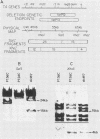
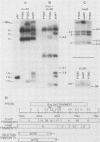
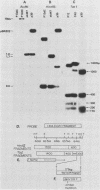
Abstract
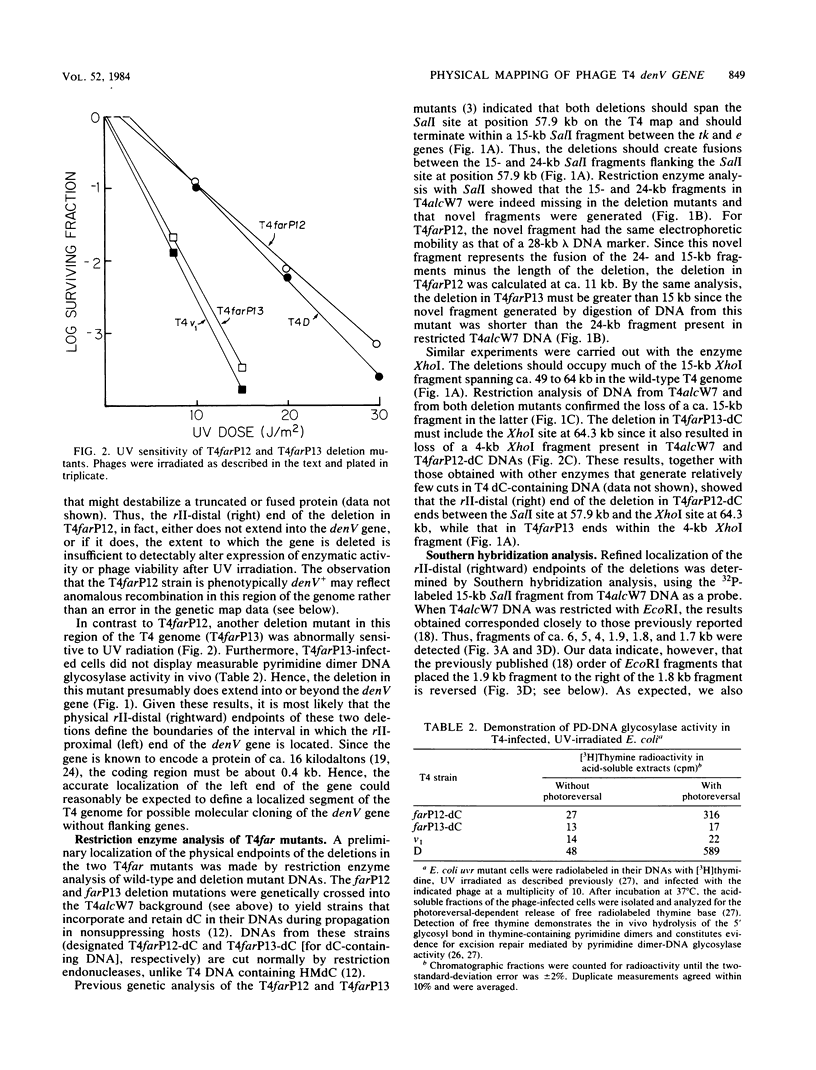
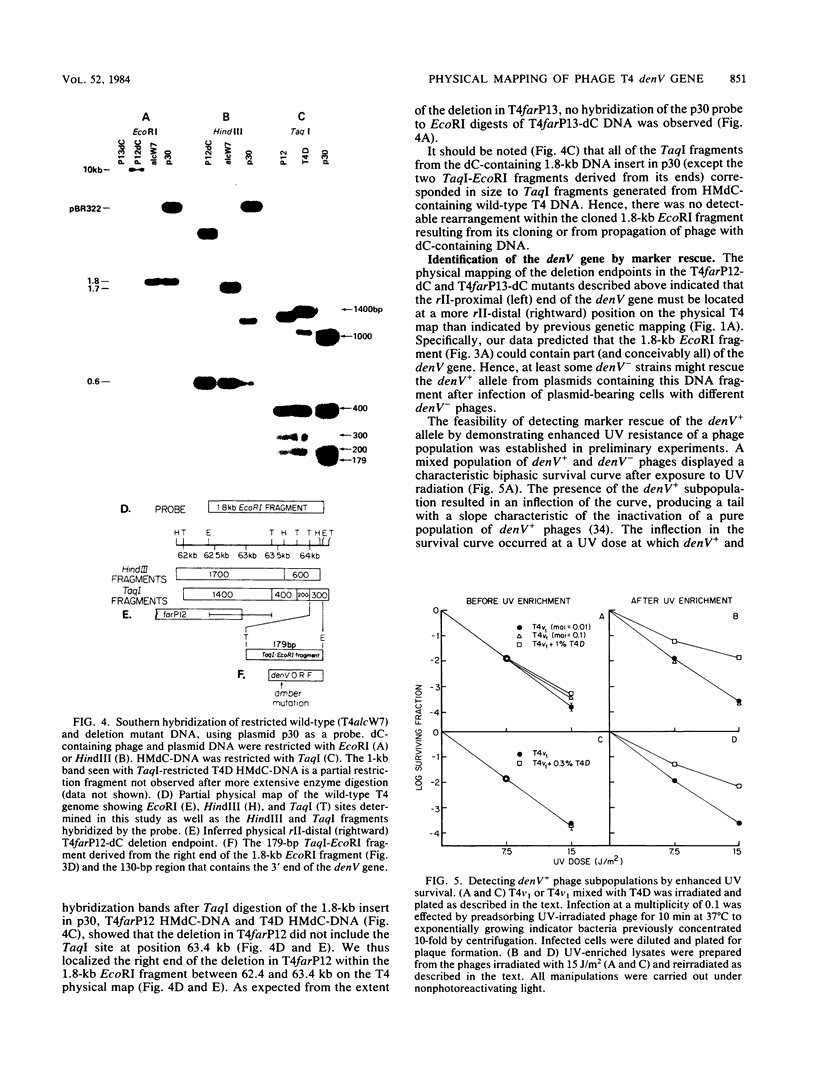
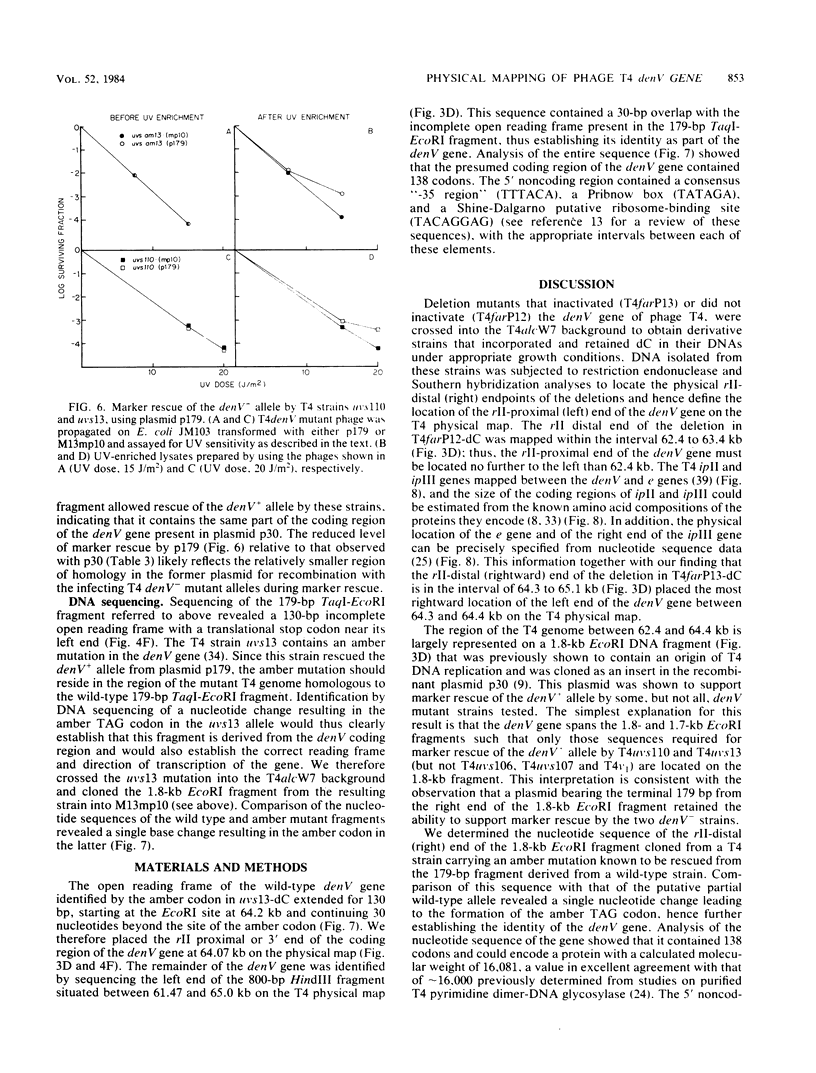
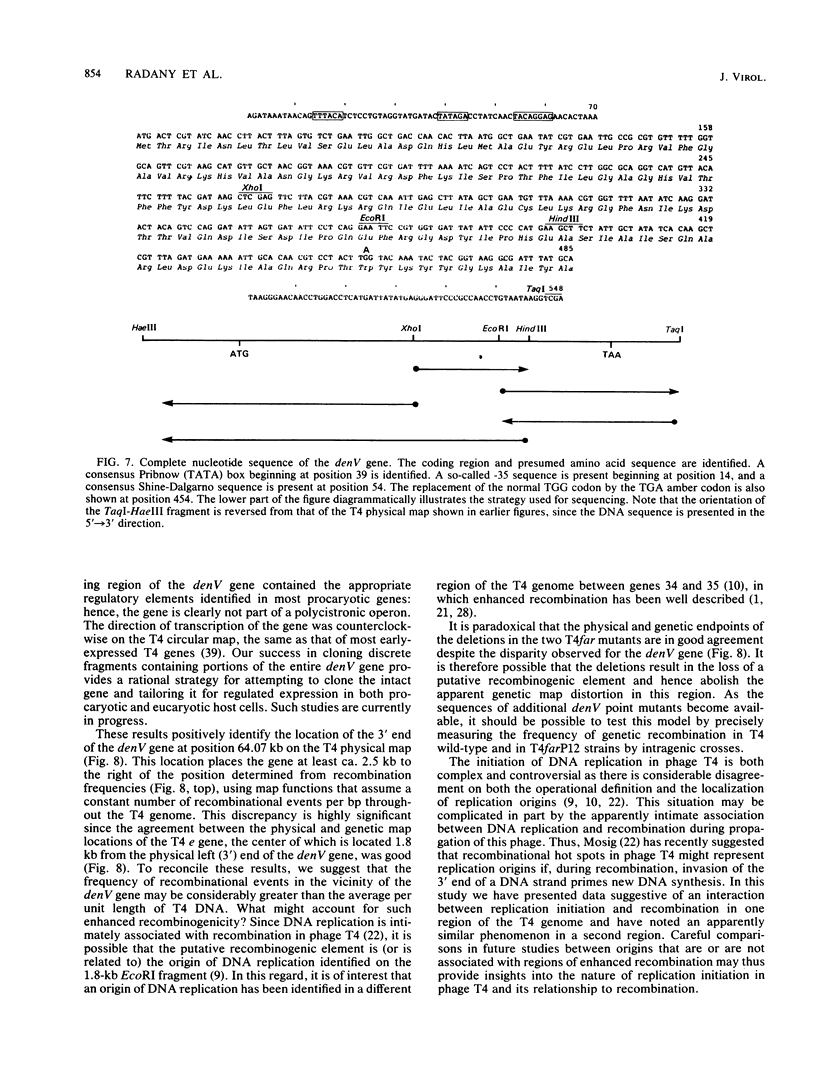
Phage T4 deletion mutants that are folate analog resistant (far) and contain deletions in the region of the T4 genome near denV have been isolated previously. We showed that one of these mutants (T4farP12) expressed normal denV gene activity, whereas another mutant (T4farP13) was defective in the denV gene. The rII-distal (right) physical endpoints of these deletions defined the limits of the interval in which the rII-proximal (left) endpoint of the denV gene should be located. The deletion endpoints were identified by restriction and Southern hybridization analyses of phage derivatives containing deoxycytidine instead of hydroxymethyldeoxycytidine in their DNAs. The results of these analyses localized the rII-proximal (left) end of the denV gene to a region between 62.4 and 64.3 kilobases on the T4 physical map. denV+ phage resulted from marker rescue with two of five denV- alleles tested, using plasmids containing a 1.8-kilobase fragment from this region or a 179-base-pair terminal fragment derived from it. Sequencing of the 179-base-pair fragment from wild-type DNA showed a 130-base-pair open reading frame with its termination codon at the rII-proximal end. Confirmation that this open reading frame is part of the denV coding sequence was obtained by identifying a TAG amber codon in the homologous DNA derived from a denV amber mutant strain. This mutant strain rescued the denV+ allele from plasmids