Abstract
The cellular slime mold Dictyostelium discoideum is an attractive system for studying the roles of microtubule-based motility in cell development and differentiation. In this work, we report the first molecular characterization of kinesin-related proteins (KRPs) in Dictyostelium. A PCR-based strategy was used to isolate DNA fragments encoding six KRPs, several of which are induced during the developmental program that is initiated by starvation. The complete sequence of one such developmentally regulated KRP (designated K7) was determined and found to be a novel member of the kinesin superfamily. The motor domain of K7 is most similar to that of conventional kinesin, but unlike conventional kinesin, K7 is not predicted to have an extensive α-helical coiled-coil domain. The nonmotor domain is unusual and is rich in Asn, Gln, and Thr residues; similar sequences are found in other developmentally regulated genes in Dictyostelium. K7, expressed in Escherichia coli, supports plus end–directed microtubule motility in vitro at a speed of 0.14 μm/s, indicating that it is a bona fide motor protein. The K7 motor is found only in developing cells and reaches a peak level of expression between 12 and 16 h after starvation. By immunofluorescence microscopy, K7 localizes to a membranous perinuclear structure. To examine K7 function, we prepared a null cell line but found that these cells show no gross developmental abnormalities. However, when cultivated in the presence of wild-type cells, the K7-null cells are mostly absent from the prestalk zone of the slug. This result suggests that in a population composed largely of wild-type cells, the absence of the K7 motor protein interferes either with the ability of the cells to localize to the prestalk zone or to differentiate into prestalk cells.
INTRODUCTION
Since the prototype kinesin was purified from squid axoplasm (Lasek and Brady, 1985; Schroer et al., 1985; Vale et al., 1985), dozens of related proteins (here referred to as kinesins or kinesin-related proteins [KRPs]) have been identified in organisms ranging from fungi to mammals (Moore and Endow, 1996). KRPs share a conserved mechanochemical motor domain responsible for ATP hydrolysis and microtubule binding. Outside of the 350-amino acid-long motor domain, KRPs show great sequence diversity, but regions predicted to mediate homodimerization via formation of coiled-coil structures are often present (Goldstein, 1993). Sequence diversity among KRPs outside of the motor domain is thought to play a key role in their functional diversity by specifying which proteins and cargo associate with the motor (Goldstein, 1993). Kinesins have been shown to drive the movement of vesicles in fast axonal transport (Hall and Hedgecock, 1991; Hurd and Saxton, 1996; Hurd et al., 1996), the movement of vesicles between the Golgi and the ER (Lippincott-Schwartz and Cole, 1995; Lippincott-Schwartz et al., 1995), and the movement of other organelles, including secretory vesicles, lysosomes, mitochondria, and nuclei (Hirokawa, 1996; Lopez, 1996; Cottingham and Hoyt, 1997; DeZwaan, et al., 1997; Pereira et al., 1997). Kinesins have also been shown to be involved in the organization, assembly, and operation of the mitotic spindle (Barton and Goldstein, 1996; Walczak and Mitchison, 1996) as well as in the function of the kinetochores, which connect chromosomes to microtubules (Brown et al., 1996).
Several studies have also implicated kinesins in developmental processes. Microtubules are known to be necessary for the proper localization of Vg1 RNA in Xenopus oocytes (Yisraeli et al., 1990), and it has been proposed that microtubule-based motors are involved in this and other cases of RNA localization (for review, see Wilhelm and Vale, 1993). A role in development has been demonstrated for the kinesin-related protein Xklp1 in Xenopus, which is required for both spindle assembly and for the aggregation of germ plasm in early Xenopus embryos (Robb et al., 1996). In Drosophila, COS2 is a distant kinesin relative, which is thought to tether a signaling complex consisting of the fused kinase and the cubitus interruptus transcription factor to microtubules (Robbins, et al., 1997; Sisson, et al., 1997). The access of the complex to the nucleus is in turn believed to be regulated by the hedgehog gene product, which may reverse the binding of COS2 to microtubules.
Dictyostelium has many advantages as a model system for the study of the cytoskeleton (for reviews, see Schleicher and Noegel, 1992; Schleicher et al., 1995) and developmental processes (for reviews, see Firtel, 1996; Loomis, 1996). Like yeast, it offers the possibility of combining biochemical and genetic approaches, but in addition, it shows a full spectrum of cellular functions, including chemotaxis and multicellular development. Dictyostelium has been used extensively in the study of actin-based myosin motors (Novak et al., 1995; Jung et al., 1996; Temesvari et al., 1996; Wessels et al., 1996). Work on microtubule-based motors has included the characterization of cytoplasmic dynein (Koonce and McIntosh, 1990; Koonce et al., 1992, 1994) and the purification of a protein with the properties of a kinesin motor from vegetative cells (McCaffrey and Vale, 1989). The following is the first report on the molecular biology of kinesin motors in this organism.
Here, we describe the identification of six KRPs, several of which are induced during the Dictyostelium developmental program. One motor (termed K7) was extensively characterized; its complete sequence, developmental expression pattern, motor activity, intracellular localization, and null phenotype are reported in this work. K7 is a novel plus end–directed kinesin motor with an unusual structure in the nonmotor domain. Transcription is developmentally regulated, and the protein localizes to what appears to be a membranous perinuclear structure. In pure culture, the mutant null cells show no obvious phenotype. However, when mixed with wild-type cells, the K7-null cells are absent from the prestalk zone of the slug. This result reveals a developmental defect in an environment where mutant and wild-type cells are both present.
MATERIALS AND METHODS
Cloning of Dictyostelium KRPs
Fully degenerate oligonucleotides (kindly provided by Dr. Mark Rose, Princeton University, Princeton, NJ) corresponding to the highly conserved kinesin peptides IFAYGQT and LVDLAGSE (Stewart et al., 1991) were used to prime a PCR using genomic Dictyostelium DNA as a template. Two PCR products (K2 and K4) were cloned and used as probes to screen a cDNA λ-gt11 library (Clontech, Palo Alto, CA) made from cells that had been starved for 4 h (developmental time points are referred to as hours after starvation, e.g., “T4”). Two KRP cDNAs were isolated using each probe: K6 and K8 using K2, and K3 and K7 using K4. Southern and Northern blots were hybridized at 65°C in phosphate buffer (250 mM NaPO4, 250 mM NaCl, 10% wt/vol PEG 8000, 1 mM EDTA, pH 7.2) as described (Amasino, 1986).
The entire 3.7-kb K7 insert was sequenced, and it was found to be a mixed clone of two unrelated cDNAs: one end of the insert encodes a KRP, whereas the other end encodes a protein with a high degree of homology to the mitochondrial ribosomal protein S14. This insert is thought to be the product of a cloning artifact. The breakpoint between the two parts of the insert was located in a region containing repeats of the trinucleotide AAC (nucleotides 2640–2700).
To find additional cDNAs encoding K7, a λ-Zap cDNA library made from T8–T12 cells (kindly provided by Dr. R. Firtel, University of California, San Diego, CA) was screened using the fragment of the original insert corresponding to the KRP and ending at an EcoRV site upstream of the breakpoint. A cDNA was obtained that spanned the breakpoint in the original K7 clone but still appeared to terminate prematurely. The final piece of the K7 3′ end was obtained by RT-PCR using a primer based on the 3′ end of the λ-Zap clone. Total RNA was extracted from T12 cells (RNeasy kit; Qiagen, Santa Clarita, CA) and used as a template for RT-PCR (3′-AmpliFINDER rapid amplification of cDNA ends 3′ RT-PCR kit; Clontech, Palo Alto, CA) which yielded a product corresponding to the 3′ end of the K7 transcript. Percent identity between protein sequences was calculated using the program Bestfit of the Wisconsin Package (Genetics Computer Group, Madison, WI). Multiple sequence alignment was done with the program Pileup (Genetics Computer Group). Coiled-coil predictions were made using the worldwide web version of the program Paircoil (Berger et al., 1995) available at http://ostrich.lcs.mit.edu/cgi-bin/score.
