Abstract
The Saccharomyces cerevisiae Ste11p protein kinase is a homologue of mammalian MAPK/extracellular signal-regulated protein kinase kinase kinases (MAPKKKs or MEKKs) as well as the Schizosaccharomyces pombe Byr2p kinase. Ste11p functions in several signaling pathways, including those for mating pheromone response and osmotic stress response. The Ste11p kinase has an N-terminal domain that interacts with other signaling molecules to regulate Ste11p function and direct its activity in these pathways. One of the Ste11p regulators is Ste50p, and Ste11p and Ste50p associate through their respective N-terminal domains. This interaction relieves a negative activity of the Ste11p N terminus, and removal of this negative function is required for Ste11p function in the high-osmolarity glycerol (HOG) pathway. The Ste50p/Ste11p interaction is also important (but not essential) for Ste11p function in the mating pathway; in this pathway binding of the Ste11p N terminus with both Ste50p and Ste5p is required, with the Ste5p association playing the major role in Ste11p function. In vitro, Ste50p disrupts an association between the catalytic C terminus and the regulatory N terminus of Ste11p. In addition, Ste50p appears to modulate Ste11p autophosphorylation and is itself a substrate of the Ste11p kinase. Therefore, both in vivo and in vitro data support a role for Ste50p in the regulation of Ste11p activity.
INTRODUCTION
In eukaryotic cells, the MAPK cascade module is an important and highly conserved signaling element. MAPK cascades mediate the transduction of many signals from the cell surface to the nucleus to permit responses to cues from the extracellular environment. A typical MAPK module consists of three highly conserved protein kinases: a MAPK, a MAPKK (or MAPK/extracellular signal-regulated kinase [ERK] kinase [MEK]), and a MAPKKK (or MEKK). The MAPK module transduces signals through sequential activation of these kinases by phosphorylation. MAPK is activated by the dual-specificity serine/threonine tyrosine kinase MAPKK, which in turn is activated by the serine/threonine kinase MAPKKK (for reviews, see Robinson and Cobb, 1997; Banuett, 1998). The MAPKKK becomes activated in response to a signal generated by various membrane proteins, including G-protein–coupled seven-transmembrane receptors, two-component His-Asp phosphorelay sensors, receptor–tyrosine kinases, and integral membrane sensor proteins (for reviews, see Herskowitz, 1995; Banuett, 1998).
The budding yeast Saccharomyces cerevisiae has several MAPK cascades that regulate responses to osmotic stress, pheromones, the perturbation of cell wall integrity, pseudohyphal growth conditions, and sporulation signals (for review, see Herskowitz, 1995). In response to pheromone binding to their cognate seven-transmembrane receptors, haploid cells undergo cellular changes, including transcriptional activation of mating-specific genes, cell cycle G1 arrest, and morphological changes. The MAPK module controlling the pheromone response pathway consists of Ste11p (MAPKKK), Ste7p (MAPKK), and Fus3p (MAPK). The sequential activation of the protein kinases in the module has been studied extensively; however, the signals ultimately controlling activation of Ste11p (MAPKKK), the first protein kinase in the MAPK module, remain unclear. Ste11p is an in vitro substrate of Ste20p, a kinase that functions upstream of Ste11p in the mating pathway, but the regulatory significance of this phosphorylation remains to be determined (Wu et al., 1995). Ste5p has been shown to associate with all three protein kinases of the MAPK module (Choi et al., 1994; Marcus et al., 1994; Printen and Sprague, 1994), and with the Gβγ subunits of the heterotrimeric G-protein (Whiteway et al., 1995). Hence, Ste5p may be involved directly in the transmission of the signal from the pheromone receptor to Ste11p (Feng et al., 1998) and may facilitate sequential activation of the protein kinases in the module by aligning them together properly. Ste50p has been shown to be required for sustaining or propagating the pheromone response signal and to interact with the N-terminal domain of Ste11p by two-hybrid analysis (Rad et al., 1992; Xu et al., 1996); however, the biological significance of these interactions remains unknown. Intriguingly, Ste4p of Schizosaccharomyces pombe, which has limited sequence similarity to Ste50p, is also required for the function of the Byr2p MAPKKK, the Ste11p homologue in sexual developmental processes in S. pombe (Tu et al., 1997).
