Abstract
The bacteriophage phi X174 strain ins6 constructed previously was used to investigate the maximum genome size that could be packaged into the icosahedral phage without concomitant loss of phage viability. The J-F intercistronic region of ins6, which already contains an insert of 117 base pairs with a unique PvuII site, was enlarged further by insertion of HaeIII restriction fragments of the plasmid pBR322 into that PvuII site. By using a biochemical approach for the site-specific mutagenesis as well as selection of mutant genomes, a series of mutants was isolated with genomes of up to 5,730 nucleotides, 6.4% larger than that of the wild-type DNA. Phages with genomes larger than 5,550 nucleotides were highly unstable and were rapidly outgrown by spontaneously occurring deletion mutants. The data predict that genomes of at least 6,090 nucleotides could be constructed and, most likely, packaged, but the resulting phages would not grow well. We speculate that the volume of the phage capsid is not the limiting factor of genome size or is not the only limiting factor.
Full text
PDF


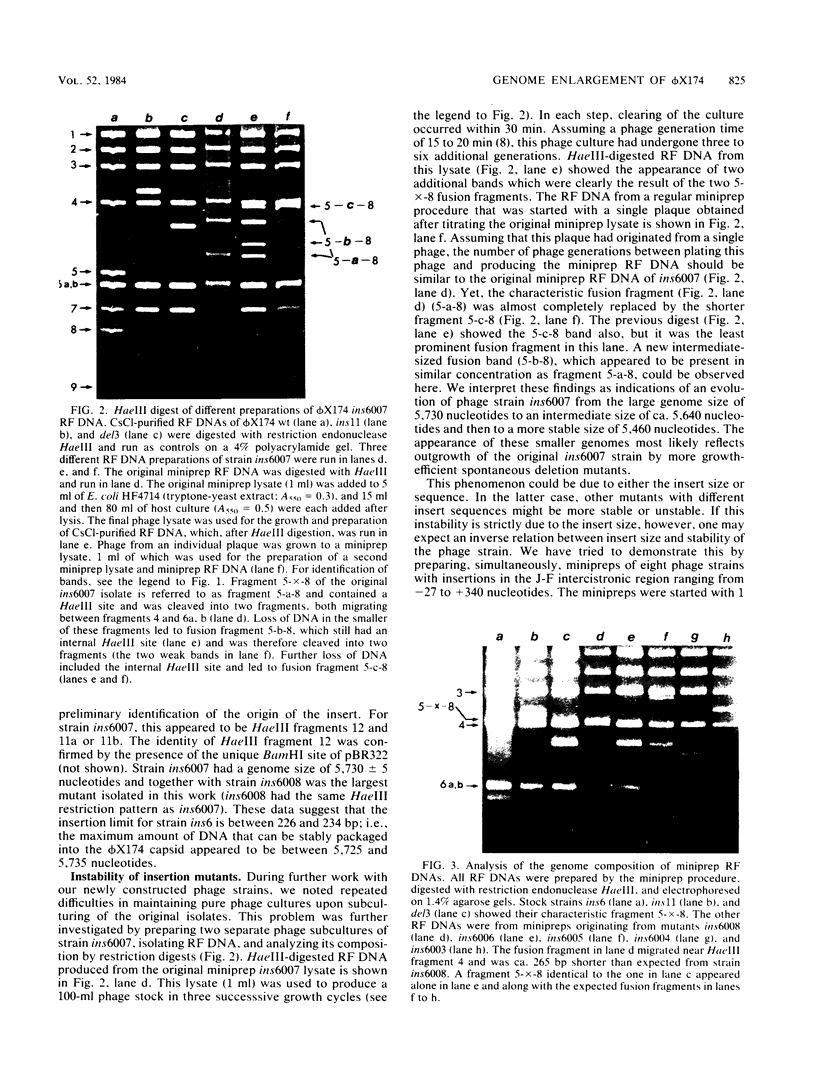


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aoyama A., Hamatake R. K., Hayashi M. Morphogenesis of phi X174: in vitro synthesis of infectious phage from purified viral components. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7285–7289. doi: 10.1073/pnas.78.12.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbow R. M., Hutchison C. A., Fabricant J. D., Sinsheimer R. L. Genetic Map of Bacteriophage phiX174. J Virol. 1971 May;7(5):549–558. doi: 10.1128/jvi.7.5.549-558.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardies S. C., Patient R. K., Klein R. D., Ho F., Reznikoff W. S., Wells R. D. Construction and mapping of recombinant plasmids used for the preparation of DNA fragments containing the Escherichia coli lactose operator and promoter. J Biol Chem. 1979 Jun 25;254(12):5527–5534. [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M., Müller U. R. Role for the J-F intercistronic region of bacteriophages phi X174 and G4 in stability of mRNA. J Virol. 1983 Oct;48(1):186–196. doi: 10.1128/jvi.48.1.186-196.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U. R. Enzymatic construction and selection of bacteriophage G4 mutants with modifications of a DNA secondary structure in the J-F intercistronic region. J Virol. 1983 Oct;48(1):170–179. doi: 10.1128/jvi.48.1.170-179.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U. R., Wells R. D. Intercistronic regions in phi X174 DNA. I. Construction of mutants with altered intercistronic regions between genes J and F. J Mol Biol. 1980 Jul 25;141(1):1–24. doi: 10.1016/s0022-2836(80)80026-x. [DOI] [PubMed] [Google Scholar]
- Müller U. R., Wells R. D. Intercistronic regions in phi X174 DNA. II. Biochemical and biological analysis of mutants with altered intercistronic regions between genes J and F. J Mol Biol. 1980 Jul 25;141(1):25–41. doi: 10.1016/s0022-2836(80)80027-1. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Molecular weight estimation and separation of ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry. 1968 Feb;7(2):668–674. doi: 10.1021/bi00842a023. [DOI] [PubMed] [Google Scholar]
- Romantschuk M. L., Müller U. R. Mutations in the J-F intercistronic region of bacteriophages phi X174 and G4 affect the regulation of gene expression. J Virol. 1983 Oct;48(1):180–185. doi: 10.1128/jvi.48.1.180-185.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Hardies S. C., Horn G. T., Klein B., Larson J. E., Neuendorf S. K., Panayotatos N., Patient R. K., Selsing E. RPC-5 column chromatography for the isolation of DNA fragments. Methods Enzymol. 1980;65(1):327–347. doi: 10.1016/s0076-6879(80)65043-5. [DOI] [PubMed] [Google Scholar]
- van der Avoort H. G., van Arkel G. A., Weisbeek P. J. Cloned bacteriophage phi X174 DNA sequence interferes with synthesis of the complementary strand of infecting bacteriophage phi X174. J Virol. 1982 Apr;42(1):1–11. doi: 10.1128/jvi.42.1.1-11.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ende A., Teertstra R., Weisbeek P. J. Initiation and termination of the bacteriophage phi X174 rolling circle DNA replication in vivo: packaging of plasmid single-stranded DNA into bacteriophage phi X174 coats. Nucleic Acids Res. 1982 Nov 11;10(21):6849–6863. doi: 10.1093/nar/10.21.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]