Gene Disruption
The K7 gene was disrupted following a previously described strategy (de Hostos et al., 1993) designed to replace the endogenous copy of the gene with a modified copy interrupted by an antibiotic resistance cassette. Using the first 2 kb of the K7 cDNA as a template, 5′ and 3′ end fragments (corresponding to nucleotides 32–1073 and 1502–2723, respectively) containing appropriate restriction sites were generated by PCR and subcloned. A cassette conferring blasticidin resistance (kindly provided by Dr. H. Adachi, University of Tokyo, Tokyo, Japan; Adachi et al., 1994) was cloned between the two K7 fragments to complete the gene disruption construct. Plasmid DNA was digested to release the gene disruption construct from the vector and transformed into cells by electroporation as described (de Hostos et al., 1993). Transformants were selected in DD-broth 20 media (Manstein et al., 1995) containing 5 μg/ml blasticidin (ICN, Costa Mesa, CA).
Antibodies
A PCR-generated fragment encoding the first 520 amino acids of K7 was cloned into the EcoRI site of the maltose-binding protein (MBP) expression vector pMAL-p2 (New England BioLabs, Beverly, MA). The K7/MBP fusion protein was expressed in Escherichia coli and purified by affinity chromatography over an amylose matrix as specified by the manufacturer (New England BioLabs). Recombinant protein was eluted with maltose in PBS and used for immunization of a rabbit (Cocalico Biologicals, Reamstown, PA). Antiserum was used at a dilution of 1:1000–3000 for Western blotting, using alkaline phosphatase-coupled secondary antibodies (Harlow and lane, 1988). To reduce background labeling in later Western blots and immunofluorescence labeling experiments, anti-K7/MBP antiserum was preadsorbed with strips of nitrocellulose previously soaked in a mixture of an extract of E. coli expressing MBP and Dictyostelium T12 K7-knock-out (KO) cells.
For immunofluorescence, cells were grown on glass coverslips. The coverslips with cells were rinsed in Soerensen phosphate buffer (Malchow et al., 1972) and then submerged in buffer to a depth of ∼2 mm for 6–12 h. After development, the coverslips were tapped dry and submerged in dehydrated methanol containing 1% formaldehyde at −15°C (Fukui et al., 1987). After 10 min, the cover slips were removed, allowed to dry, rinsed with 0.5% Tween in Tris-buffered saline (TNT; Sambrook et al., 1989), and blocked with 5% BSA in TNT. Preadsorbed antiserum diluted 1:30–100 in BSA-TNT was added and incubated for 30 min at 37°C. Cells were washed twice with TNT for 10 min and then incubated with FITC-labeled anti-rabbit antibodies (Sigma, St. Louis, MO) under the same conditions as the primary antiserum. After labeling the cells were washed as before, except that DAPI (1 μg/ml; Sigma) was included in an additional 5-min wash. The coverslips were rinsed in distilled water, air dried, and mounted using Vectashield medium (Vector Laboratories, Burlingame, CA). Cells were observed in a Zeiss (Oberkochen, Germany) Axiophot microscope, and images were captured on slide film (Sensia ASA 400; Fuji, Tokyo, Japan). In the experiments involving cells expressing green fluorescent protein (GFP) targeted to the ER, the cells were fixed under an agar overlay (Fukui et al., 1987), and anti-K7/MBP antibodies were followed by Texas Red-labeled anti-rabbit antibodies (Vector Laboratories). Anti-Kar2p antibodies (Ng and Walter, 1996) were kindly provided by Drs. Davis Ng and Peter Walter (University of California, San Fancisco, CA). Antinuclear pore antibodies (Snow et al., 1987) were kindly provided by Drs. R. Mahajan and L. Gerace (Scripps Research Institute, La Jolla, CA).
Developmental Time Course
Cells were grown in DD-broth 20 media to a density of 5 × 106 cells/ml and then collected by centrifugation, washed once in phosphate buffer (Malchow et al., 1972), and resuspended in the same buffer at a density of 2 × 108 cells/ml. One milliliter of cells was placed on a phosphate buffer-agar plate for development. Developing cells were collected from the plates at the appropriate time points and either lysed in SDS gel-loading buffer for protein electrophoresis or subjected to RNA extraction (RNeasy). Protein samples containing 1 × 106 cell equivalents were subjected to SDS-PAGE (Laemmli, 1970) through 8% gels and then transferred to nitrocellulose using a semidry blotting apparatus (E & K, Saratoga, CA). Ten micrograms of RNA for each time point were analyzed by Northern blotting as described above. As a hybridization probe, the EcoRV fragment of the K7 cDNA corresponding to the N terminus of the protein was radioactively labeled by random priming using the RediPrime system (Amersham, Arlington Heights, IL).
Expression of K7-GFP in E. coli
The same K7 fragment that was used for the generation of a K7/MBP antigen (residues 1–520) was cloned into the EcoRI site of the histidine tag expression vector pET23a (Novagen, Madison, WI). The coding sequence of the Aequorea victoria green fluorescent protein (S65T GFP mutant; Heim et al., 1995) was cloned into the HincII and XhoI sites of the vector. GFP was included in the construct to make the fusion protein suitable for future single-molecule fluorescence experiments (see Pierce et al., 1997); the green color of the fusion protein also serves as a convenient marker during purification. BL21(DE3) E. coli cells transformed with the expression construct were grown to an OD600 of 1–2 at 37°C in low-salt TPM medium (amounts per liter: 20 g tryptone, 15 g yeast extract, 4 g NaCl, 2 g Na2HPO4, 1 g NaH2PO4, 2 g glucose, 0.1 g ampicillin, pH 7.0). Cultures were cooled to 23°C, induced by the addition of 0.2 mM isopropyl-1-thio-β-d-galactopyranoside, and grown for an additional 14 h at 23°C. Cells were harvested by centrifugation and stored at −80°C. Cell pellets from 1 l of culture were thawed and resuspended in 25 ml of lysis buffer (50 mM NaPO4, pH 8.0, 250 mM NaCl, 20 mM imidazole, 250 mM NaCl, 1 mM MgCl2, 0.5 mM ATP, 10 mM β-mercaptoethanol [β-ME]) containing leupeptin (1 μg/ml), pepstatin (1 μg/ml), chymostatin (1 μg/ml), aprotinin (1 μg/ml), and 0.25 μg/ml Pefabloc (Boehringer Mannheim, Indianapolis, IN) and disrupted in a French press. The lysate was clarified by centrifugation for 30 min at 28,000 × g, and the supernatant was incubated with Ni-nitrilotriacetic acid resin (Qiagen) for 1 h at 4°C (1.5-ml bed volume of resin per 50 ml of supernatant). The mixture was then transferred to a disposable column, and the resin was washed with 50 ml of 50 mM NaPO4 (pH 6), 250 mM NaCl, 1 mM MgCl2, 0.1 mM ATP, and 10 mM β-ME. K7-GFP was eluted with 50 mM NaPO4, 500 mM imidazole, 250 mM NaCl, 1 mM MgCl2, 0.1 mM ATP, and 10 mM β-ME (pH 7.2). The peak fractions were then diluted 20-fold into column buffer [25 mM Na-piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8, 2 mM MgCl2, 1 mM EGTA, 1 mM DTT, and 0.1 mM ATP] and further purified by FPLC chromatography on a mono-Q column (Pharmacia, Piscataway, NJ) using a 20-ml 0–1.0 M NaCl gradient in column buffer. K7-GFP eluted as a sharp band centered at 290 mM NaCl.
In Vitro Motility Assay
Polarity-marked microtubules were prepared and an in vitro motility assay was performed essentially as described (Howard and Hyman, 1993), except that microtubules were added in BRB12 buffer [12 mM K-Na-piperazine-N,N′-bis(2-ethanesulfonic acid) pH 6.8, 2 mM MgCl2, 1 mM EGTA, 1 mM ATP[ containing 2 mg/ml casein and 20 μM taxol (Molecular Probes, Eugene, OR) and a coupled enzyme oxygen scavenger system (Harada et al., 1990). Recombinant motor protein eluted from the mono-Q purification step was loaded directly into a flow cell and allowed to bind to the glass surface. Microtubules were introduced into the chamber and imaged with a Zeiss Axiophot microscope. Images were obtained using a silicon intensifier target camera (Hamamatsu Photonics, Bridgewater, NJ) and recorded to sVHS tape. Individual frames were grabbed from the video using an Apple Power Macintosh running Adobe (San Diego, CA) Premiere. Velocities declined on prolonged illumination when using fluorescent microtubules; therefore velocity measurements reported were performed using unlabeled microtubules imaged by differential interference contrast and contrast enhanced using an Argus 20 image processor (Hamamatsu).