S. cerevisiae cells detect and respond to high extracellular osmolarity by activating the high-osmolarity glycerol (HOG) MAP kinase cascade, which is essential for the survival of yeast cells in high-osmolarity environments (Boguslawski and Polazzi, 1987; Brewster et al., 1993). The activation of this MAP kinase pathway increases the synthesis of glycerol, which leads to an increased internal glycerol concentration and therefore increased internal osmolarity, to compensate for the external hyperosmolarity. Because glycerol appears to be the major osmolyte used by yeast, this MAP kinase pathway is referred as the HOG pathway. Activation of the HOG MAPK cascade is achieved by two membrane sensor proteins, Sln1p and Sho1p, which detect extracellular hyperosmolarity (Maeda et al., 1994, 1995).
Sln1p, along with two other proteins, Ypd1p and Ssk1p, comprises a “two-component” osmosensor (Maeda et al., 1994; Posas et al., 1996), which works by a multistep phosphorelay mechanism. Under high-osmolarity conditions, Sln1p histidine kinase activity is suppressed, leading to accumulation of unphosphorylated Ssk1p (Maeda et al., 1994; Posas et al., 1996). The unphosphorylated Ssk1p activates the Ssk2p and Ssk22p MAPKKKs by inducing their autophosphorylation (Posas and Saito, 1998). Once activated, Ssk2p and Ssk22p phosphorylate and turn on the Pbs2p MAPKK, which in turn activates the Hog1p MAPK by phosphorylation.
The other branch for the activation of the Pbs2p MAPKK is mediated by the Sho1p osmosensor, which has four predicted transmembrane domains at its N terminus (Maeda et al., 1995; Posas and Saito, 1997). Thus the Pbs2p MAPKK can be regulated by two different branches, both sensing hyperosmolarity but acting independently of the other (Maeda et al., 1994; Maeda et al., 1995; Posas and Saito, 1997). The SLN1 branch involves the two-component His-Asp phosphorelay system and regulates Ssk2p and Ssk22p; the SHO1 branch uses a membrane osmosensor and activates the Ste11p MAPKKK. Any one of the three activated MAPKKKs (Ssk2p, Ssk22p, and Ste11p) can phosphorylate and turn on the Pbs2p MAPKK, which in turn activates the Hog1p MAPK to induce responses to hyperosmotic stress. How the Sho1p osmosensor leads to the activation of Ste11p MAPKKK is unknown. Recently, Ste50p has been shown to be necessary for the HOG pathway activation mediated by the SHO1–STE11 branch (O’Rourke and Herskowitz, 1998; Posas et al., 1998), and the physical association of Ste50p and Ste11p through their N-terminal domains has been found essential for the pathway activation (Posas et al., 1998).
Here we dissect genetically and biochemically the Ste50p–Ste11p association and show that this interaction is necessary both for the activation of the Ste11p–Sho1p dependent HOG pathway and for proper signaling in the pheromone response pathway. Ste50p shares a role in the activation and proper functioning of Ste11p in the pheromone response pathway with Ste5p. We also show that the Ste11p N-terminal regulatory region physically associates with its C-terminal catalytic kinase domain, and this interaction can be displaced by Ste50p. In addition, we show that Ste50p modulates Ste11p autophosphorylation in vitro and establish that Ste50p is an in vitro substrate of the Ste11p kinase. It is likely that the Ste50p–Ste11p association is part of a multistep activation process in vivo, because in cells Ste50p appears to interact with Ste11p constitutively.
MATERIALS AND METHODS
Materials
Restriction endonucleases and DNA-modifying enzymes were obtained from Roche Molecular Biochemicals (Hertforshire, UK), Life Technologies (Gaithersburg, MD), Amersham Pharmacia Biotechnology (Oakville, Ontario, Canada), and New England Biolabs (Beverly, MA). Taq thermostable DNA polymerase was purchased from Roche Molecular Biochemicals. Acid-washed glass beads (450–600 μm), synthetic α-mating factor, protease inhibitors, and BSA were purchased from Sigma (St. Louis, MO). α-mating factor was dissolved in 90% methanol at a concentration of 1.0 mg/ml and stored at −20°C. Plasmid pGEX-4T-3, pGEX-2TK, glutathione-Sepharose beads, glutathione, and protein A/G Sepharose beads were obtained from Pharmacia LKB Biotechnology (Dorval, Québec, Canada). Nitrocellulose membranes were from Xymotech (Montreal, Québec, Canada). The monoclonal anti-myc (9E10) monoclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA), antibodies against maltose binding protein (MalBP) and amylose agarose beads were from New England Biolabs, and the antibody against GST was described previously (Wu et al., 1997). Horseradish peroxidase-conjugated secondary antibodies were from Bio-Rad (Mississauga, Ontario, Canada). The enhanced chemiluminescence (ECL) assay system was purchased from Amersham.