containing the wild-type sequence. An adjacent overlapping restriction fragment was also cloned, permitting determination of the remaining denV gene sequence. Based on these results, the 3' end of the coding region of the denV locus was mapped to kilobase position 64.07 on the T4 physical map, and the 5' end was mapped to position 64.48.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beckendorf S. K., Wilson J. H. A recombination gradient in bacteriophage T4 gene 34. Virology. 1972 Nov;50(2):315–321. doi: 10.1016/0042-6822(72)90382-0. [DOI] [PubMed] [Google Scholar]
- Chace K. V., Hall D. H. Characterization of new regulatory mutants of bacteriophage T4. II. New class of mutants. J Virol. 1975 Apr;15(4):929–945. doi: 10.1128/jvi.15.4.929-945.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Linn S. DNA N-glycosylases and UV repair. Nature. 1980 Sep 18;287(5779):203–208. doi: 10.1038/287203a0. [DOI] [PubMed] [Google Scholar]
- Friedberg E. C., King J. J. Endonucleolytic cleavage of UV-irradiated DNA controlled by the V+ gene in phage T4. Biochem Biophys Res Commun. 1969 Nov 6;37(4):646–651. doi: 10.1016/0006-291x(69)90859-6. [DOI] [PubMed] [Google Scholar]
- Gordon L. K., Haseltine W. A. Comparison of the cleavage of pyrimidine dimers by the bacteriophage T4 and Micrococcus luteus UV-specific endonucleases. J Biol Chem. 1980 Dec 25;255(24):12047–12050. [PubMed] [Google Scholar]
- HARM W. Mutants of phage T4 with increased sensitivity to ultraviolet. Virology. 1963 Jan;19:66–71. doi: 10.1016/0042-6822(63)90025-4. [DOI] [PubMed] [Google Scholar]
- Isobe T., Black L. W., Tsugita A. Primary structure of bacteriophage T4 internal protein II and characterization of the cleavage upon phage maturation. J Mol Biol. 1976 Apr 5;102(2):349–365. doi: 10.1016/s0022-2836(76)80059-9. [DOI] [PubMed] [Google Scholar]