Preparation of Dictyostelium Membranes
Cells were starved in suspension culture for 12 h at a density of 1 × 107 cells/ml of phosphate buffer. Crude membranes were prepared essentially as described (Goodloe-Holland and Luna, 1987). The cells were resuspended in lysis buffer supplemented with a protease inhibitor mix (Complete, Boehringer Mannheim) and 1 mM DTT. The cells were broken by passage though a BioNeb cell disrupter (Glas-Col, Terre Haute, IN) at 150 psi of N2. The homogenate was centrifuged for 20 min at 38,000 × g to obtain a supernatant and crude membrane pellet. Protein samples containing 1 × 106 cell equivalents were subjected to SDS-PAGE (Laemmli, 1970) through 8% gels and Western blotting.
Construction of a GFP Marker for the ER
The coding sequence of the GFP S65T mutant was modified using PCR by the addition to its 3′ end of codons coding for the yeast ER retention signal HDEL (Pelham et al., 1988) and cloned into the KpnI and XbaI sites of the vector pDXA-3H (Manstein et al., 1995), which carries the marker neoR and expresses cloned inserts under the control of the constitutive actin-15 promoter. To provide a translation start site and a leader peptide, a DNA fragment encoding the first 38 amino acids of contact site A (Noegel et al., 1986) was amplified by PCR from a cDNA library (de Hostos, unpublished results) made in the vector λ-YES (Elledge et al., 1991) and cloned into HindIII and KpnI sites upstream of the modified GFP. The construct was cotransformed by electroporation (de Hostos et al., 1993) into AX2 cells with the pREP helper plasmid (Manstein et al., 1995), and transformants were selected and maintained in DD-broth 20 media containing 20 μg/ml Geneticin (Life Technologies, Gaithersburg, MD).
Cell-tracking Experiments
Mixing experiments with fluorescently labeled tracer cells were conducted essentially as described (Knecht and Shelden, 1995; Xu, et al., 1996). K7-null and wild-type AX2 cells (106 cells each) were labeled by resuspending in 200 μl of phosphate buffer containing 50 μM Cell Tracker Green (Molecular Probes) and shaking at room temperature for 20 min. The cells were washed twice in phosphate buffer and added to 5% of a mixture with unlabeled AX2 cells. Cells (4 × 106) from the mixture were spread at high density on phosphate agar plates containing 2% activated charcoal to provide contrast during microscopy. Cell aggregates were photographed at T12.
RESULTS
Identification of Six Dictyostelium Genes Encoding Kinesin-related Proteins
The cloning of sequences encoding KRPs from Dictyostelium was carried out in two steps. First, using Dictyostelium genomic DNA as a substrate, PCRs using fully degenerate primers corresponding to highly conserved peptides in the kinesin motor domain (Stewart et al., 1991) resulted in the amplification of two distinct products encoding KRPs. These PCR products (termed K2 and K4) were then used as probes to screen a cDNA library made from cells in early development (4 h of starvation or T4). In this manner, two different KRP cDNAs were isolated with each probe: K6 and K8 using the K2 probe, and K3 and K7 using the K4 probe. The cDNAs isolated ranged in size from 300 to 3.7 kb. No additional cDNAs corresponding to K2 and K4 were isolated in this screen.
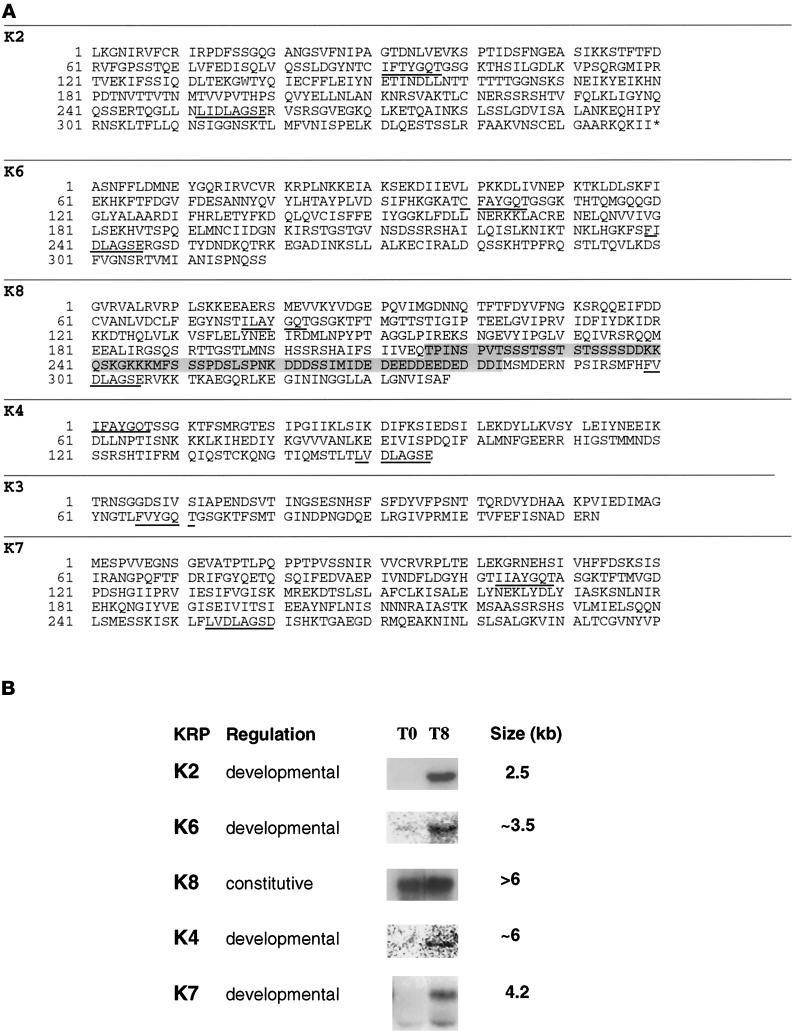
The predicted amino acid sequences of all of the fragments obtained are shown in Figure 1A. Codon usage in the predicted reading frames of all fragments showed the pronounced bias toward A/T-rich codons, which is characteristic of Dictyostelium coding sequences. In addition, all of the DNA fragments isolated except K2 have been found to hybridize to YAC clones of Dictyostelium DNA (A. Kuspa, personal communication). A nearly full-length cDNA corresponding to K2, however, has been isolated (Mayorga and de Hostos, unpublished data) by screening a development phase cDNA library (kindly provided by S. Liu and A. Kuspa, Baylor College of Medicine, Houston, TX). The various KRP fragments were used to probe Northern blots of RNA from vegetative and T8 cells; the summary of these experiments is shown in Figure 1B. No hybridization was detectable with the K3 probe but the presence of K3 sequences in the Dictyostelium genome has been confirmed by high-stringency hybridization to YACs and Southern blotting of digested Dictyostelium DNA. Taken together, these results indicate that all of the KRP-coding fragments isolated represent genuine Dictyostelium sequences and are not derived from contaminating sources.
Figure 1.
Kinesin-related proteins in Dictyostelium. (A) The highly conserved P loop (IFAYGQT) and N-3 loop (LVDLAGSE) are underlined. The genes encoding the Dictyostelium KRPs have been mapped to YAC clones (Kuspa et al., 1992) and renamed according to Dictyostelium genetic nomenclature as follows: ksnB (K2), ksnC (K3), ksnD (K4), ksnF (K6), ksnG (K7), and ksnH (K8). The sequences have been submitted to GenBank and have the following accession numbers: K2 (AF015712), K3 (AF015714), K4 (AF015713), K6 (U69984), K7 (U41289), and K8 (U69985). (B) Expression patterns of Dictyostelium kinesins. KRP fragments were used to probe Northern blots of RNA from vegetative (T0) and developing (T8) cells. Exposure times varied from blot to blot. Transcripts are classified as “developmental” if they are expressed primarily in T8 cells and “constitutive” when comparable amounts are detectable at both time points.