Yeast Strains and Manipulations
Yeast media, culture conditions, and manipulations of yeast strains were as described previously (Rose et al., 1990). Yeast transformations with circular or linearized plasmid DNA were performed after treatment of yeast cells with lithium acetate (Rose et al., 1990). The yeast strains used in this study are listed in Table 1.
Table 1.
Yeast strains used in this study
| Strains | Relevant genotype | Source |
|---|---|---|
| TM141 | MATa ura3 leu2 trp1 his3 | H. Saito |
| TM254 | MATa ura3 leu2 his3 ssk2Δ::LEU2 ssk22Δ::LEU2 | H. Saito |
| W303-1A | MATa ade2 ura3 his3 leu2 trp1 can1 | R. Rothstein |
| W303-1B | MATα ade2 ura3 his3 leu2 trp1 can1 | R. Rothstein |
| YCW241 | W303-1A FUS1::LacZ::LEU2 | This study |
| YCW315 | W303-1A ste50Δ::TRP1 | This study |
| YCW316 | W303-1B ste50Δ::TRP1 | This study |
| YCW340 | MATa ura3 leu2 his3 trp1 ssk2Δ::LEU2 ssk22Δ::LEU2 ste11Δ::KanR | This study |
| YCW362 | MATa ura3 leu2 his3 trp1 ssk2Δ::LEU2 ssk22Δ::LEU2 | This study |
| YCW360 | MATα ura3 leu2 his3 ssk2Δ::LEU2 ssk22Δ::LEU2 ste50Δ::TRP1 | This study |
| YCW365 | MATa ura3 leu2 his3 ssk2Δ::LEU2 ssk22Δ::LEU2 ste50Δ::TRP1 | This study |
| YCW555 | MATa ura3 leu2 his3 ssk2Δ::LEU2 ssk22Δ::LEU2 ste11Δ::KanR ste50Δ::TRP1 | This study |
| YCW335 | MATa his3 leu2 trp1 ura3 ste50Δ::TRP1 sst1::hisG FUS1::LacZ::LEU2 | This study |
| YCW338 | MATa his3 leu2 trp1 ura3 sst1::hisG FUS1::LacZ::LEU2 | This study |
| DC17 | MATα his1 | J. Hicks |
Plasmid Constructions
Yeast expression constructs carrying different fragments of STE50 were generated by PCR (Saiki et al., 1988) using appropriate primers. The PCR products were digested with BamHI and EcoRI and cloned into the BamHI–EcoRI-digested yeast expression vector pYEX-4T-2 (Clontech, Palo Alto, CA) to yield the following plasmids, which express various Ste50p fragments as GST fusion proteins: pCW205 (aa 27–346), pCW207 (aa 115–346), pCW208 (aa 148–346), pCW209 (aa 187–346), pCW213 (aa 1–218), pCW214 (aa 1–130), pVL57 expressing full-length Ste50p, and pCW215 expressing Ste50p with an internal in-frame deletion of aa 131–218 inclusive. To generate STE11 under the control of the GAL1 promoter, STE11 was amplified by PCR using primers 5′-CGGGATCCGTCGACATGCATAAAGAGAGACCA-3′ (OCWS11N) and 5′-GGACTAGTGGTACCTGTTTCTTCGTGCTTCC-3′ (OCWS11C). The PCR products were digested with BamHI and KpnI and cloned into pRD56 (a GAL1–GST fusion vector, kindly provided by Dr. M. Peter, Swiss Institute for Experimental Cancer Research) to create pVL101. This plasmid was digested with KpnI, blunt-ended, and then cut with SalI, and the purified fragment containing STE11 was cloned into pRS313GAL to yield pCW183. pCW183 was then digested with ClaI and religated to yield pCW184, which expresses Ste11p lacking the N-terminal amino acids 1–130. To make the GAL1/10 promoter-controlled, myc-tagged version of STE11, the GAL1/10 promoter region as a BamHI–ClaI fragment from pRS313GAL and STE11 as a ClaI–XbaI fragment from pNC245 (Rhodes et al., 1990) was ligated into BamHI–XbaI-digested pCW194, which is a pRS313 derivative lacking the multiple cloning sites from XhoI to EcoRI inclusive. This three-way ligation created plasmid pCW198 (GAL-STE11ΔSAM). Plasmid pCW198 was digested with EcoRI and EspI, blunt-ended with T4-DNA polymerase, and religated to yield pCW233 (GAL-STE11ΔSAMEE). pSL1654, carrying STE11–1, was kindly provided by Dr. G. F. Sprague (University of Oregon). Plasmid pVL23 expresses full-length Ste5p as GST fusion protein under the control of CUP1 promoter.