- King G. J., Huang W. M. Identification of the origins of T4 DNA replication. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7248–7252. doi: 10.1073/pnas.79.23.7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. S., Hanawalt P. C. Expression of the denV gene of bacteriophage T4 cloned in Escherichia coli. Proc Natl Acad Sci U S A. 1981 May;78(5):2796–2800. doi: 10.1073/pnas.78.5.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E. Reactivation of Irradiated Bacteriophage by Transfer of Self-Reproducing Units. Proc Natl Acad Sci U S A. 1947 Sep;33(9):253–264. doi: 10.1073/pnas.33.9.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOROWITZ H. J. Absorption effects in volume irradiation of microorganisms. Science. 1950 Mar 3;111(2879):229–229. doi: 10.1126/science.111.2879.229-a. [DOI] [PubMed] [Google Scholar]
- McMillan S., Edenberg H. J., Radany E. H., Friedberg R. C., Friedberg E. C. den V gene of bacteriophage T4 codes for both pyrimidine dimer-DNA glycosylase and apyrimidinic endonuclease activities. J Virol. 1981 Oct;40(1):211–223. doi: 10.1128/jvi.40.1.211-223.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileham A. J., Murray N. E., Revel H. R. Gamma-T4 hybrid bacteriophage carrying the thymidine kinase gene of bacteriophage T4. J Virol. 1984 May;50(2):619–622. doi: 10.1128/jvi.50.2.619-622.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton K., Durphy M., Taylor R., Friedberg E. C. The ultraviolet endonuclease of bacteriophage T4. Further characterization. J Biol Chem. 1975 Apr 25;250(8):2823–2829. [PubMed] [Google Scholar]
