Abstract
Oligodeoxyribonucleotide mutagenesis has been used to produce a G----A mutation at nucleotide 557 of the phi X174 genome. This changes the ribosome-binding sequence GAGG of gene E to GAAG without affecting the amino acid, glutamine, encoded by the overlapping gene D. The phi X174rb(E)557 mutant does not lyse infected Escherichia coli C and therefore results in the accumulation of a large number intracellular mature phage particles. Thus, the mutation inactivates production of the gene E lytic product, presumably by blocking translation of gene E, without affecting other phage functions.
Full text
PDF
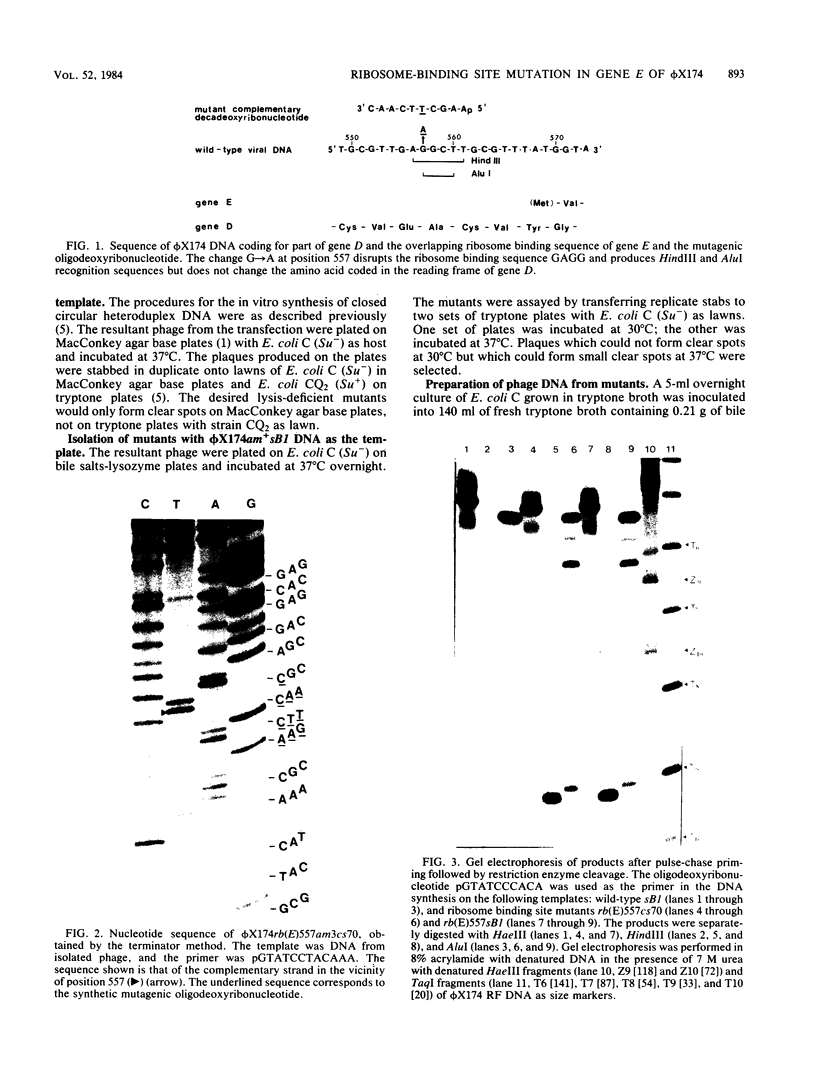

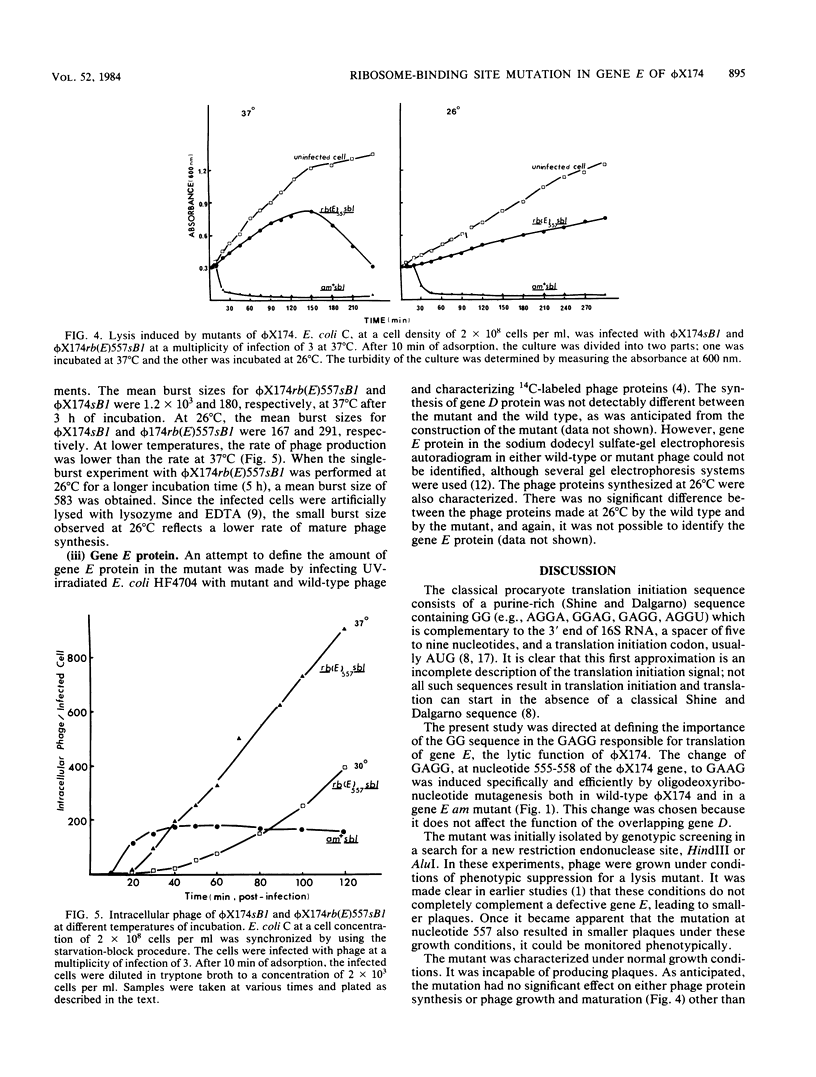

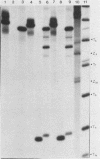
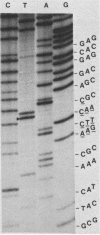
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrell B. G., Air G. M., Hutchison C. A., 3rd Overlapping genes in bacteriophage phiX174. Nature. 1976 Nov 4;264(5581):34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- Dowell C. E. Cold-sensitive mutants of bacteriophage phi-X-174. I. A mutant blocked in the eclipse function at low temperature. Proc Natl Acad Sci U S A. 1967 Sep;58(3):958–961. doi: 10.1073/pnas.58.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Buzash-Pollert E., Studier F. W. Mutations of bacteriophage T7 that affect initiation of synthesis of the gene 0.3 protein. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2741–2745. doi: 10.1073/pnas.75.6.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam S., Astell C. R., Smith M. Site-specific mutagenesis using oligodeoxyribonucleotides: isolation of a phenotypically silent phi X174 mutant, with a specific nucleotide deletion, at very high efficiency. Gene. 1980 Dec;12(1-2):129–137. doi: 10.1016/0378-1119(80)90023-2. [DOI] [PubMed] [Google Scholar]