To make STE11 tagged with c-myc and under the control of its own promoter, STE11 was amplified by PCR with primers OCW95 5′-GCGGATCCGTCGACCTTTGATACAGCCTCGG-3′ and OCWS11C 5′- GGACTAGTGGTACCTGTTTCTTCGTGCTTCC-3′, and the PCR product was digested with BamHI and ClaI. A 1.1 kb fragment was purified and ligated together with the ClaI–XbaI fragment of pNC245 into pCW194 to yield plasmid pCW199 (STE11WT-myc). Similarly, to make STE11 deleted for aa 26–129, the 5′-flanking sequence of STE11 was amplified by PCR with primers OCW95 and OCW96 5′-GCATCGATGTCTGTTCCATGTATATTTC-3′, digested with BamHI and ClaI, and ligated with the ClaI–XbaI fragment of pNC245 into pCW194 to yield pCW204 (STE11ΔSAM-myc). Plasmids pCW199 and pCW204 were then digested with EcoRI–EspI, blunt-ended with T4-DNA polymerase, and religated to yield pCW227 (STE11ΔEE-myc) and pCW223 (STE11ΔSAMEE-myc), respectively.
Several Escherichia coli expression constructs used in this study expressed GST fusions. These were pVL56 expressing full-length Ste50p, pCW166 expressing aa 1–218 of Ste50p, pCW167 expressing aa 115–346 of Ste50p, and pCW228 expressing aa 385–738 of Ste11p with myc tag. Other constructs expressed MalBP fusions. These were pCW164 expressing aa 1–386 of Ste11p, pCW165 expressing aa 1–386 of Ste11–1p, pCW168 expressing 148–364 of Ste50p, and pCW169 expressing aa 1–218 of Ste50p.
Preparation of Fusion Proteins and Yeast Cell Extracts
The GST fusion proteins were expressed in E. coli strain UT5600 (New England Biolabs), extracted, bound to glutathione-Sepharose beads, and eluted with glutathione as described previously (Wu et al., 1995). The eluted proteins were then concentrated and washed with storage buffer (50 mM Tris-HCl, pH 7.5, 200 mM KCl, 1 mM DTT, and 10% glycerol) by centrifugation using the Centricon-30 system (Amicon, Beverly, MA) and stored at −80°C. The MalBP fusion proteins were expressed in E. coli and purified essentially according to the procedure recommended by the manufacturer (New England Biolabs), except that the lysis buffer was supplemented with 0.1% Triton X-100. Amylose agarose beads-bound proteins were extensively washed before use in the binding assays. For yeast GST fusion expression constructs, cells were induced with galactose for GAL1 promoter-driven GST fusion vectors or with the addition of 0.5 mM CuSO4 for CUP1 promoter-driven GST fusion vectors. Total cell extracts were prepared as described previously (Wu et al., 1995), except that cells were disrupted using a bead beater (Biospec Products, Bartlesville, OK) for 3 min at 4°C. The extracts were clarified by centrifugation in a microfuge at 10,000 × g for 10 min at 4°C, and total protein concentration was determined by the Bradford reaction using the Bio-Rad protein determination reagents following the manufacturers instructions. The supernatant fractions were supplemented with 10% glycerol and stored at −80°C. Purification of yeast GST fusion proteins was performed with yeast extracts and glutathione-Sepharose beads according to the procedure described previously for purification of E. coli GST fusion proteins (Wu et al. 1995). Eluted proteins were washed and concentrated with Centricon 30 (Amicon) and stored at −80°C.
Coprecipitation, Resin Binding, and Kinase Assays
For coprecipitation assays, yeast cell extracts from cells that expressed GST fusion proteins were incubated with 60 μl (50% vol/vol) of glutathione-Sepharose beads in a final volume of 0.5–1 ml of lysis buffer supplemented with 0.1% BSA for 2 h at 4°C. Beads were then collected by centrifugation (800 × g, 2 min), washed extensively in lysis buffer, then resuspended in sample loading buffer and subjected to SDS-PAGE. Immunoblotting analyses were performed using anti–c-myc monoclonal antibody 9E10 and polyclonal anti-GST antibody and visualized with the ECL detection system.
