′-ATGCGCGGTGGCGGCCGCTCTAGA-3′) and SOE4 (5′-ATGTCATGGGAGCTCTGGTTAGCTCACTCAT-TAG-3′) (PCR2). The recombinant DNA fragment, containing the endogenous MPS2 promoter and the 9mycN-MPS2 ORF, was amplified using the overlap between PCR1 and PCR2 as a primer and oligonucleotides SOE1 and SOE4. The SacI–XhoI fragment from the final PCR product was cloned into pRS306 (integrative, URA3) or pRS304 (integrative, TRP1) vectors yielding p20 and p23, respectively. In p20 and p23, 9xmycN-MPS2 is expressed from its endogenous promoter.
p30: pRS306-GFP-3xmycN-MPS2.
The GFP3 sequence from plasmid pYGFP3 (kindly provided by Brendan Cormack, Johns Hopkins Medical School, Baltimore, MD) was amplified by PCR using oligonucleotides GFP1 (5′-CCCAGCTTTACTAGTATGTCTAAAGGTGAAGAATTATTC-3′) and GFP2 (5′-CCAAAAAGCACTAGTTTTGTACAATTCATCCATACCATG-3′). The SpeI PCR fragment was cloned into p20 digested with XbaI.
pRS306–9xmycC-MPS2.
Nine copies of the myc tag were introduced at the termination codon of MPS2. The 9xmyc sequence from plasmid 9xmycFZ01 (a gift from G. Hermann and Janet Shaw, University of Utah, Salt Lake City, UT) was amplified by PCR with primers 9xMycNXbaI and 9xMycCXbaI. The PCR fragment was digested with XbaI and ligated into a unique SpeI site created by PCR immediately before the termination codon in MPS2 (plasmid pRS314.B). A 2.1-kb SacI/KpnI fragment from pRS314 was subcloned into pRS306. The function of the fusion protein was verified by the ability of pRS314-9xmycC-MPS2 to rescue the mps2–1 temperature-sensitive phenotype and the mps2Δ::KanMX4 strain.
Western Blot Analysis
Protein extracts were made from 25-ml aliquots of exponentially growing cells (see Figure legends for strains). Cells were pelleted and transferred to an Eppendorf tube in 150 μl of cold lysis buffer (50 mM Tris-HCl, pH 8, 0,3% Na deoxycholate, 1% Triton X-100, 0.2% SDS, 50 mM NaCl, 5 mM EDTA, 2 mg/ml each of pepstatin A, aprotinin, leupeptin, chymostatin, and Pfablock plus 1 mM PMSF). An equal volume of glass beads was added, and the cells were lysed by vortexing. Extracts were then clarified by 15 min of centrifugation at 4°C, and the supernatant was collected. Total cell protein (40 μg) was denatured by boiling in SDS sample buffer and then separated on 10% SDS polyacrylamide gels. Proteins were transferred to nitrocellulose membranes (Hybond ECL, Amersham). 9xmycN-Mps2p was detected using the 9E10 anti-myc monoclonal antibody (Santa Cruz Biotechnology,Santa Cruz, CA) at a concentration of 0.1 μg/ml, and Pds1p-HA was detected using a 1:1000 dilution of 12CA5 anti-HA ascites fluid. Cdc28p, Sec61p, and hexokinase were identified using polyclonal antibodies (van Tuinen and Riezman, 1987; Wilkinson et al., 1996). To detect Dpm1p, the 5C5-A7 monoclonal antibody (Molecular Probes, Eugene, OR) was used at a concentration of 4 μg/ml.
Subcellular Fractionation
Subcellular fractionation was performed essentially as described (Harris and Waters, 1996). MCL120 cells expressing 9xmycN-Mps2p from the MPS2 promoter were grown in 250 ml of YPD at 24°C to an OD600 nm of 0.5–0.8, harvested by centrifugation, and washed two times in spheroplast buffer (50 mM Tris-HCl, pH 7.5, 1.2 M sorbitol, 10 mM NaN3, 40 mM β-mercaptoethanol). Cells were resuspended in spheroplast buffer at 40 0D U/ml. Zymolyase 100T (ICN, Costa Mesa, CA) was added to 300 μg/ml, and cells were incubated at 30°C for 30 min with occasional mixing. The spheroplasts were washed two times in spheroplast buffer and resuspended in 0°C lysis buffer (20 mM HEPES/KOH, pH 7.4, 100 mM K-acetate, 5 mM Mg-acetate, 1 mM EDTA 1 mM DTT) plus a protease inhibitor mixture (2 μg/ml each of pepstatin A, aprotinin, leupeptin, chymostatin, and Pfablock plus 1 mM PMSF) to the same volume in which they were spheroplasted. Cells were lysed in a 5-cm3 Dounce homogenizer (25–30 strokes on ice), and then the lysates were centrifuged at 4°C for 3 min at 1000 × g. The resulting supernatant (crude extract) was centrifuged at 4°C for 15 min at 13,000 × g to generate pellet (P13) and supernatant (S13) fractions. The S13 fraction was centrifuged at 4°C for 60 min at 100,000 × g in a Beckman (Palo Alto, CA) TL-100 centrifuge generating pellet (P100) and supernatant (S100) fractions. Protein concentrations were determined using the Bradford assay (Bradford, 1976).
Mps2p was solubilized from the P13 fraction essentially as described (Ruohola and Ferro-Novick, 1987). The pellet was homogenized in lysis buffer, divided into aliquots, and centrifuged at 13,000 × g for 15 min. The pellet for each sample was resuspended in lysis buffer (control) or in lysis buffer containing either 1 M NaCl, 50 mM Tris (pH 7.5)-10 mM EDTA, 0.2 M Na2CO3 (pH 11), 1% Triton X-100, or 6 M urea (other combined treatments are indicated in the Figure legends). The mixtures were incubated on ice for 10 min and then centrifuged at 13,000 × g for 15 min. The supernatant was removed, and the pellet was resuspended in an equal volume of lysis buffer. Equivalent samples of the supernatant and pellet fractions were analyzed by immunoblotting.
Cytological Techniques
Flow cytometry analysis of cells was performed as described using the DNA stain propidium iodide (Winey et al., 1991). Stained cells were analyzed using a Becton Dickinson FACScan flow cytometer using CELL QUEST software packages to collect and analyze the data (BDIS, San Jose, CA).