- Mosig G. A map of distances along the DNA molecule of phage T4. Genetics. 1968 Jun;59(2):137–151. doi: 10.1093/genetics/59.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y., Sekiguchi M. Physical association of pyrimidine dimer DNA glycosylase and apurinic/apyrimidinic DNA endonuclease essential for repair of ultraviolet-damaged DNA. Proc Natl Acad Sci U S A. 1981 May;78(5):2742–2746. doi: 10.1073/pnas.78.5.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y., Yamashita K., Sekiguchi M. Purification and characterization of normal and mutant forms of T4 endonuclease V. J Biol Chem. 1982 Mar 10;257(5):2556–2562. [PubMed] [Google Scholar]
- Owen J. E., Schultz D. W., Taylor A., Smith G. R. Nucleotide sequence of the lysozyme gene of bacteriophage T4. Analysis of mutations involving repeated sequences. J Mol Biol. 1983 Apr 5;165(2):229–248. doi: 10.1016/s0022-2836(83)80255-1. [DOI] [PubMed] [Google Scholar]
- Radany E. H., Friedberg E. C. A pyrimidine dimer-DNA glycosylase activity associated with the v gene product of bacterophage T4. Nature. 1980 Jul 10;286(5769):182–185. doi: 10.1038/286182a0. [DOI] [PubMed] [Google Scholar]
- Radany E. H., Friedberg E. C. Demonstration of pyrimidine dimer-DNA glycosylase activity in vivo: bacteriophage T4-infected Escherichia coli as a model system. J Virol. 1982 Jan;41(1):88–96. doi: 10.1128/jvi.41.1.88-96.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel H. R. Molecular cloning of the T4 genome: organization and expression of the tail fiber gene cluster 34--38. Mol Gen Genet. 1981;182(3):445–455. doi: 10.1007/BF00293934. [DOI] [PubMed] [Google Scholar]