For resin binding with protein expressed in E. coli, cell lysates containing GST fusion proteins (0.1–1 μg) were mixed with cell lysates containing MalBP fusion proteins (0.1–1 μg), incubated for 1 h at 4°C, and then incubated with 60 μl (50% vol/vol) of either glutathione-Sepharose beads or amylose agarose beads for 30 min at 4°C. The beads were collected by centrifugation, washed extensively, resuspended in sample buffer, and subjected to SDS-PAGE. Immunoblotting analyses were performed with polyclonal anti-MalBP and anti-GST antibodies.
In vitro kinase assays were performed essentially as described (Wu et al. 1995), except that the protein mixtures were preincubated in 20 μl of kinase buffer for 30 min at 4°C before starting the kinase reactions by addition of 20 μl of kinase buffer containing 20 μM [γ-32P]ATP (2.5 × 104 Ci/mol), and the reactions were incubated at 30°C for 15 min.
Yeast Mating and Other Assays
Plate mating tests were performed as described (Leberer et al., 1992). Quantitative mating assays were carried out by a filter assay as described (Leberer et al., 1997). β-galactosidase activities were measured as described (Leberer et al., 1992), with Miller units defined as (OD420 × 1000)/(OD600 × t × V), measuring t in minutes and V in milliliters. Halo assays to test cell growth inhibition in response to α-mating factor were performed as described (Leberer et al., 1997).
Photomicroscopy
Cells were grown as indicated, sonicated, and fixed with formaldehyde at a final concentration of 3.7% with 150 mM NaCl. Cells were viewed with a microscope equipped with Nomarski optics, and microscopic photographs were acquired with a 100× objective using a Micro Max camera (Princeton Instruments, Princeton, NJ) with Northern Eclipse imaging software (Empix Imaging, Mississauga, Ontario, Canada), and processed using Adobe Photoshop for Macintosh.
RESULTS
A Hyperactive Allele of Ste11p Kinase Bypasses the Requirement for Ste50p in the Mating and Osmotic Responses
Ste50p plays a role in the mating pheromone response pathway. Cells with a ste50 deletion are partially defective in both pheromone-induced transcriptional activation and cell cycle arrest and have a reduced efficiency of diploid formation (Rad et al., 1992; Xu et al., 1996). We investigated whether a constitutively active allele of STE11, STE11–1, identified by its ability to activate the transcription of mating-specific genes in the absence of mating pheromone (Stevenson et al., 1992), could bypass the need for Ste50p. Ste50p expressed as GST fusion construct was fully functional to complement all defects caused by ste50Δ, including mating, transcriptional activation, and cell cycle arrest. As shown in Figure 1, STE11–1 was partially able to rescue the defect in pheromone response caused by ste50Δ as judged by the level of pheromone-induced transcriptional activation of FUS1::LacZ as well as by cell cycle arrest (our unpublished results). In addition, GAL promoter-driven overexpression of Ste11p could fully rescue the pheromone response defect caused by ste50Δ, consistent with the previous finding that overexpression of Ste11p rescues the mating defect caused by the deletion of STE50 (Xu et al., 1996).
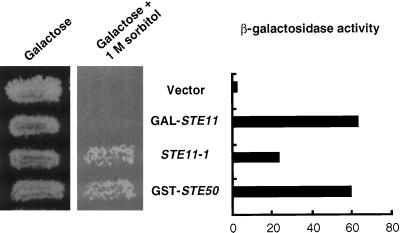
Figure 1.
Constitutively active Ste11p bypasses the requirement for Ste50p both in the HOG and the pheromone response pathways. Left, STE11–1, but not the overexpression of wild-type STE11, suppressed the hyperosmosensitivity caused by ssk2Δ ssk22Δ ste50Δ. Yeast strain YCW365 (ssk2Δ ssk22Δ ste50Δ) transformed with plasmid pVL101 (GAL-STE11), plasmid pSL1654 (STE11–1), plasmid pVL57 (GST-STE50), or the cloning vector (vector) were grown on galactose-containing selective medium or grown on galactose medium with 1 M sorbitol at 30°C for 3 d. Right, Overexpression of STE50 or overexpression of wild-type STE11, and to a lesser extent, expression of STE11–1 restored pheromone-induced FUS1 expression to a ste50Δ strain. Yeast strain YCW335 (MATa ste50Δ sst1::hisG FUS1::LacZ::LEU2) was transformed with the same plasmids as indicated and assayed for pheromone-dependent FUS1 transcriptional induction by measuring β-galactosidase activity (Miller units).