Whole-cell fluorescence microscopy (see Figure 6) was carried out essentially as described by Chial et al. (1998), developed from Rout and Kilmartin (1990). Briefly, cells were grown at 24°C to an OD600 between 0.2 and 0.8. Cells were pelleted and fixed in 3.7% formaldehyde for 5 min. Cells were spheroplasted in Solution A (1.2 M sorbitol, 100 mM KPO4, pH 7.5) containing 4.8 μg/ml Zymolyase 100T (ICN) for 45 min at 30°C. Cells were rinsed with PBSA (10 mg/ml NaCl, 0.2 mg/ml KCl, 1.43 mg/ml KH2PO4), then resuspended in 50–100 μl of Solution A. Cells (10 μl) were spotted onto a polylysine-treated multi-well slide, then submerged in methanol followed by acetone. Slides were treated with blocker (PBSA, 10 mg/ml BSA, 0.1% Tween-20) for 5 min, followed by incubation in a 1:400 dilution of α-myc antibody overnight at 4°C. The myc antibody was detected using either a Texas Red-conjugated sheep anti-mouse antibody diluted 1:400 or an FITC-conjugated sheep anti-mouse antibody diluted 1:200 (both from Amersham), and the DNA was stained with DAPI. Standard fluorescence microscopy was performed using a Leica DMRXA/RF4/V automated microscope equipped with a digital camera (SensiCam CCD camera; Cooke, Tonawanda, NY). Images were acquired and deconvolved using the Slidebook software package (Intelligent Imaging Innovations, Denver, CO).
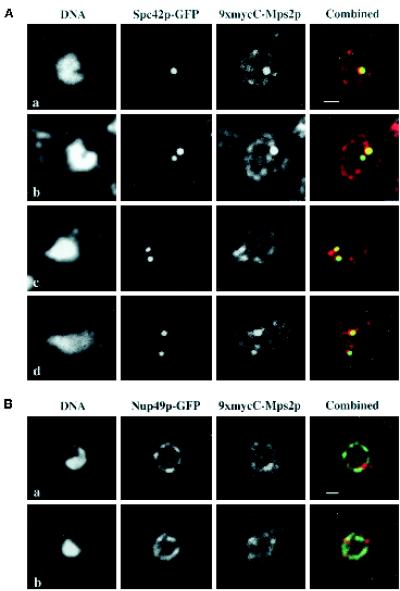
Figure 6.
9xmycC-Mps2p colocalizes with Spc42p-GFP, but not Nup49p-GFP. (A) A strain expressing both 9xmycC-Mps2p and Spc42p-GFP (SMY28–2b) was prepared for indirect immunofluorescence microscopy (see MATERIALS AND METHODS). The Spc42p-GFP was visualized via autofluorescence of GFP, the 9xmycC-Mps2p was detected using a Texas Red-conjugated 2° antibody, and the DNA was visualized with DAPI. Four different cells are shown at a high magnification to show the detail of colocalization. The bright spots of 9xmycC-Mps2p (red) and the Spc42p-GFP (green) signals overlap, as seen in the combined image (yellow). The Spc42-GFP and 9xmycC-Mps2p signals at the lower SPBs in images b and d do not completely overlap in these focal planes; however, Spc42p and Mps2p are not expected to be in the same exact position in the organelle. In addition, the amount of overlap depends on the ability to indirectly label Mps2p at any given SPB. (B) Two different cells from a strain expressing both 9xmycC-Mps2p and Nup49p-GFP (SMY1892) are shown at a high magnification. There are bright spots of 9xmycC-Mps2p fluorescence observed along the nuclear periphery that are devoid of Nup49p-GFP signal. All panels in this Figure represent a single optical plane that was deconvolved using the Slidebook software package (see MATERIALS AND METHODS). Bar, 1 μm in both panels.
For visualizing GFP-Mps2p in living cells, a 2 ml aliquot of cells grown in YPD at an OD600 of 0.5–1.0 was collected and washed in water. Cells were resuspended in 50 μl of Dabco solution (24.5 mg/ml diazabicyclo 2–2-2 octane in PBS, 75% glycerol). The DNA was visualized with Hoechst at a final concentration of 5 μg/ml for 5 min.
Immunoelectron microscopy was performed as described previously (Winey et al., 1995; Ding et al., 1997; Chial et al., 1998). Cells were high-pressure frozen in a Balzers HPM-010 High Pressure Freezer (Bal-Tec, Middebury, CT). Thin sections were labeled with an affinity-purified anti-GFP rabbit polyclonal antibody (a kind gift from Jason Kahana and Pam Silver, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA) and reacted with goat anti-rabbit 15-nm colloidal gold conjugate (Ted Pella, Redding, CA), followed by rinses and staining with uranyl acetate and lead citrate.
RESULTS
Isolation of the MPS2 Gene
The MPS2 gene was isolated from a yeast genomic library constructed in the centromeric plasmid YCp50 (Rose and Fink, 1987) by screening transformants of mps2–1 (Wx178–1a; Table 1) for those able to grow at the restrictive temperature of 37°C. The insert in one of the two rescuing plasmids was subcloned, identifying a region of 3.4 kb required for complementation. A nested deletion series was used for sequence analysis of the 3.4-kb insert, and the minimal complementing fragments were found to contain a single ORF that has been identified as YGL075c by the Yeast Genome Project.
To confirm that YGL075c was truly the gene mutated in mps2–1 strains, the original 3.4-kb insert was placed in the integrative plasmid pRS306 (URA3). The insert in pRS306-MPS2 directed integration at the MPS2 locus as determined by the meiotic linkage of the integrated marker (URA3) to mps2–1. To further verify that YGL075c was indeed MPS2, the gene was amplified from the mps2–1 strain (WX178–1a) and sequenced. A single base change was detected at nucleotide 114 leading to an amino acid change of a glutamic acid to lysine residue at position 39 in the protein (Figure 1A). We confirmed that the base change found in the mps2–1 strain was the cause of the temperature-sensitive phenotype, and not a polymorphism, by cloning the region containing the mutation into a yeast integrative vector to create pRS306-mps2-1. Targeted integration at the MPS2 locus, with excision of the wild-type copy of the gene, conferred temperature sensitivity that is complemented by wild-type MPS2 on a plasmid, but not by a cross to the mps2–1 strain (our unpublished results). These results indicate that YGL075c is the gene mutated in the mps2–1 strain.