- Gillam S., Jahnke P., Astell C., Phillips S., Hutchison C. A., 3rd, Smith M. Defined transversion mutations at a specific position in DNA using synthetic oligodeoxyribonucleotides as mutagens. Nucleic Acids Res. 1979 Jul 11;6(9):2973–2985. doi: 10.1093/nar/6.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam S., Jahnke P., Smith M. Enzymatic synthesis of oligodeoxyribonucleotides of defined sequence. J Biol Chem. 1978 Apr 25;253(8):2532–2539. [PubMed] [Google Scholar]
- Gillam S., Smith M. Site-specific mutagenesis using synthetic oligodeoxyribonucleotide primers: I. Optimum conditions and minimum ologodeoxyribonucleotide length. Gene. 1979 Dec;8(1):81–97. doi: 10.1016/0378-1119(79)90009-x. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Phillips S., Edgell M. H., Gillam S., Jahnke P., Smith M. Mutagenesis at a specific position in a DNA sequence. J Biol Chem. 1978 Sep 25;253(18):6551–6560. [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Sinsheimer R. L. The process of infection with bacteriophage phi-X174. X. Mutations in a phi-X Lysis gene. J Mol Biol. 1966 Jul;18(3):429–447. doi: 10.1016/s0022-2836(66)80035-9. [DOI] [PubMed] [Google Scholar]
- Pollock T. J., Tessman E. S., Tessman I. Identification of lysis protein E of bacteriophage phiX174. J Virol. 1978 Oct;28(1):408–410. doi: 10.1128/jvi.28.1.408-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnegg B., Hofschneider P. H. Mutant of phi X174 accessible to host-controlled modification. J Virol. 1969 May;3(5):541–542. doi: 10.1128/jvi.3.5.541-542.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. S., Gold L., Shinedling S. T., Colkitt M., Hunter L. R., Pribnow D., Nelson M. A. Analysis in vivo of translational mutants of the rIIB cistron of bacteriophage T4. J Mol Biol. 1981 Jul 5;149(3):405–432. doi: 10.1016/0022-2836(81)90479-4. [DOI] [PubMed] [Google Scholar]
- Steitz J. A., Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]