All of the other probes except K8 hybridized to transcripts that were significantly enriched in RNA from T8 cells. The signal obtained from hybridization to the K4 and K6 probes was particularly weak, indicating a very low level of expression at this stage of development. The developmental regulation of K2 and K4 provides a possible explanation for why corresponding cDNAs were not identified in screening the cDNA library, which is made from cells at an early stage of development (T4). K8 was found to be expressed constitutively and but showed a small induction in developing cells.
The amino acid sequences deduced from the various DNA fragments show similarities to KRPs from a variety of kinesin subfamilies. The K2 fragment shows strong homology to a family of C-terminal kinesins (KatA–KatC) from the plant Arabidopsis thaliana (Mitsui et al., 1993, 1994). The sequence of the nearly full-length K2 cDNA has confirmed the C-terminal location of the K2 motor domain (Mayorga and de Hostos, unpublished data), which has 52% identity with the motor domain of KatA (Mitsui et al, 1993; Liu et al., 1996). The K8 sequence is unusual in that it contains a segment ∼67 residues long, which has no counterpart in other kinesins. This segment is located in a region that corresponds to a surface loop (L10) in the crystal structure of kinesin motor domains (Kull et al., 1996; Sablin et al., 1996), but its function is unknown.
K7 Is an Unusual Kinesin-related Protein
One of the developmentally regulated kinesin-related genes was chosen for further study. The complete 4.2-kb K7 cDNA sequence was assembled from three separate fragments and found to encode a ∼140 kDa protein that represents a novel member of the kinesin superfamily. The derived sequence is shown in Figure 2. The K7 motor domain shows the highest degree of similarity to conventional kinesin from Neurospora (32% identity; Steinberg and Schliwa, 1995) and other members of the conventional kinesin family (e.g., human kinesin heavy chain; Navone et al., 1992). However, the sequence outside of the motor domain does not show any amino acid similarity with conventional kinesin or any other member of the kinesin superfamily. Thus, the sequence data indicate that K7 is novel kinesin superfamily member that does not clearly belong to any identified kinesin subfamily.
Figure 2.
Derived polypeptide sequence of K7. An asterisk indicates the position in the original clone corresponding to a breakpoint between the K7 cDNA and an extraneous sequence. Four conserved kinesin nucleotide binding loops and one microtubule binding loop (NYVPYRDSK) are underlined. Regions of the polypeptide sequence predicted to form a coiled-coiled structure are double underlined. Clusters of Asn, Glu, and Thr residues are shown in italics. Pro residues in the central nonhelical core are shown in bold. A back arrowhead indicates the C terminus of the motor domain fragment used for immunization and in vitro motility studies.
The nonmotor domain of K7 is unusual in several respects. First, unlike conventional kinesin and several other types of kinesin motors, K7 is predicted to adopt a coiled-coil conformation in two relatively short segments that are widely separated from one another (Figure 2, double underlined). Based on a structure prediction using the program Paircoil (Berger et al., 1995), the nonmotor domain (beginning approximately after residue 350) can be divided into a central non-α helical core (residues 424-1089) that has 22 scattered Pro residues, flanked by two segments predicted to have a largely coiled-coil conformation (residues 390–423 and 1090–1218). The nonhelical central domain of K7 is also different from any other known kinesin in that it contains clustered repeats of Asn, Thr, or Gln residues (Figure 2, italicized). Most of the poly-Gln repeats and many of the residues in the clusters of Asn and Thr are encoded by the trinucleotide AAC in the alternate reading frames CAA, AAC, and ACA, respectively. The significance of the nucleotide repeats and the amino acid clusters is not known, but similar repeats have been identified in other developmentally regulated genes in Dictyostelium (Shaw et al., 1989). The structures adopted by these repeat elements are unknown.
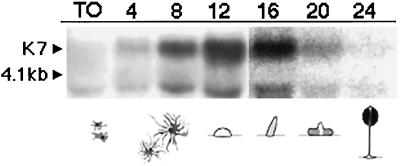
Figure 3 shows a K7 Northern blot of RNA extracted from cells at different times after development was induced by starvation. A transcript of ∼4.2 kb is detectable in developing cells, with the highest levels detectable between 12 and 16 h of development. During this time window, wild-type cells are between the finger and slug stages of development (Soll, 1987). An additional band of ∼4 kb is also detectable in these blots and shows approximately the same pattern of expression as the major 4.2-kb transcript, except at T0, when only the 4-kb band is seen. The nature of the 4-kb band is unknown, but it could represent a degradation product of the main 4.2-kb transcript or an alternatively processed transcript of the K7 gene.
Figure 3.
Expression of K7 mRNA during development. The probe hybridizes to a transcript (arrowhead) migrating just above the 4.1-kb ribosomal RNA band in developing cells (T12) but not in vegetative cells (T0). The transcript is expressed most strongly between 12 and 16 h of development, corresponding to the segment of development between finger or slug and early culminate stages. An additional band of ∼4 kb is also detectable in these blots, migrating just below the 4.1-kb ribosomal RNA. The figure is a composite of two separate blots corresponding to T0–T12 and T16–T24, respectively.
K7 Is a Plus End–directed Motor
A fragment of K7 encoding residues 1–520 was cloned in frame with the sequence of the GFP into a histidine tag pET expression vector. The fragment of K7 expressed includes the first putative coiled-coil domain and the beginning of the non-α-helical domain of the protein. Expressed fragments consisting of the first 448 amino acids of Drosophila kinesin heavy chain have been shown to form stable dimers and to have the same speed, directionality, and microtubule tracking properties of full-length kinesins (Stewart, et al., 1993; Berliner, et al., 1995). Thus it is likely that the K7 fragment will have motor properties very similar to those of the full-length motor.
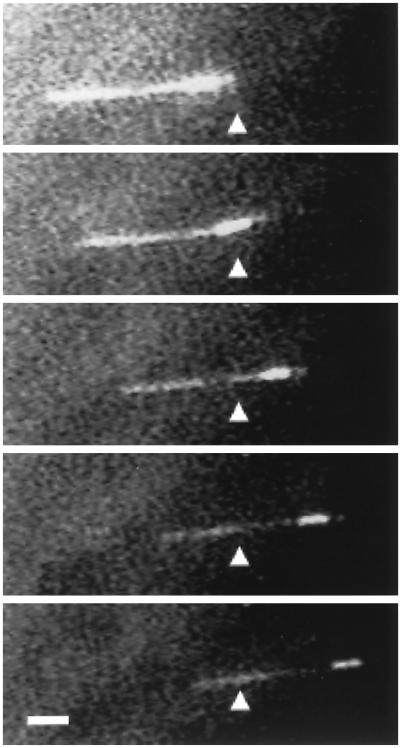
The recombinant fusion protein was purified and tested in an in vitro motility assay using polarity-labeled microtubules that contain a highly fluorescent segment marking the microtubule minus end (Howard and Hyman, 1993). The recombinant K7 fusion protein supported the movement of the microtubules with their brighter minus ends leading (Figure 4), demonstrating that K7 is a plus end–directed motor. The labeled microtubules moved at a speed of 0.14 ± 0.02 μm/s (n = 23), which is approximately half the speed (0.32 μm/s) of human kinesin heavy chain (K560) fused to GFP under similar conditions (Case et al., 1997). Thus, K7 is a bona fide microtubule motor protein.
Figure 4.
K7 is a plus end–directed motor. A polarity-labeled microtubule is shown in this fluorescence image to be moving from left to right, with the bright minus end of the microtubule leading. The triangle serves as a point of reference. Bar, 1 μm. Frames were taken approximately every 10 s, but velocities declined during prolonged fluorescence illumination; therefore, speed measurements were performed by imaging microtubules by video-enhanced differential interference contrast.