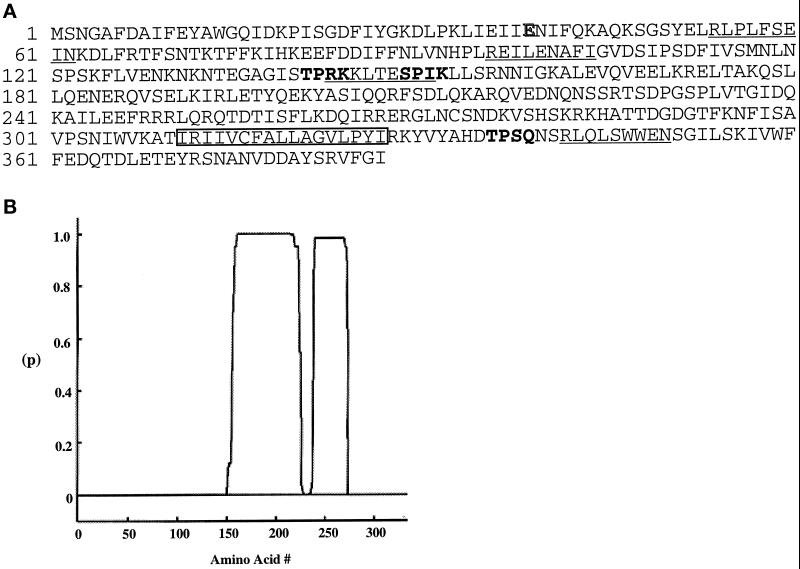
Figure 1.
Features of the MPS2 encoded protein. (A) Deduced amino acid sequence of the MPS2 encoded protein. The putative transmembrane segment (residues 311–327) is boxed. Potential Cdc28p phosphorylation sites are indicated in bold letters, and potential destruction box sequences are underlined. The point mutation found in the mps2–1 allele at amino acid 39 is highlighted (E to K). (B) Coiled-coil probability plot. The x-axis is the position of Mps2p amino acids and the y-axis is the probability (p) of being in a coiled-coil using the COILS program (http://www.ch.embnet.org/software/COILS form.html), based on the Lupas algorithm (Lupas et al., 1991).
The MPS2 gene was also identified in a search for proteins involved in the onset of anaphase. The cim3–1 and cim5–1 mutations affect two different regulatory subunits of the 26S proteasome (Ghislain et al., 1993). Both mutants arrest division at the G2/M transition with unsegregated chromosomes at the restrictive temperature of 37°C, suggesting that one or more proteins must be proteolyzed for cells to enter into anaphase. We searched for proteins involved in this proteasome-mediated metaphase/anaphase transition by screening for yeast genes whose overexpression would inhibit growth in the cim5–1 mutant, but not the wild-type strain, at the cim5–1 semipermissive temperature of 30°C (see MATERIALS AND METHODS). At this temperature, the cim5 mutant population of cells contains a higher than normal fraction of cells containing a 2N DNA content, suggesting that inefficient degradation of mitotic substrates of the proteasome was limiting the rate of cell division (Ghislain et al., 1993). We reasoned that the overexpression of a gene encoding such a substrate, or a protein involved in this process, might inhibit the growth of cells containing these mutant proteasomes. The overexpression of the MPS2 protein was toxic in these cells, suggesting that Mps2p may fit these criteria.
The predicted Mps2p sequence is 387 amino acids long and shows no strong sequence similarity to other proteins in the available databases (Figure 1A). Mps2p has a predicted molecular mass of 44,587 Da and a theoretical pI of 8.4. The Mps2p sequence contains a 17 amino acid hydrophobic stretch (residues 311–327) as a putative transmembrane domain (Kyte and Doolittle, 1982; Klein et al., 1985), three potential Cdc28 phosphorylation sites S/TPXK/R/Q (Songyang et al., 1994), and four potential destruction box sequences RXXLXXXXN/I (Glotzer et al., 1991). In addition, the Mps2p sequence reveals two regions capable of forming coiled-coils by the algorithm of Lupas (Lupas et al., 1991) (Figure 1B).
MPS2 Is an Essential Gene
To test whether MPS2 encodes an essential gene product, part of the chromosomal copy of MPS2 was replaced by homologous recombination with the HIS3 gene (see MATERIALS AND METHODS). Sporulated diploid yeast strains heterozygous for the null allele of MPS2 segregated a lethal mutation in tetrads inspected a few days after tetrad dissection (AM610, Table 1). Furthermore, the HIS3 marker for the null allele was not recovered in these tetrads, showing that it cosegregated with the lethal phenotype. The null allele could be recovered from sporulated diploids only when they contained a plasmid-borne copy of MPS2 (pRS316-MPS2). Furthermore, the mps2 null strains that contained a URA3-marked, plasmid-borne copy of MPS2 could not grow in the presence of 5-FOA, which induces plasmid loss, indicating that MPS2 is essential; however, on continued incubation (10–14 d) at 23°C, the dissection plates of diploids heterozygous for the null allele of MPS2 would contain very slow growing “pinhead” colonies that were found to contain the nutritional marker for the null allele (Figure 2A). The appearance of these colonies was variable from strain to strain and cross to cross, but approximately half of the spore clones containing a null allele eventually formed one of these very small colonies. On microscopic inspection of the dissected spores that did not form a small visible colony, most were found to have germinated and accumulated tens or hundreds of cells before ceasing to grow further. Some of the null-containing spore clones could be cultured, and PCR analysis of genomic DNA from these cells confirmed that they only contained the null allele of MPS2 and had not acquired a wild-type copy of the MPS2 gene (our unpublished results). Other null alleles of MPS2, in which the entire gene was replaced, were constructed throughout the course of this work, and all behaved identically.
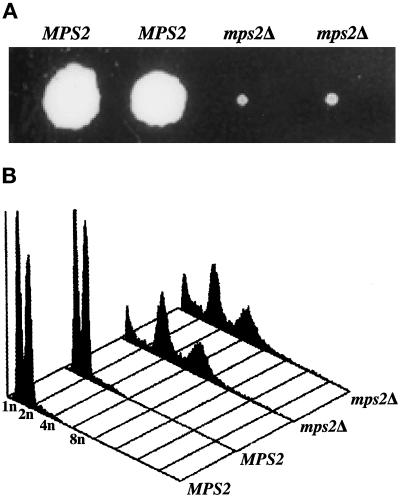
Figure 2.
Deletion of MPS2 results in abnormal ploidy. (A) A representative tetrad from the mps2Δ(116–387)::HIS3/MPS2 strain (AM610) grown for 14 d at 23°C. (B) Histogram of relative DNA content from each of the spore clones shown in A.