Recently, it has been shown that Ste50p also functions in the osmotic response pathway (O’Rourke and Herskowitz, 1998; Posas et al., 1998). We crossed a strain with ssk2Δ and ssk22Δ mutations to a strain with a ste50Δ mutation and identified cells with the ssk2Δ ssk22Δ ste50Δ triple mutation that were unable to grow on high-osmolarity medium. In contrast, cells with a ste11Δ or a ste50Δ or a ste11Δ ste50Δ double mutation could grow on the high-osmolarity medium (our unpublished results), and thus the role of Ste50p in the regulation of the HOG MAPK pathway was likely to be exerted via Ste11p and not via Ssk2p and Ssk22p. The osmosensitive phenotype caused by deletion of STE50 could be bypassed by expression of the hyperactive STE11–1 allele, whereas overexpression of wild-type Ste11p did not rescue the osmosensitive phenotype of the ste50 deletion (Figure 1). Thus the constitutively active STE11–1 allele can bypass the function of Ste50p in both the pheromone response pathway and the HOG pathway activation, whereas Ste11p overexpression appears effective only in bypassing the requirement for Ste50p in the mating response pathway.
Molecular Characterization of Ste50p Defines Domains that Are Required for Its Function
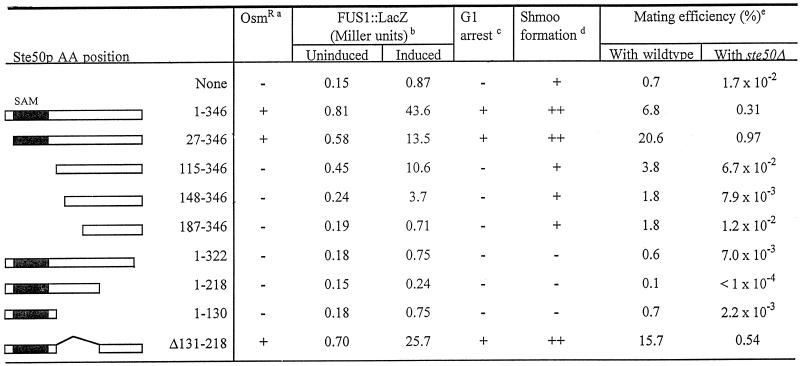
To study the role of Ste50p in yeast cells, we characterized its functional domains by deletion analysis. Full-length Ste50p was fused to GST to allow subsequent biochemical purification. The resulting chimeric molecule was fully able to complement the ste50Δ mutation in the mating pathway as measured by mating, pheromone-induced transcriptional activation, and cell cycle arrest, as well as in the HOG pathway as measured by rescuing osmosensitivity in an ssk2Δ ssk22Δ strain background. A series of deletion mutants of Ste50p were then constructed and examined for their ability to replace the function of wild-type Ste50p. As summarized in Table 2, the first 26 amino acid residues at the N terminus are dispensable for the function of Ste50p in both the HOG pathway and the pheromone response pathway. Amino acid residues 27–115 contain elements that appear to be essential for Ste50p function in the HOG pathway as well as for pheromone-induced cell cycle arrest but are less important for pheromone-induced transcriptional activation. This region of Ste50p contains a conserved motif called a SAM domain. SAM domains are found in many signal transduction molecules from various organisms and have been implicated in protein–protein interaction (Ponting, 1995; Schultz et al., 1997). Further deletion from the N terminus of the protein made a nonfunctional Ste50p that is phenotypically identical to a null mutant. The C terminus was also essential for the activity of Ste50p because deletion of as few as 24 amino acid residues abolished the function in both the pheromone response and HOG pathways. The central region of the molecule, however, appeared to be dispensable, because Ste50p with an in-frame deletion of 88 amino acid residues (aa 131–218 inclusive) was fully active in both pathways. Interestingly, several C-terminally truncated, nonfunctional mutants of Ste50p were found to have a dominant inhibitory phenotype. In the pheromone response pathway, these C-terminally truncated mutants generated a more extreme phenotype than the null mutation itself; that is, cells were more resistant to morphological changes caused by pheromone treatment and more resistant to cell cycle arrest (Figure 2A). In the HOG pathway, expression of these deletion mutants caused osmosensitivity in strains that were normally resistant (Fig-ure 2B).