K7 Shows Perinuclear Localization
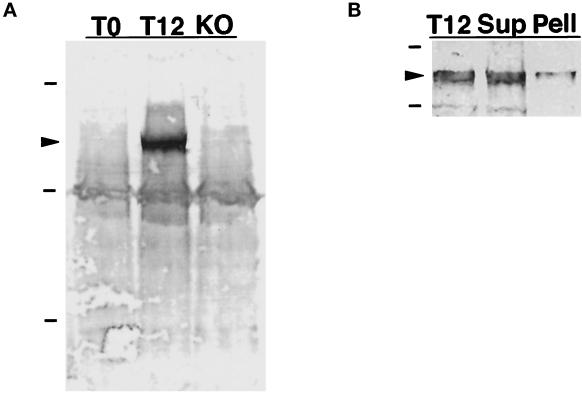
Antiserum was raised against K7 residues 1–520, which were to fused to the C terminus of the MBP. The crude anti-K7/MBP antiserum reacted strongly with a protein band present only in extracts from developing cells but also showed significant reactivity against various proteins in both vegetative and developing cells. This background reactivity was reduced significantly, but not completely, by preadsorption of the antiserum with nitrocellulose filters previously soaked in a crude lysate of the Dictyostelium K7-null strain (see below) and of E. coli expressing MBP. A Western blot in Figure 5A shows that the anti-K7/MBP antiserum reacts strongly against a ∼140 kDa protein in cells starved for 12 h (T12) cells but not in vegetative cells (T0) or in starved cells in which the K7 gene had been knocked out (KO; see below). These results are consistent with the size of the protein predicted by the K7 cDNA sequence as well as with the mRNA expression pattern of K7 in vegetative and starved cells (Figure 3). In addition to the prominent 140-kDa band, the antiserum reacts weakly with a band of ∼116 kDa that is present in all three lanes, as well as other minor components.
Figure 5.
Immunodetection of K7 in Western blots. (A) Preadsorbed antiserum raised against a K7/MBP fusion protein was used to label a Western blot of total Dictyostelium proteins. The antiserum recognizes a band of ∼140 kDa (arrowhead) in proteins from cells that have been starved for 12 h (T12) but not in proteins expressed in vegetative cells (T0) or in K7 KO cells. Dashes indicate molecular mass standards of 205, 116, and 66 kDa, respectively. (B) Western blot of total proteins from T12 cells, and equivalent amounts of protein found in 38,000 × g supernatant (Sup) and pellet (Pell) fractions.
Using the above antiserum, the subcellular localization of K7 was determined by immunofluorescence microscopy (Figure 6). In developing cells, K7 was detected in what appears to be a membranous and in some cases vesiculated (Figure 6a) structure around the nucleus. The overall pattern is distinct from that reported for the Golgi apparatus of Dictyostelium (Weiner, et al., 1993) but is similar to that expected for the nuclear envelope. To determine whether the structure labeled could be the nuclear envelope, antibodies against mammalian nuclear pore proteins (Snow et al., 1987) and against the ER-resident protein Kar2p (BiP homolog) from yeast (Ng and Walter, 1996) were tested but did not label Dictyostelium cells.
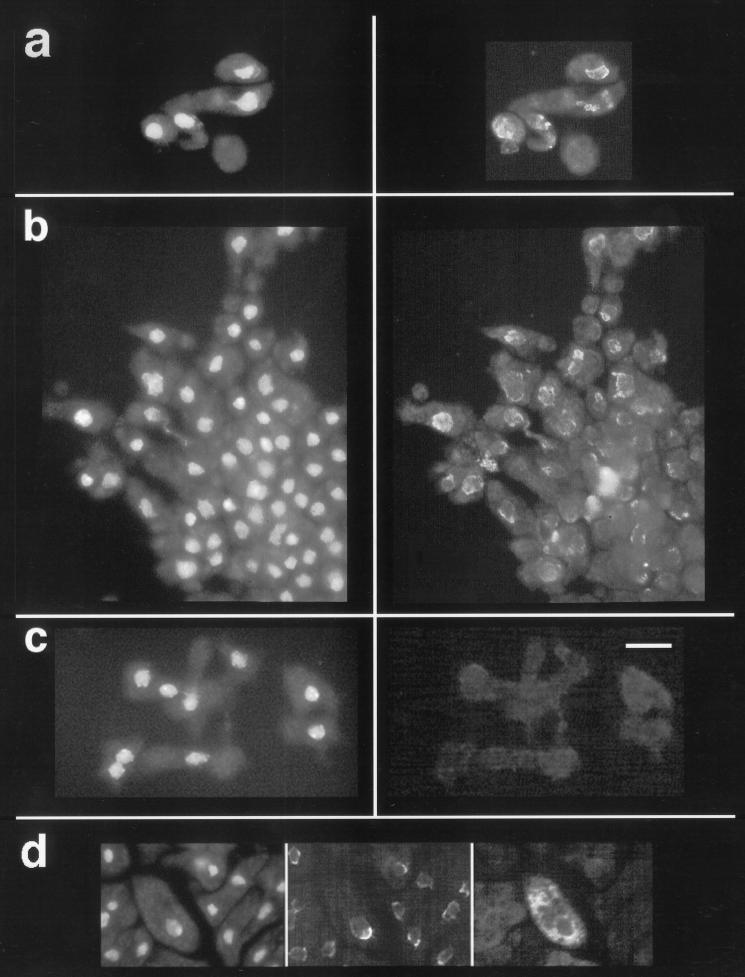
Figure 6.
Immunofluorescence localization of K7. In a–c, the frames on the left are images of cells stained with DAPI, which reveals the nuclei. The frames on the right show that the anti-K7/MBP antiserum labels a membranous perinuclear structure. In a, what appear to be vesiculations of the nuclear envelope are brightly labeled by the antiserum. The frames in b show cells that were streaming toward an aggregate forming at the bottom right corner of the images. K7-null cells in c show only a background level of immunolabeling. (d) In the area of the nucleus, the fluorescence of GFP targeted to the ER and nuclear envelope is similar to the labeling pattern obtained with the anti-K7/MBP antiserum. The frames are, from left to right, images of DAPI staining, labeling with anti-K7/MBP antiserum, and GFP fluorescence. Bar, 10 μm.
To generate a tag for the nuclear envelope, we modified the GFP (GFP S65T mutant; Heim et al., 1995) so that it would be targeted to the ER–nuclear envelope membrane complex. This was accomplished by introducing an N-terminal signal sequence derived from the contact site A cell adhesion glycoprotein (Noegel et al., 1986) and a C-terminal sequence encoding the peptide HDEL, which functions as the ER retention signal in yeast (Pelham et al., 1988) and other lower eukaryotes. Cells transformed with the GFP-ER construct showed a fluorescence pattern consistent with a distribution within the ER and nuclear envelope membrane system (Figure 6d). Green fluorescent vesicles can be prepared from these cells. These results strongly suggest that the peptide HDEL functions as an ER retention signal in Dictyostelium. Figure 6d also shows that the perinuclear portion of the fluorescence in the GFP-ER–expressing cells is similar to the labeling pattern of K7 in the same cells. This similarity is consistent with our hypothesis that the membranous structure labeled by the anti-K7/MBP antiserum is either the nuclear envelope or a membranous structure close to the nucleus.
To determine whether K7 is associated with a membrane fraction, Dictyostelium cells were broken, and soluble and sedimentable fractions were prepared. The Western blot in Figure 5b shows that the majority of K7 is soluble and not enriched in a crude membrane pellet where nuclei are expected. Residual K7 is stripped by further purification of the pellet fraction by washing with buffer and centrifugation through a sucrose gradient. These results indicate that K7 is released into a soluble fraction after cell lysis and is not tightly associated with a membrane fraction under these conditions. Similarly, although membrane staining patterns have been described for conventional kinesin (Hollenbeck, 1989; Lippincott-Schwartz et al., 1995; Lin et al., 1996), the majority of this motor is in the soluble fraction after cell lysis (Hollenbeck, 1989; Niclas et al., 1994).