The ability to culture some of the strains containing the null allele of MPS2 allowed partial characterization of these strains. They were found to grow very slowly, with doubling times of 10–20 h. In addition, mps2 null strains are temperature-sensitive at 34°C. Unlike strains containing the mps2–1 mutation, which arrest as large budded cells with good viability on return to permissive temperature (Winey et al., 1991), strains containing a mps2 null allele did not show a uniform cell cycle arrest and rapidly lose viability at the nonpermissive temperature (our unpublished results). In the course of this analysis, we found by analyzing their DNA content that the surviving null strains were all apparent tetraploids (Figure 2B), with the exception of one diploid strain. There is also a peak at <1N that may correspond to either hypoploid cells or to debris from dead cells. The survival mechanism for the strains containing a null allele of MPS2 is not known.
Mps2p Is Present throughout the Cell Cycle
As described previously, Mps2p contains four potential destruction boxes (Figure 1A). The destruction box motif is required for the ubiquitination and subsequent degradation of several proteins, including Pds1p, Ase1p, and the mitotic cyclins, in mitosis and in the G1 phase of the cell cycle by the anaphase-promoting complex (APC)–proteasome pathway (Amon et al., 1994; Cohen-Fix et al., 1996; Juang et al., 1997). Degradation of these proteins is important for the initiation of chromosome segregation and for the exit from mitosis. Because Mps2p is toxic when overexpressed in a proteasome mutant, we reasoned that Mps2p could be a substrate for the proteasome. We tested whether Mps2p, like other APC substrates, is an unstable protein in α-factor–arrested cells. Although MPS2 mRNA levels are relatively constant through the cell cycle (Cho et al., 1998), we placed MPS2, tagged with nine copies of the myc epitope, under the control of a tetracycline-repressible promoter (Gari et al., 1997), whose expression is negligible in the presence of high antibiotic concentrations in the growth medium (5 μg/ml) and low in the presence of lower antibiotic concentrations (50 or 100 ng/ml). An exponentially growing culture of cells expressing this low, constitutive level of 9xmycN-Mps2p was treated with α-factor to arrest cells in G1 phase before Start, a stage of the cell cycle in which the APC is very active (Amon et al., 1994). The quantity of 9xmycN-Mps2p was compared in the exponentially growing and α-factor–arrested cells by immunoblotting of total cell extracts (Figure 3A). No decrease in 9xmycN-Mps2p levels was seen in the G1-arrested cells, suggesting that Mps2p is unlikely to be degraded by the APC–proteasome pathway.
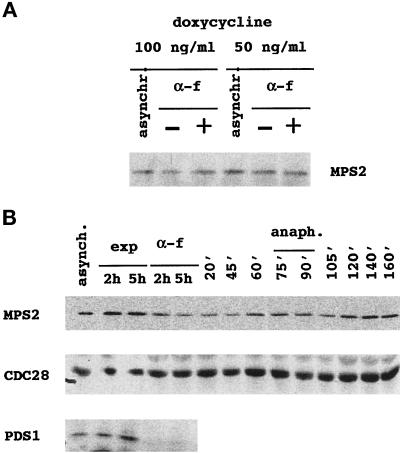
Figure 3.
Mps2p levels show little fluctuation during the cell cycle. (A) An overnight culture of MCL 94 cells containing pTet-9xmycN-MPS2 grown in the presence of 5 μg/ml doxycycline (repressing conditions) was diluted in the same selective medium containing 50 or 100 ng/ml doxycycline to allow a low constitutive expression from the tet promoter. Cells were grown overnight to early log phase at 30°C (asynchr.) and then divided into two separate cultures: one culture was left untreated (−) and the other was arrested in the G1 phase with α-factor (0.2 μM final concentration) for 2 h (+). Equal quantities of total cell protein were then subjected to SDS-PAGE, and proteins were transferred to membranes to determine the levels of 9xmycN-Mps2p by immunoblot analysis. 9xmycN-Mps2p is absent when cells are grown in the presence of fully repressing concentrations of doxycycline (5 μg/ml) or from cells that do not contain the 9xmycN-MPS2 plasmid. The G1 arrest was verified by flow cytometric analysis. (B) Wild-type cells expressing both 9xmycN-Mps2p and Pds1-HAp (MCL123) were grown to early log phase at 24°C (asynchr.) and then divided into two separate cultures: one culture was left untreated (exp) and the other was arrested in the G1 phase with α-factor (0.2 μM) for 2 or 5 h. After 2 h, >90% of the cells were already arrested in G1 phase as determined by flow cytometry analysis, as were cells treated with α-factor for 5 h. After this time, cells were released from the arrest by washing out the α-factor. Samples were taken at the indicated time points and analyzed for 9xmycN-Mps2p, Cdc28p, and Pds1-HAp by immunoblot analysis. Cell and nuclear morphology and DNA content were analyzed by propidium iodide staining.
We also tested whether the level of Mps2p oscillates during the cell cycle. For this experiment, 9xmycN-MPS2 was placed under the control of its endogenous promoter and integrated at the URA3 locus in a mps2Δ::HIS3 strain that initially contained a plasmid-borne copy of MPS2. To follow Pds1p levels under the same conditions, the wild-type PDS1 gene was replaced with a construct encoding a hemagglutinin epitope-tagged version of Pds1p, PDS1-HA (Cohen-Fix et al., 1996). Cells simultaneously expressing 9xmycN-Mps2p and Pds1-HAp were released synchronously from α-factor arrest. Cell morphology and 9xmycN-Mps2p, Pds1-HAp, and Cdc28p levels were determined at various timepoints after release from the α-factor arrest (Figure 3B). Levels of 9xmycN-Mps2p were roughly constant compared with the unvarying Cdc28p, except during the G1-S phases, in which 9xmycN-Mps2p appeared to decrease slightly. In contrast, Pds1-HAp completely disappeared from G1-arrested cells, as described previously (Cohen-Fix et al., 1996). We conclude that the level of 9xmycN-Mps2p does not oscillate significantly in the cell cycle of S. cerevisiae; however, we cannot exclude the possibility that the tagged version of Mps2p is somehow resistant to this type of regulation.