Table 2.
Functional domains of Ste50p
a The hyperosmolarity-resistant phenotype of yeast cells YCW365 (ssk2Δ ssk22Δ ste50Δ) transformed with different STE50 alleles was scored for growth (+) on selective media with 1.5 M sorbitol after 4 d at 30°C.
β-galactosidase activity was determined with yeast cells of YCW335 (ste50Δ::TRP1 sst1::hisG FUS1::LacZ::LEU2) transformed with different alleles of STE50, either treated with 1 μM of α-factor (induced) or not (uninduced) for 1.5 h, and the activity was expressed as Miller units (see MATERIALS AND METHODS). Data represent the mean value of two to three independent experiments (the estimate of the error for each value was < 25% of the mean value).
Cell cycle arrest was assessed with yeast cells as in noteb for their ability to arrest (+) on selective media with 1 μM of α-factor after a 24 h incubation at 30°C.
Yeast cells as in noteb were treated with 1 μM α-factor, and times required for shmoo formation were determined. ++ indicates shmoo formation within 2 h after α-factor treatment, + indicates shmoo formation 5 h after the treatment, and − indicates no shmoo formation after 8 h.
Mating was performed by mixing exponentially growing cells of YCW315 (ste50Δ) bearing different alleles of STE50 with fivefold excess of tester cells of either DC17 (his1) or YCW316 (ste50Δ) and incubating for 4 h at 30°C (MATERIALS AND METHODS). Mating efficiency was defined as (the number of diploid cells)/(the number of the experimental haploid cells) × 100. Data represent the mean of two to three independent experiments.
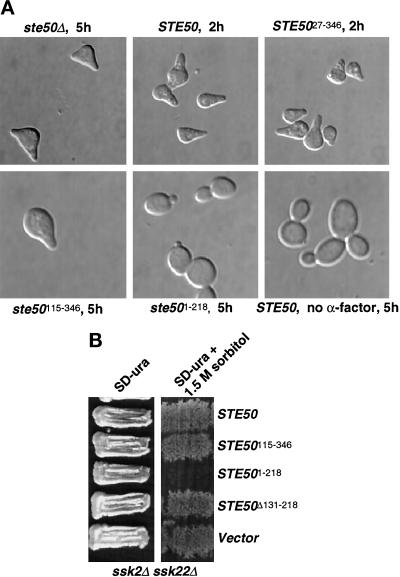
Figure 2.
Requirement of the N and C termini of Ste50p in the pheromone response and the HOG pathways. (A) Both N- and C-terminal domains of Ste50p are required for the pheromone-induced morphogenesis. Yeast strain YCW335 (MATa ste50Δ sst1) was transformed with wild-type Ste50p as a GST fusion construct (pVL57) or various Ste50p deletion mutants as GST fusion constructs (pCW205, encoding Ste50p27–346; pCW207, encoding Ste50p115–346; and pCW213, encoding Ste50p1–218) or with the cloning vector expressing GST only. Transformants were grown to exponential phase in medium with 0.5 mM CuSO4, and cultures were split and either treated with 1 μM α-factor or left untreated. The treated cultures were monitored every 0.5 h, and the time indicated was the time required for clear pheromone-mediated morphological changes to occur. Wild-type and Ste50p27–346 looked to have the same response after 2 h. Ste50p115–346 took 5 h to respond, and the Ste50p1–218 was totally unresponsive and looked like untreated cells after 5 h. (B) The C-terminally truncated allele of Ste50p had dominant inhibitory effect in the HOG pathway. Yeast strain YCW362 (ssk2Δ ssk22Δ) was transformed with plasmids pVL57, pCW207, pCW213, and pCW215 (encoding Ste50pΔ131–218) or the cloning vector, grown on selective medium, and challenged with high-osmolarity selective medium containing 1.5 M sorbitol at 30°C for 3 d.