Disruption of the K7 Gene
A gene disruption construct was made by inserting a cassette conferring resistance to blasticidin (Adachi et al., 1994) between PCR-generated fragments from the first 2 kb of the original K7 cDNA. Clones of cells in which the K7 gene had been inactivated were found at a high frequency (>80%). In RNA from two KO cell lines after 12 h of development (T12), neither the main 4.2-kb transcript nor the minor 4-kb band was detectable (Figure 7A). In these cells, K7 was also not detectable by immunoblotting (Figure 5A) or immunofluorescence (Figure 6c). Disruption of the K7 gene in these cell lines was confirmed by Southern blotting; a single fragment was detected, which was 1 kb greater in size compared the fragment detected in DNA from wild-type cells (Figure 7B). This size shift is expected from a gene replacement event. The K7-null cells grew well and appeared normal in morphology during vegetative growth. In the course of development, the null cells formed fingers, slugs, and fruiting bodies with a normal appearance.
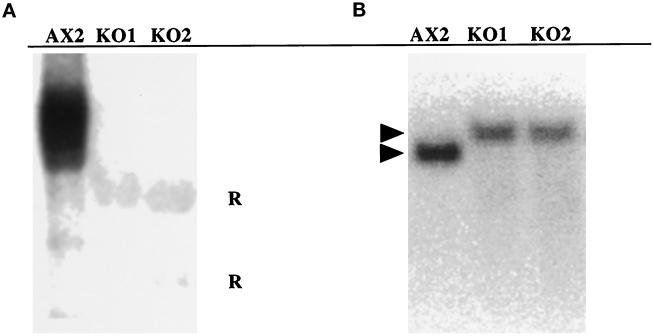
Figure 7.
Characterization of K7 KO cells. (A) Northern blot of RNA isolated from the AX2 wild type and the two K7 mutants (KO1 and KO2) at T12 was hybridized to a K7 probe. After prolonged exposure, the RNA from the KO cells shows only background hybridization to the ribosomal RNA (R). (B) Southern blot of EcoRV-restricted genomic DNA isolated from the same strains was hybridized to the K7 probe. The band hybridizing to the K7 probe shows a 1-kb increase in size, which is expected from the gene replacement event. The lack of hybridization to additional bands indicates that additional copies of the K7 gene disruption construct did not integrate elsewhere in the genome.
To determine whether the K7-null cells are at some disadvantage when developing together with wild-type cells, mixing experiments with fluorescently labeled tracer cells (Knecht and Shelden, 1995; Xu et al., 1996) were conducted. K7-null and wild-type AX2 cells were labeled by incubation with a cell-tracking dye and added as 5% of a mixture with unlabeled AX2 cells. The mixtures were plated on phosphate plates and photographed after 12 h of development. Figure 8 shows that the distribution of labeled AX2 cells is uniform throughout the body of the slugs, but labeled K7-null cells are mostly absent from the front of the slug. The anterior portion of the slug contains prestalk cells that go on to form the stalk of the Dictyostelium fruiting body. Similar results were obtained when K7-null cells were detected histochemically after transformation with a plasmid containing a constitutively expressed lacZ gene. These results suggest that the absence of the K7 motor protein impairs the ability of the mutant cells to localize properly in the slug or affects the developmental fate of the mutant cells in a population composed largely of wild-type cells.
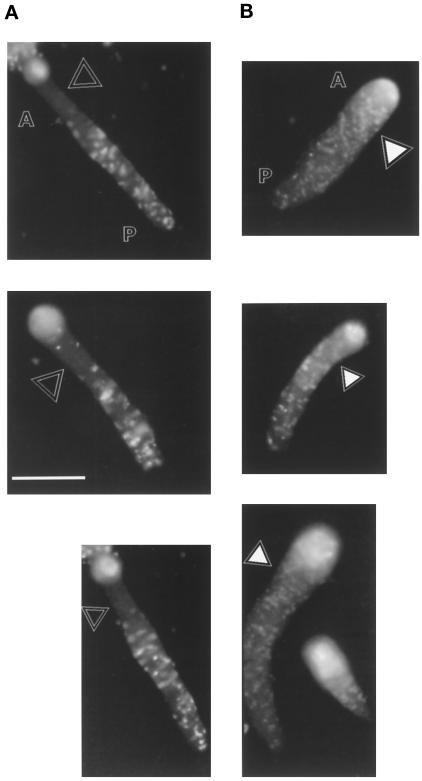
Figure 8.
Localization of K7-null cells in slugs. Mutant cells (A) or AX2 cells (B) were labeled with a fluorescent dye for tracking within an aggregate consisting of 95% wild-type cells. The open triangles show that labeled K7-null cells are mostly absent form the anterior (A) portion of the slugs. The filled triangles show that AX2 cells are not excluded from the front of the slug and are evenly in the anterior and posterior (P) portions. The bright spot at the tip of the slugs in both mixtures is thought to be an artifact due to the fiberoptic properties of the slug (Fisher, 1997; Wallraff and Wallraff, 1997) and not to represent an accumulation of labeled cells. Bar, 0.5 mm.
DISCUSSION
In this study, we report the characterization of a novel plus end–directed kinesin motor protein (K7) that plays a role in Dictyostelium development. The sequence of the K7 motor domain is most homologous to that of conventional kinesin, but the unusual sequence of the nonmotor domain indicates that K7 does not belong to an existing kinesin subfamily. The most intriguing element in the nonmotor domain of K7 is a large (∼74 kDa) nonhelical region that is rich in Pro and characterized by clusters of either Asn, Gln, or Thr residues. These clusters are mostly encoded by repeats of the trinucleotide CAA. Similar repeats have been found in a diverse group of developmentally regulated proteins in Dictyostelium (Shaw et al., 1989), but their significance is unknown. It will be interesting to explore whether the function of these repeats is important at the RNA or at the protein level; the K7-null strain will be useful in addressing this question.
K7-null cells demonstrate a conditional developmental defect. In pure cultures, K7-null cells appear to develop normally. However, when mutant cells are mixed with wild-type cells, the K7-null cells are mostly absent from the prestalk region of the slug. In a wild-type slug, the front 20% of the body consists primarily of prestalk cells (Jermyn, et al., 1996). The absence of K7-null cells from the front of the slug suggests that K7-null cells have a problem in the localization or differentiation of prestalk cells in the mixed aggregate. This finding also demonstrates the importance of conducting mixing experiments to uncover subtle phenotypic differences in Dictyostelium gene KO cell lines.
There are two possible explanations for the aberrant localization of K7-null cells in the mixed aggregate. The first possibility is that K7-null cells have a defect in motility that makes it more difficult for them to crawl in the aggregate and reach the appropriate position in the body of a slug. However, if the cells have such a defect, it appears to manifest itself only when the mutant cells must compete with wild-type cells for position within the slug. Myosin II mutants display an inability to penetrate aggregating streams when these are composed mostly of wild-type cells. This results in their exclusion from fruiting bodies and is thought to be caused by impaired motility and a weakened cell cortex (Knecht and Shelden, 1995; Xu et al., 1996). To address the possibility that the K7-null cells have a problem in motility within the wild-type slug, it will be informative to trace the paths of labeled cells within the slug by time-lapse confocal microscopy as was done for the myosin II mutants (Knecht and Shelden, 1995; Xu et al., 1996).
The other possible explanation for the K7-null phenotype is that these cells do not differentiate properly into prestalk cells in the mixed aggregate. However, K7-null cells in pure culture seem to form prestalk and prespore cells in normal proportions, so if a defect in cell determination exists, it must only be manifested in the mixed population. To determine whether K7-null cells simply do not become prestalk cells in the mixed aggregate, the expression of prestalk-specific genes can be tested using reporter gene constructs (Bichler and Weijer, 1994; Jermyn, et al., 1996). In addition, experiments should be conducted to determine whether isolated K7-null cells have a defect in cell fate determination (Gomer and Firtel, 1987).
Immunolocalization results suggest that K7 is associated with a membranous perinuclear structure, which could be the ER–nuclear envelope complex of membranes. This type of tight perinuclear envelope localization with little associated staining of vesicles, the Golgi, or the ER is intriguing and has not been reported previously for a kinesin motor. Immunoelectron microscopy should clarify the subcellular localization of K7 and its relation to the nuclear envelope. This information will help formulate a better model for the function of this motor protein.