Mps2p Is an Integral Membrane Protein
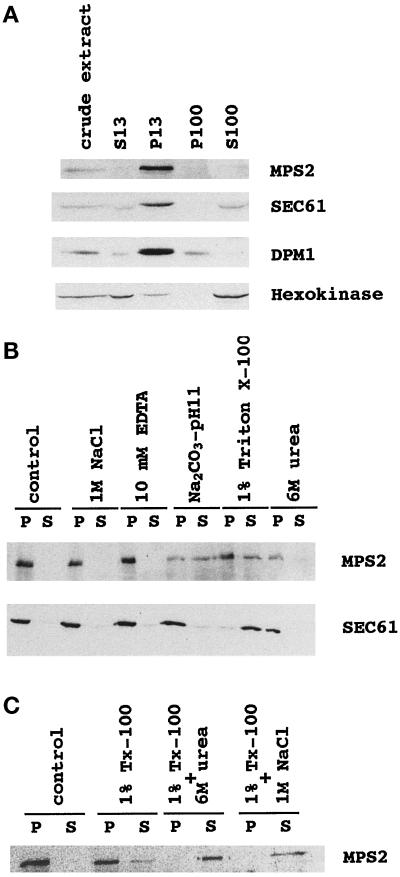
Mps2p contains a 17 amino acid hydrophobic segment that could correspond to a transmembrane domain. Biochemical fractionation experiments were undertaken in cells expressing 9xmycN-Mps2p at its normal endogenous levels (MCL120) to test whether Mps2p is a membrane protein. Extracts obtained by gentle spheroplast lysis were fractionated by differential centrifugation at 13,000 × g for 15 min followed by 60 min at 100,000 × g (see MATERIALS AND METHODS). At each stage, the supernatants and pellets were collected and analyzed by immunoblotting. Mps2p fractionated with nuclei and endoplasmic reticulum (ER) membranes in the 13,000 × g pellet (P13) and was undetectable in the supernatant or in the pellet obtained after 100,000 × g centrifugation (S100 or P100 fractions, respectively) (Figure 4A). To control for the purity of the different fractions, the same nitrocellulose membrane was reincubated with antibodies directed against Sec61p and Dpm1p, both of which are ER integral membrane proteins (Beck et al., 1990; Wilkinson et al., 1996), and with antibodies directed against the cytosolic hexokinase (van Tuinen and Riezman, 1987). Sec61p and Dpm1p cofractionated with Mps2p in the P13 fraction; hexokinase did not cofractionate.
Figure 4.
9xmycN-Mps2p fractionates as an integral membrane protein. A whole-cell extract was prepared from cells expressing 9xmycN-Mps2p from its normal promoter (MCL120) by gentle spheroplast lysis (see MATERIALS AND METHODS) and then fractionated as described in MATERIALS AND METHODS. (A) Cell fractionation: the same amount of protein from the whole-cell extract and S13, P13, P100, and S100 fractions were subjected to SDS-PAGE and immunoblotted with anti-myc, anti-Sec61p, anti-Dpm1p, and anti-hexokinase antibodies successively. (B) Solubilization of Mps2p from the P13 fraction prepared from MCL120. The pellet was extracted with lysis buffer (control) or lysis buffer containing 1 M NaCl, 50 mM Tris (pH 7.5)-10 mM EDTA, 0.2 M Na2CO3 (pH 11), 1% Triton X-100, or 6 M urea as described in MATERIALS AND METHODS. The mixtures were incubated on ice for 10 min and then centrifuged to produce a pellet fraction (P) and a supernatant fraction (S). Equivalent samples of the pellet and supernatant were analyzed by immunoblotting with anti-myc or anti-Sec61 antibodies. (C) Total solubilization of Mps2p protein from the P13. The P13 pellet was extracted with lysis buffer (control) or lysis buffer containing 1% Triton X-100, 1% Triton X-100 plus 6 M urea, or 1% Triton X-100 plus 1 M NaCl. Pellet (P) and supernatant (S) were separated and analyzed by immunoblotting.
To determine the nature of the association of 9xmycN-Mps2p with the nuclear/ER membrane fraction, the 13,000 × g pellet was extracted with various buffers and evaluated for the quantity of solubilized 9xmycN-Mps2p by immunoblotting. Agents that release peripheral membrane proteins (associated with membranes through electrostatic or hydrophobic interactions with integral membrane proteins) and soluble intranuclear proteins, such as 1 M NaCl or 6 M urea, did not solubilize 9xmycN-Mps2p from the P13 (Figure 4B). 9xmycN-Mps2p was partially solubilized by a detergent treatment (1% Triton X-100) or by an alkali treatment (0.2 M Na2CO3, pH 11). As an internal control for integral membrane protein behavior, we used Sec61p, which was completely solubilized by 1% Triton X-100 but was not solubilized by the sodium carbonate (Figure 4B). Overall, the fractionation characteristics of 9xmycN-Mps2p suggest that it is an integral membrane protein. The unusual partial extraction by alkaline sodium carbonate may be due to the rather small hydrophobic stretch of 17 amino acids containing Tyr326 in the Mps2p sequence. A negative charge would in principle be introduced on this residue at pH 11, and this ionization might be important for stripping Mps2p from the membrane. Similar extraction behavior was seen for Sss1p, an integral ER membrane protein containing a 16 amino acid hydrophobic stretch including a tyrosine residue (Esnault et al., 1994).
9xmycN-Mps2p was partially solubilized by 1% Triton X-100, whereas Sec61p, which spans the ER membrane multiple times, was completely extracted using the same conditions (Figure 4B). Partial extraction might indicate that the protein is trapped within multilayered membrane-detergent aggregates, which are destroyed by sonication; however, partial solubilization of 9xmycN-Mps2p from the P13 fraction was still observed after sonication in the presence of 1% Triton X-100 (our unpublished results). One possibility to explain this behavior is that 9xmycN-Mps2p could be physically associated with other proteins that prevent its full solubilization from membranes in the presence of detergent. To test this possibility, 9xmycN-Mps2p in the P13 was extracted with 1% Triton X-100 containing 6 M urea (to weaken hydrophobic interactions) or Triton X-100 containing 1 M NaCl (to neutralize electrostatic bonds). As shown in Figure 4C, 9xmycN-Mps2p was completely solubilized with both double treatments, suggesting that hydrophobic and electrostatic interactions could be preventing total solubilization of 9xmycN-Mps2p by detergent treatment alone.