- STAHL F. W., EDGAR R. S., STEINBERG J. THE LINKAGE MAP OF BACTERIOPHAGE T4. Genetics. 1964 Oct;50:539–552. doi: 10.1093/genetics/50.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STREISINGER G. The genetic control of ultraviolet sensitivity levels in bacteriophages T2 and T4. Virology. 1956 Feb;2(1):1–12. doi: 10.1016/0042-6822(56)90072-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seawell P. C., Smith C. A., Ganesan A. K. den V gene of bacteriophage T4 determines a DNA glycosylase specific for pyrimidine dimers in DNA. J Virol. 1980 Sep;35(3):790–796. doi: 10.1128/jvi.35.3.790-796.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Christensen L. M., Persson M. L. Evidence that the UV endonuclease activity induced by bacteriophage T4 contains both pyrimidine dimer-DNA glycosylase and apyrimidinic/apurinic endonuclease activities in the enzyme molecule. J Virol. 1981 Oct;40(1):204–210. doi: 10.1128/jvi.40.1.204-210.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Demple B. F., Deutsch W. A., Kane C. M., Linn S. Apurinic/apyrimidinic endonucleases in repair of pyrimidine dimers and other lesions in DNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4602–4606. doi: 10.1073/pnas.77.8.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Tanyashin V. I., Murray N. E. Molecular cloning of fragments of bacteriophage T4 DNA. Mol Gen Genet. 1977 Nov 14;156(2):203–214. doi: 10.1007/BF00283493. [DOI] [PubMed] [Google Scholar]
- Wood W. B., Revel H. R. The genome of bacteriophage T4. Bacteriol Rev. 1976 Dec;40(4):847–868. doi: 10.1128/br.40.4.847-868.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S., Sekiguchi M. T4 endonuclease involved in repair of DNA. Proc Natl Acad Sci U S A. 1970 Dec;67(4):1839–1845. doi: 10.1073/pnas.67.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]