In this study, we have also identified five KRPs other than K7. Based on the homology of these sequences to other motor proteins, Dictyostelium appears to have representatives of several KRP subfamilies, as well as unique kinesins. With the availability of these probes, it will be possible to characterize a broad range of kinesin-based functions in Dictyostelium.
ACKNOWLEDGMENTS
We thank Drs. Angela Barth and Michael Gustin for helpful discussions. Dr. Yoshio Fukui provided advice on microscopy. The lacZ labeling experiments were conducted in the laboratory of Dr. Adam Kuspa. Michael Berger assisted in this project during a laboratory rotation. Oliver Mayorga provided technical assistance. E.L.dH was supported by a Career Development Award from the American Heart Association and National Science Foundation grant 9600923. Additional support was provided by National Institutes of Health grant 38499 to R.D.V.
REFERENCES
- Adachi H, Hasebe T, Yoshinaga K, Ohta T, Sutoh K. Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem Biophys Res Commun. 1994;205:1808–1814. doi: 10.1006/bbrc.1994.2880. [DOI] [PubMed] [Google Scholar]
- Amasino RM. Acceleration of nucleic acid hybridization rate by polyethylene glycol. Anal Biochem. 1986;152:304–307. doi: 10.1016/0003-2697(86)90413-6. [DOI] [PubMed] [Google Scholar]
- Barton NR, Goldstein LS. Going mobile: microtubule motors and chromosome segregation. Proc Natl Acad Sci USA. 1996;93:1735–1742. doi: 10.1073/pnas.93.5.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berliner E, Young EC, Anderson K, Mahtani HK, Gelles J. Failure of a single-headed kinesin to track parallel to microtubule protofilaments. Nature. 1995;373:718–721. doi: 10.1038/373718a0. [DOI] [PubMed] [Google Scholar]
- Bichler G, Weijer C. A Dictyostelium anterior-like cell mutant reveals sequential steps in the prespore prestalk differentiation pathway. Development. 1994;120:2857–2868. doi: 10.1242/dev.120.10.2857. [DOI] [PubMed] [Google Scholar]
- Brown KD, Wood KW, Cleveland DW. The kinesin-like protein CENP-E is kinetochore-associated throughout poleward chromosome segregation during anaphase-A. J Cell Sci. 1996;109:961–969. doi: 10.1242/jcs.109.5.961. [DOI] [PubMed] [Google Scholar]
- Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 1997;90:959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos EL, Rehfuess C, Bradtke B, Waddell DR, Albrecht R, Murphy J, Gerisch G. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J Cell Biol. 1993;120:163–173. doi: 10.1083/jcb.120.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Mulligan JT, Ramer SW, Spottswood M, Davis RW. Lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc Natl Acad Sci USA. 1991;88:1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firtel RA. Interacting signaling pathways controlling multicellular development in Dictyostelium. Curr Opin Genet Dev. 1996;6:545–554. doi: 10.1016/s0959-437x(96)80082-7. [DOI] [PubMed] [Google Scholar]
- Fisher PR. Genetics of phototaxis in a model eukaryote, Dictyostelium discoideum. Bioessays. 1997;19:397–407. doi: 10.1002/bies.950190507. [DOI] [PubMed] [Google Scholar]
- Fukui Y, Yumura S, Yumura TK. Agar-overlay immunofluorescence: high-resolution studies of cytoskeletal components and their changes during chemotaxis. Methods Cell Biol. 1987;28:347–356. doi: 10.1016/s0091-679x(08)61655-6. [DOI] [PubMed] [Google Scholar]
- Goldstein LS. With apologies to Scheherazade: tails of 1001 kinesin motors. Annu Rev Genet. 1993;27:319–351. doi: 10.1146/annurev.ge.27.120193.001535. [DOI] [PubMed] [Google Scholar]
- Gomer RH, Firtel RA. Cell-autonomous determination of cell-type choice in Dictyostelium development by cell-cycle phase. Science. 1987;237:758–762. doi: 10.1126/science.3039657. [DOI] [PubMed] [Google Scholar]
- Goodloe-Holland CM, Luna EJ. Purification and characterization of Dictyostelium discoideum plasma membranes. Methods Cell Biol. 1987;28:103–128. doi: 10.1016/s0091-679x(08)61639-8. [DOI] [PubMed] [Google Scholar]
- Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- Harada Y, Sakurada K, Aoki T, Thomas DD, Yanagida T. Mechanochemical coupling in actomyosin energy transduction studied by in vitro movement assay. J Mol Biol. 1990;216:49–68. doi: 10.1016/S0022-2836(05)80060-9. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. The molecular mechanism of organelle transport along microtubules: the identification and characterization of KIFs (kinesin superfamily proteins) Cell Struct Funct. 1996;21:357–367. doi: 10.1247/csf.21.357. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ. The distribution, abundance and subcellular localization of kinesin. J Cell Biol. 1989;108:2335–2342. doi: 10.1083/jcb.108.6.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence microscopy. Methods Cell Biol. 1993;39:105–113. doi: 10.1016/s0091-679x(08)60164-8. [DOI] [PubMed] [Google Scholar]
- Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144:1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd DD, Stern M, Saxton WM. Mutation of the axonal transport motor kinesin enhances paralytic and suppresses Shaker in Drosophila. Genetics. 1996;142:195–204. doi: 10.1093/genetics/142.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermyn K, Traynor D, Williams J. The initiation of basal disc formation in Dictyostelium discoideum is an early event in culmination. Development. 1996;122:753–760. doi: 10.1242/dev.122.3.753. [DOI] [PubMed] [Google Scholar]
- Jung G, Wu X, Hammer JA., III Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J Cell Biol. 1996;133:305–323. doi: 10.1083/jcb.133.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht DA, Shelden E. Three-dimensional localization of wild-type and myosin II mutant cells during morphogenesis of Dictyostelium. Dev Biol. 1995;170:434–444. doi: 10.1006/dbio.1995.1227. [DOI] [PubMed] [Google Scholar]
- Koonce MP, Grissom PM, Lyon M, Pope T, McIntosh JR. Molecular characterization of a cytoplasmic dynein from Dictyostelium.J. Euk Microbiol. 1994;41:645–651. doi: 10.1111/j.1550-7408.1994.tb01528.x. [DOI] [PubMed] [Google Scholar]
- Koonce MP, Grissom PM, McIntosh JR. Dynein from Dictyostelium—primary structure comparisons between a cytoplasmic motor enzyme and flagellar dynein. J Cell Biol. 1992;119:1597–1604. doi: 10.1083/jcb.119.6.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce MP, McIntosh JR. Identification and immunolocalization of cytoplasmic dynein in Dictyostelium. Cell Motil Cytoskeleton. 1990;15:51–62. doi: 10.1002/cm.970150108. [DOI] [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A, Maghakian D, Bergesch P, Loomis WF. Physical mapping of genes to specific chromosomes in Dictyostelium discoideum. Genomics. 1992;13:49–61. doi: 10.1016/0888-7543(92)90201-3. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lasek RJ, Brady ST. Attachment of transported vesicles to microtubules in axoplasm is facilitated by AMP-PNP. Nature. 1985;316:645–647. doi: 10.1038/316645a0. [DOI] [PubMed] [Google Scholar]
- Lin SX, Pfister KK, Collins CA. Comparison of the intracellular distribution of cytoplasmic dynein and kinesin in cultured cells: motor protein location does not reliably predict function. Cell Motil Cytoskeleton. 1996;34:299–312. doi: 10.1002/(SICI)1097-0169(1996)34:4<299::AID-CM5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Cole NB. Roles for microtubules and kinesin in membrane traffic between the endoplasmic reticulum and the Golgi complex. Biochem Soc Trans. 1995;23:544–548. doi: 10.1042/bst0230544. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Cole NB, Marotta A, Conrad PA, Bloom GS. Kinesin is the motor for microtubule-mediated Golgi-to-ER membrane traffic. J Cell Biol. 1995;128:293–306. doi: 10.1083/jcb.128.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Cyr RJ, Palevitz BA. A kinesin-like protein, KatAp, in the cells of Arabidopsis and other plants. Plant Cell. 1996;8:119–132. doi: 10.1105/tpc.8.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF. Genetic networks that regulate development in Dictyostelium cells. Microbiol Rev. 1996;60:135–150. doi: 10.1128/mr.60.1.135-150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez LA. Factors regulating organelles transport along microtubules. Biocell. 1996;20:313–316. [PubMed] [Google Scholar]