Mps2p Is Localized at the SPB and the NE/ER
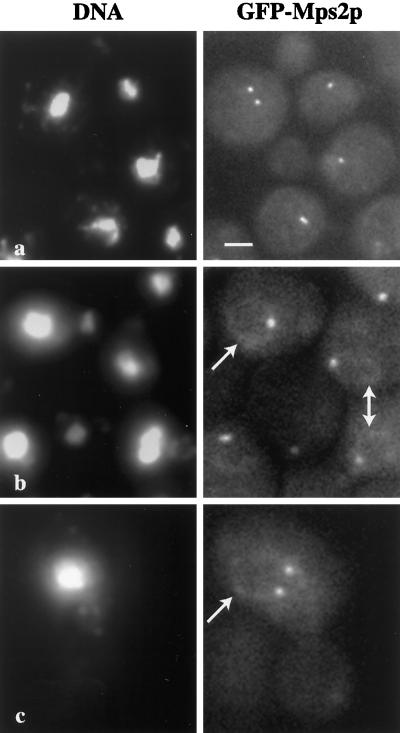
We localized tagged Mps2p in whole cells to confirm and potentially extend the 9xmycN-Mps2p fractionation data. To visualize Mps2p in living cells, Mps2p was tagged at the amino terminus with the green fluorescent protein (GFP). As shown in Figure 5, one or two spots of strong autofluorescence were visible that were coincident with the nuclear DNA (Figure 5, a–c). Additionally, we often observed a weaker, perinuclear GFP-Mps2p signal (Figure 5, b and c, arrows). The number, intensity, and location of the brighter spots of fluorescence suggested that they correspond to SPBs. We confirmed this localization by two methods. First, we used whole-cell indirect immunofluorescence deconvolution microscopy to determine whether Mps2p colocalized with a known SPB component, Spc42p (Donaldson and Kilmartin, 1996). We used a strain that contains both Mps2p epitope tagged at the C terminus with 9xmyc (9xmycC-Mps2p; see MATERIALS AND METHODS) and Spc42p-GFP (Chial et al., 1998; Schutz and Winey, 1998). Similar to the GFP-Mps2p, 9xmycC-Mps2p localized in one or two brighter spots around the nucleus and also at the nuclear periphery (Figure 6A, 9xmycC-Mps2p). In these cells, the brighter spots of 9xmycC-Mps2p localization (red) and the Spc42-GFP (green) signals were coincident (Figure 6A, combined; overlap is yellow). It should be noted that the genetic behavior of 9xmycC-MPS2 is somewhat different from that of wild-type MPS2. Cells expressing 9xmycC-Mps2p can increase in ploidy spontaneously (haploid to diploid, an mps2–1 phenotype) but are not temperature sensitive (our unpublished results), suggesting a very mild defect in Mps2p function. Nonetheless, the localization pattern is similar to that of GFP-Mps2p, for which cells stably remain haploid. Furthermore, we observe Mps2p at the SPB during various stages of the cell cycle, suggesting that Mps2p is present at the SPB throughout the cell cycle.
Figure 5.
Localization of GFP-Mps2p in living cells. Several cells are shown in a, in which GFP-Mps2p (strain MCL175) localizes in one or two discrete foci coincident with the nuclear DNA. The cell with two distinct spots of fluorescence is a small budded cell. The cells shown in b reveal that GFP-Mps2p also localizes to the nuclear periphery. Several cells are shown with one bright spot and a fainter signal at the nuclear periphery (arrows). A higher magnification of a budded cell is shown in c with two bright spots and a perinuclear localization. The GFP-Mps2p was visualized via autofluorescence of GFP, and the DNA was visualized with Hoechst. A single plane of focus is shown. Bar, 1 μm.
We also explored whether the perinuclear localization of 9xmycC-Mps2p corresponded to NPCs. For this experiment, we used a strain expressing both 9xmycC-Mps2p and Nup49p-GFP. Nup49p is a well characterized NPC protein, or nucleoporin, that exhibits a characteristic punctate, nuclear rim localization pattern by whole-cell immunofluorescence microscopy (Grandi et al., 1995; Bucci and Wente, 1997). We observed very little overlap between 9xmycC-Mps2p and Nup49-GFP in any of the cells that we examined. An example of this is shown in Figure 6B. There are discrete areas of colocalization between the two proteins (yellow), but their localization patterns are quite distinct, suggesting that Mps2p is not a nucleoporin. In addition, there are bright spots of 9xmycC-Mps2p at the nuclear periphery that are devoid of Nup49p signal, indicative of where the SPB is located. The signal of 9xmycC-Mps2p and GFP-Mps2p at the nuclear periphery is weaker than at the SPB. On the basis of this observation and the fractionation of 9xmycN-Mps2p with membranes, we propose that Mps2p is a minor membrane component of the NE.
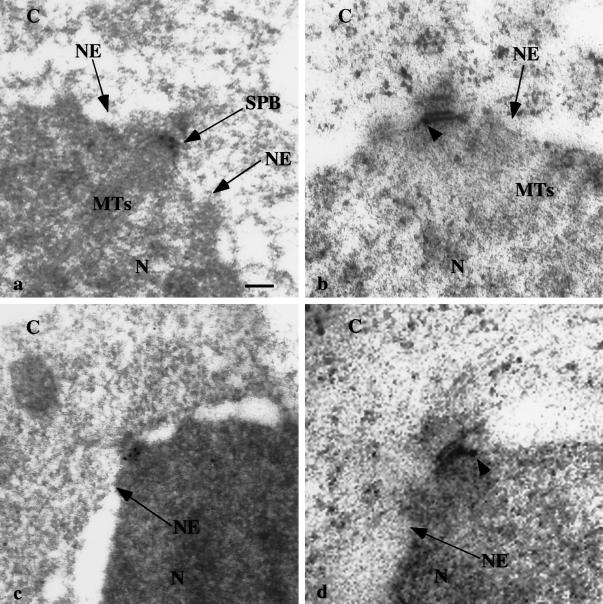
The second method that was used to confirm the SPB localization was immunoelectron microscopy of cells expressing GFP-Mps2p. As shown in Figure 7, the SPBs from four different cells were decorated with one (b) to five (c) gold particles, with little to no background (of 12 SPBs examined, all were decorated with ≥1 gold particle). We did not observe a significant number of gold particles in the NE; however, this could be due to lower levels of Mps2p in the NE than at the SPB (see DISCUSSION). We conclude from these experiments that Mps2p tagged with either GFP or 9xmyc is a component of the SPB and the NE.
Figure 7.
GFP-Mps2p localizes to the SPB. Cells expressing GFP-Mps2p were prepared for immunoelectron microscopy using anti-GFP antibodies and colloidal gold-conjugated secondary antibodies. Thin sections of four different cells are shown. C, cytoplasm; N, nucleoplasm; NE, nuclear envelope; MTs, microtubules; SPB, spindle pole body. Arrowheads show the localization of individual gold particles in b and d. Gold particles were observed in all 12 SPBs examined. Bar, 0.1 μm.
DISCUSSION
We report here that the MPS2 gene encodes a novel 44-kDa integral membrane protein localized at SPBs and the NE. MPS2 has been previously reported to be required for SPB duplication (Winey et al., 1991). Here, we provide evidence that MPS2 is linked to the ubiquitin–proteasome pathway.
Although we consider the MPS2 gene to be essential, some of the spore clones containing a null allele were able to give rise to a colony containing very slow growing cells with abnormal ploidy. The fact that defects in SPB duplication can lead to increases in ploidy via a monopolar mitosis is well documented (Winey et al., 1991). This event has also been observed at the permissive temperature for strains containing conditional alleles of CDC31 and KAR1 (Schild et al., 1981; Rose and Fink, 1987). It seems possible that the surviving mps2 null strains are not perfect tetraploids, but are aneuploid in a manner that increases or decreases the dosage of some gene(s), which allows for the nulls to survive.
The fact that MPS2 was identified in a screen for possible regulatory or substrate proteins in the proteasome pathway is intriguing. On the basis of the genetic interaction between CIM5 and MPS2, we sought to determine whether Mps2p was a potential substrate for the ubiquitin–proteasome pathway by examining protein levels throughout the cell cycle; however, with Mps2p tagged at its amino terminus with nine copies of the myc epitope and expressed from its normal promoter (Figure 3), we were unable to see any major fluctuations in Mps2p levels during the cell cycle, nor were we able to see any proteolysis of the protein in cells arrested in the G1 phase of the cell cycle with α-factor, a situation in which destruction box proteins are rapidly degraded.
At the present time, the mechanism of how cim5–1 cells die in the presence of overexpressed Mps2p remains unknown. Interestingly, it was reported recently that GFP-Cim5p–labeled proteasomes are enriched in the NE/ER in living yeast cells, indicating that proteasomal degradation is concentrated at this compartment in yeast (Enenkel et al., 1998). Because Mps2p is also localized at the NE, it is possible that Mps2p may interact with the proteasome, although it does not appear that Mps2p is a proteasome substrate. If so, overexpression of Mps2p may inhibit the normal function or localization of the proteasome such that the overexpression becomes toxic in cim5–1 cells containing a poorly functioning proteasome. In addition to the interaction between MPS2 and CIM5, the only other known gene that links SPB duplication to the ubiquitin–proteasome pathway is PCS1 (McDonald and Byers, 1997). Like CIM5, PCS1 encodes a proteasome 19S cap subunit (Russell et al., 1996). PCS1 function in SPB duplication appears to be required before MPS2 function, in that SPB duplication is blocked at an earlier stage in pcs1 mutants cells than in mps2–1 cells (Winey et al., 1991; McDonald and Byers, 1997). Taken together, these observations strengthen the idea that SPB duplication may be regulated by the ubiquitin–proteasome pathway.
It is clear that Mps2p has a primary role in insertion of the nascent SPB into the NE. First, mps2–1 cells exhibit a monopolar spindle because of a defective SPB. The nascent, defective SPB is not properly inserted into the NE and lies on the cytoplasmic side of the envelope. Moreover, the SPB appears to lack the inner (nuclear) plaque and its associated nuclear microtubules, which are observed in functional SPBs. Second, tagged Mps2p localizes to the SPB as demonstrated by fluorescence microscopy and immunoelectron microscopy. Third, YGL075c/Mps2p was identified by mass spectrometry in highly purified preparations of SPBs (Wigge et al., 1998), although these authors did not cytologically localize the YGL075c protein in this study. Taken together, these data suggest that Mps2p is directly involved in insertion of the nascent SPB into the NE, as originally proposed (Winey and Byers, 1993).
We have suggested previously that the NDC1 protein also carries out this insertion event (Winey et al., 1993). The NDC1 gene function appears to be required at the same step in SPB duplication based on the execution point and the same terminal SPB morphology. Ndc1p is a membrane protein and is a shared component of NPCs and SPBs (Chial et al., 1998). The work presented here extends the parallels between NDC1 and MPS2. Both genes encode essential membrane proteins that localize to SPBs; however, unlike Ndc1p, Mps2p does not appear to be a component of NPCs. The caveat to this is that Mps2p is expressed at a much lower level than Ndc1p (compare Figures 5–7 with Chial et al., 1998). This is most evident in the weak nuclear periphery signal (Figures 5 and 6) and a lack of gold particles found in the NE (Figure 7). Although we cannot rule out the possibility that Mps2p could be transiently associated with NPCs, Mps2p fails all criteria used to show that Ndc1p is a component of NPCs.
On the basis of the SPB localization of Mps2p and Ndc1p, one possible scenario could be that Mps2p and Ndc1p are required as membrane components for inserting the nascent SPB into the NE. There is only one other known SPB component with a putative transmembrane domain, Spc105p, which appears to localize to the nucleoplasmic face of the SPB and to the nuclear microtubules (Wigge et al., 1998). It is not known whether mutations in this protein cause an SPB duplication defect. In the future, the ultrastructural details of assembling a new SPB will be important for a more complete understanding of Mps2p and Ndc1p function. In summary, MPS2 encodes an integral membrane protein that localizes to SPBs and the NE. Future experiments with MPS2 and the genes with which it interacts may reveal the specific function of Mps2p and provide us with new insights into the role of membrane proteins in SPB assembly.
ACKNOWLEDGMENTS
We are indebted to Marie-Claude Marsolier and Ivan Le Masson for their invaluable help with constructs. We thank Lipika Roy for sequencing the mps2–1 mutation. We are grateful to Tom Giddings, Jr. and Mary Morphew for the immunoelectron microscopy. We thank Michel Ghislain for the initial suggestion for the cim5 screen and for his interest and collaboration in the project. We are grateful to Orna Cohen-Fix and Doug Koshland, Kim Nasmyth, Pam Silver, Aaron Straight, Valerie Doye, and Heidi Chial for strains, plasmids, and discussions, and to Andrea Castillo and Heidi Chial for critical reading of this manuscript. M.C.M.-C. was supported by postdoctoral research fellowships from the European Union (Human Capital and Mobility Program) and the Ministerio Español de Educacíon. This work was also supported by a European Union BioMed 2 network on the proteasome. S.M. was supported by a National Institutes of Health (NIH) postdoctoral research fellowship (GM-18473); A.M. was supported by grants from the Howard Hughes Medical Institute, the Ford Foundation, and an NIH Training Grant. This work was also supported by the National Science Foundation (YIA MCB-9357033, M.W.) and the American Cancer Society (CB-197, M.W.). Deconvolution microscopy in The Department of Molecular, Cellular, and Developmental Biology was made possible, in part, by a gift from Virginia and Mel Clark.