- Malchow D, Nagele B, Schwartz H, Gerisch G. Membrane-bound cyclic AMP phosphodiesterase in chemotactically responding cells of Dictyostelium discoideum. Eur J Biochem. 1972;28:136–142. doi: 10.1111/j.1432-1033.1972.tb01894.x. [DOI] [PubMed] [Google Scholar]
- Manstein DJ, Schuster HP, Morandini P, Hunt DM. Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene. 1995;162:129–134. doi: 10.1016/0378-1119(95)00351-6. [DOI] [PubMed] [Google Scholar]
- Mitsui H, Nakatani K, Yamaguchi-Shinozaki K, Shinozaki K, Nishikawa K, Takahashi H. Sequencing and characterization of the kinesin-related genes katB and katC of Arabidopsis thaliana. Plant Mol Biol. 1994;25:865–786. doi: 10.1007/BF00028881. [DOI] [PubMed] [Google Scholar]
- Mitsui H, Yamaguchi-Shinozaki K, Shinozaki K, Nishikawa K, Takahashi H. Identification of a gene family (kat) encoding kinesin-like proteins in Arabidopsis thaliana and the characterization of secondary structure of KatA. Mol Gen Genet. 1993;238:362–368. doi: 10.1007/BF00291995. [DOI] [PubMed] [Google Scholar]
- McCaffrey G, Vale RD. Identification of a kinesin-like microtubule-based motor protein in Dictyostelium discoideum. EMBO J. 1989;8:3229–3234. doi: 10.1002/j.1460-2075.1989.tb08482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Endow SA. Kinesin proteins: a phylum of motors for microtubule-based motility. Bioessays. 1996;18:207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- Navone F, Niclas J, Hom-Booher N, Sparks L, Bernstein HD, McCaffrey G, Vale RD. Cloning and expression of a human kinesin heavy chain gene: interaction of the COOH-terminal domain with cytoplasmic microtubules in transfected CV-1 cells. J Cell Biol. 1992;117:1263–1275. doi: 10.1083/jcb.117.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niclas J, Navone F, Hom-Booher N, Vale RD. Cloning and localization of a conventional kinesin motor expressed exclusively in neurons. Neuron. 1994;12:1059–1072. doi: 10.1016/0896-6273(94)90314-x. [DOI] [PubMed] [Google Scholar]
- Ng DT, Walter P. ER membrane protein complex required for nuclear fusion. J Cell Biol. 1996;132:499–509. doi: 10.1083/jcb.132.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noegel A, Gerisch G, Stadler J, Westphal M. Complete sequence and transcript regulation of a cell adhesion protein from aggregating Dictyostelium cells. EMBO J. 1986;5:1473–1476. doi: 10.1002/j.1460-2075.1986.tb04384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak KD, Peterson MD, Reedy MC, Titus MA. Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis. J Cell Biol. 1995;131:1205–1221. doi: 10.1083/jcb.131.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR, Hardwick KG, Lewis MJ. Sorting of soluble ER proteins in yeast. EMBO J. 1988;7:1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AJ, Dalby B, Stewart RJ, Doxsey SJ, Goldstein LS. Mitochondrial association of a plus end-directed microtubule motor expressed during mitosis in Drosophila. J Cell Biol. 1997;136:1081–1090. doi: 10.1083/jcb.136.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce DW, Hom-Booher N, Vale RD. Imaging individual green fluorescent proteins. Nature. 1997;388:338. doi: 10.1038/41009. [DOI] [PubMed] [Google Scholar]
- Robb DL, Heasman J, Raats J, Wylie C. A kinesin-like protein is required for germ plasm aggregation in Xenopus. Cell. 1996;87:823–831. doi: 10.1016/s0092-8674(00)81990-x. [DOI] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schleicher M, Andre B, Andreoli C, Eichinger L, Haugwitz M, Hofmann A, Karakesisoglou J, Stockelhuber M, Noegel AA. Structure/function studies on cytoskeletal proteins in Dictyostelium amoebae as a paradigm. FEBS Lett. 1995;369:38–42. doi: 10.1016/0014-5793(95)00579-x. [DOI] [PubMed] [Google Scholar]
- Schleicher M, Noegel AA. Dynamics of the Dictyostelium cytoskeleton during chemotaxis. New Biol. 1992;4:461–472. [PubMed] [Google Scholar]
- Schroer TA, Brady ST, Kelly RB. Fast axonal transport of foreign synaptic vesicles in squid axoplasm. J Cell Biol. 1985;101:568–752. doi: 10.1083/jcb.101.2.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw DR, Richter H, Giorda R, Ohmachi T, Ennis HL. Nucleotide sequences of Dictyostelium discoideum developmentally regulated cDNAs rich in (AAC) imply proteins that contain clusters of asparagine, glutamine, or threonine. Mol Gen Genet. 1989;218:453–459. doi: 10.1007/BF00332409. [DOI] [PubMed] [Google Scholar]
- Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll DR. Methods for manipulating and investigating developmental timing in Dictyostelium discoideum. Methods Cell Biol. 1987;28:413–431. doi: 10.1016/s0091-679x(08)61660-x. [DOI] [PubMed] [Google Scholar]
- Steinberg G, Schliwa M. The Neurospora organelle motor: a distant relative of conventional kinesin with unconventional properties. Mol Biol Cell. 1995;6:1605–1618. doi: 10.1091/mbc.6.11.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RJ, Pesavento PA, Woerpel DN, Goldstein LS. Identification and partial characterization of six members of the kinesin superfamily in Drosophila. Proc Natl Acad Sci USA. 1991;88:8470–8474. doi: 10.1073/pnas.88.19.8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RJ, Thaler JP, Goldstein LS. Direction of microtubule movement is an intrinsic property of the motor domains of kinesin heavy chain and Drosophila ncd protein. Proc Natl Acad Sci USA. 1993;90:5209–5213. doi: 10.1073/pnas.90.11.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temesvari LA, Bush JM, Peterson MD, Novak KD, Titus MA, Cardelli JA. Examination of the endosomal and lysosomal pathways in Dictyostelium discoideum myosin I mutants. J Cell Sci. 1996;109:663–673. doi: 10.1242/jcs.109.3.663. [DOI] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ. Kinesin-related proteins at mitotic spindle poles: function and regulation. Cell. 1996;85:943–946. doi: 10.1016/s0092-8674(00)81295-7. [DOI] [PubMed] [Google Scholar]
- Wallraff E, Wallraff HG. Migration and bidirectional phototaxis in Dictyostelium discoideum slugs lacking the actin cross-linking 120 kDa gelation factor. J Exp Biol. 1997;200:3213–3220. doi: 10.1242/jeb.200.24.3213. [DOI] [PubMed] [Google Scholar]
- Weiner OH, Murphy J, Griffiths G, Schleicher M, Noegel AA. The actin-binding protein comitin (p24) is a component of the Golgi apparatus. J Cell Biol. 1993;123:23–34. doi: 10.1083/jcb.123.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels D, Titus M, Soll DR. A Dictyostelium myosin I plays a crucial role in regulating the frequency of pseudopods formed on the substratum. Cell Motil Cytoskeleton. 1996;33:64–79. doi: 10.1002/(SICI)1097-0169(1996)33:1<64::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Vale RD. RNA on the move: the mRNA localization pathway. J Cell Biol. 1993;123:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XS, Kuspa A, Fuller D, Loomis WF, Knecht DA. Cell-cell adhesion prevents mutant cells lacking myosin II from penetrating aggregation streams of Dictyostelium. Dev Biol. 1996;175:218–226. doi: 10.1006/dbio.1996.0109. [DOI] [PubMed] [Google Scholar]
- Yisraeli JK, Sokol S, Melton DA. A two-step model for the localization of maternal mRNA in Xenopus oocytes: involvement of microtubules and microfilaments in the translocation and anchoring of Vg1 mRNA. Development. 1990;108:289–298. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]