Abbreviations used:
- APC
anaphase-promoting complex
- GFP
green fluorescent protein
- NE
nuclear envelope
- NPC
nuclear pore complex
- SPB
spindle pole body
REFERENCES
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck PJ, Orlean P, Albright C, Robbins PW, Gething MJ, Sambrook JF. The Saccharomyces cerevisiae DPM1 gene encoding dolichol-phosphate-mannose synthase is able to complement a glycosylation-defective mammalian cell line. Mol Cell Biol. 1990;10:4612–4622. doi: 10.1128/mcb.10.9.4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bucci M, Wente SR. In vivo dynamics of nuclear pore complexes in yeast. J Cell Biol. 1997;136:1185–1199. doi: 10.1083/jcb.136.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullitt E, Rout MP, Kilmartin JV, Akey CW. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Shriver K, Goetsch L. The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae. J Cell Sci. 1978;30:331–352. doi: 10.1242/jcs.30.1.331. [DOI] [PubMed] [Google Scholar]
- Chial H, Rout M, Giddings T, Winey M. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J Cell Biol. 1998;143:1789–1800. doi: 10.1083/jcb.143.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Kilmartin JV. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Mulligan JT, Ramer SW, Spottswood M, Davis RW. Lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc Natl Acad Sci USA. 1991;88:1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C, Lehmann A, Kloetzel PM. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 1998;17:6144–6154. doi: 10.1093/emboj/17.21.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault Y, Feldheim D, Blondel MO, Schekman R, Kepes F. SSS1 encodes a stabilizing component of the Sec61 subcomplex of the yeast protein translocation apparatus. J Biol Chem. 1994;269:27478–27485. [PubMed] [Google Scholar]
- Friedman DB, Sundberg HA, Huang EY, Davis TN. The 110-kDa spindle pole body component of Saccharomyces cerevisiae is a phosphoprotein that is modified in a cell cycle-dependent manner. J Cell Biol. 1996;132:903–914. doi: 10.1083/jcb.132.5.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gordon C, McGurk G, Dillon P, Rosen C, Hastie ND. Defective mitosis due to a mutation in the gene for a fission yeast 26S protease subunit. Nature. 1993;366:355–357. doi: 10.1038/366355a0. [DOI] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 1995;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink G. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–933. [PubMed] [Google Scholar]
- Harris SL, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Horton RM. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- Hyams JS, Borisy GG. Nucleation of microtubules in vitro by isolated spindle pole bodies of the yeast Saccharomyces cerevisiae. J Cell Biol. 1978;78:401–414. doi: 10.1083/jcb.78.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang YL, Huang J, Peters JM, McLaughlin ME, Tai CY, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lauze E, Stoelcker B, Luca FC, Weiss E, Schutz AR, Winey M. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Kornbluth S. Regulatory roles of cyclin dependent kinase phosphorylation in cell cycle control. Curr Opin Cell Biol. 1996;8:795–804. doi: 10.1016/s0955-0674(96)80080-9. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- McDonald HB, Byers B. A proteasome cap subunit required for spindle pole body duplication in yeast. J Cell Biol. 1997;137:539–553. doi: 10.1083/jcb.137.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Knop M, Schiebel E. Spc98p directs the yeast gamma-tubulin complex into the nucleus and is subject to cell cycle-dependent phosphorylation on the nuclear side of the spindle pole body. Mol Biol Cell. 1998;9:775–793. doi: 10.1091/mbc.9.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramer SW, Elledge SJ, Davis RW. Dominant genetics using a yeast genomic library under the control of a strong inducible promoter. Proc Natl Acad Sci USA. 1992;89:11589–11593. doi: 10.1073/pnas.89.23.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Fink GR. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruohola H, Ferro-Novick S. Sec53, a protein required for an early step in secretory protein processing and transport in yeast, interacts with the cytoplasmic surface of the endoplasmic reticulum. Proc Natl Acad Sci USA. 1987;84:8468–8472. doi: 10.1073/pnas.84.23.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Sathyanarayana UG, Johnston SA. Isolation and characterization of SUG2. J Biol Chem. 1996;271:32810–32817. doi: 10.1074/jbc.271.51.32810. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Laboratory; 1989. [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schild D, Ananthaswamy HN, Mortimer RK. An endomitotic effect of a cell cycle mutation of Saccharomyces cerevisiae. Genetics. 1981;97:551–562. doi: 10.1093/genetics/97.3-4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz AR, Winey M. New alleles of the yeast MPS1 gene reveal multiple requirements in spindle pole body duplication. Mol Biol Cell. 1998;9:759–774. doi: 10.1091/mbc.9.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Botstein D. A gene required for the separation of chromosomes on the spindle apparatus in yeast. Cell. 1986;44:65–76. doi: 10.1016/0092-8674(86)90485-x. [DOI] [PubMed] [Google Scholar]
- Vallen EA, Scherson TY, Roberts T, van Zee K, Rose MD. Asymmetric mitotic segregation of the yeast spindle pole body. Cell. 1992;69:505–515. doi: 10.1016/0092-8674(92)90451-h. [DOI] [PubMed] [Google Scholar]
- van Tuinen E, Riezman H. Immunolocalization of glyceraldehyde-3-phosphate dehydrogenase, hexokinase, and carboxypeptidase Y in yeast cells at the ultrastructural level. J Histochem Cytochem. 1987;35:327–333. doi: 10.1177/35.3.3546482. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson BM, Critchley AJ, Stirling CJ. Determination of the transmembrane topology of yeast Sec61p, an essential component of the endoplasmic reticulum translocation complex. J Biol Chem. 1996;271:25590–25597. doi: 10.1074/jbc.271.41.25590. [DOI] [PubMed] [Google Scholar]
- Winey M, Byers B. Assembly and functions of the spindle pole body in budding yeast. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Hoyt MA, Chan C, Goetsch L, Botstein D, Byers B. NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J Cell Biol. 1993;122:743–751. doi: 10.1083/jcb.122.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]