Identification of Regions in the N Terminus of Ste11p Required for the Activation of the HOG Pathway and the Mating Pheromone Pathway
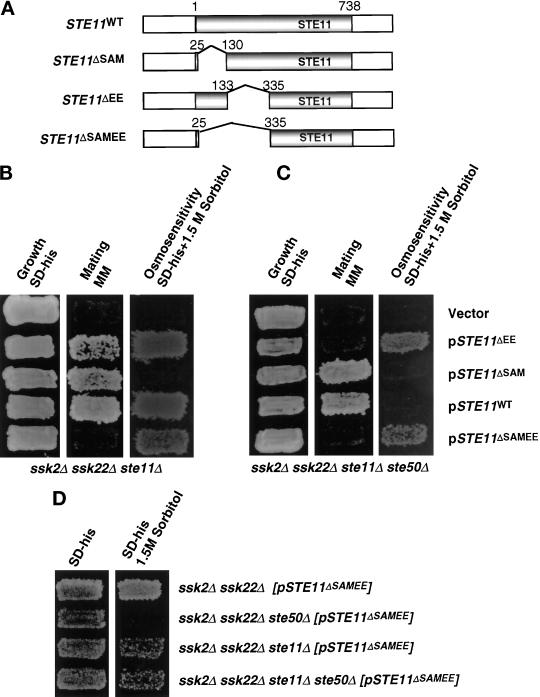
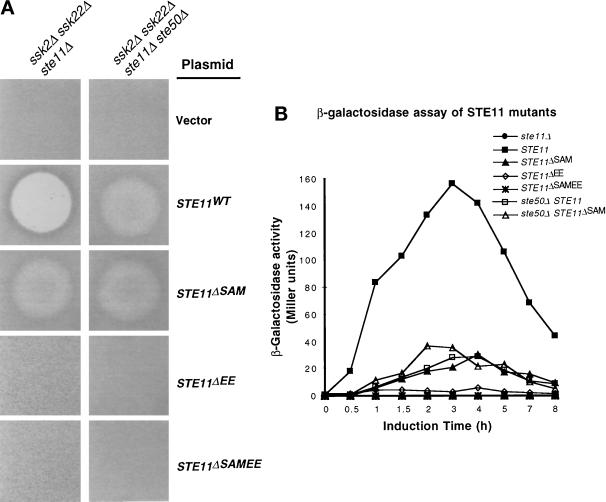
Ste50p interacts with the N terminus of Ste11p (Barr et al., 1996; Xu et al., 1996), and it has been suggested that the N terminus plays a negative regulatory role in Ste11p function (Cairns et al., 1992; Stevenson et al., 1992). Thus the N terminus of Ste11p may be the target for the regulatory function of Ste50p in signaling networks containing Ste11p. To investigate more fully the regulatory role of the N-terminal region of Ste11p, we introduced STE11 under its own promoter onto a CEN plasmid and constructed a series of in-frame deletions of the N-terminal region. We tested the function of these deletion mutants for their ability to activate the HOG pathway and to mate. Three deletion constructs of STE11 were made. STE11ΔSAM encodes Ste11p deleted for amino acid residues from 26 to 130 (which includes the SAM motif region of this protein [Ponting, 1995; Schultz et al., 1997]); STE11ΔEE encodes Ste11p with an in-frame deletion of amino acid residues 133–335 (corresponding to a deletion of the STE11 coding sequence from EcoRI to EspI sites; see MATERIALS AND METHODS); and STE11ΔSAMEE encodes Ste11p with an in-frame deletion of amino acid residues 26–335 (Figure 3A).
Figure 3.
Requirement for the N-terminal regions of Ste11p in the HOG and the pheromone response pathways. (A) The schematic representation of STE11 and its in-frame deletion constructs (see MATERIALS AND METHODS). The shaded box represents the coding region of STE11, gaps between the shaded boxes represent deletions, and the numbers on two sides of the gap indicate the aa fused after deletion of the gap region. All constructs are low-copy centromeric plasmids. (B) The effect of the N-terminal mutations in a ste11Δ strain. Yeast strain YCW340 (ssk2Δ ssk22Δ ste11Δ) was transformed either with the plasmids indicated or the cloning vector as a negative control, and the transformants were examined for growth on selective medium (left), the ability to form diploids (middle) on minimal medium (MM), and the ability to grow on high-osmolarity medium containing 1.5 M sorbitol (right). (C) The effect of the N-terminal mutations in a ste11Δ ste50Δ strain. Yeast strain YCW555 (ssk2Δ ssk22Δ ste11Δ ste50Δ) was transformed with either the plasmids indicated or the cloning vector as negative control, and examined for growth on selective medium (left), the ability to form diploids (middle) on minimal medium, and the ability to grow on high-osmolarity medium containing 1.5 M sorbitol (right). (D) Wild-type Ste11p has a dominant interfering effect on the constitutively activated allele of Ste11p in the HOG pathway. Yeast strains indicated were transformed with the constitutively active allele of STE11 construct, pSTE11ΔSAMEE, and examined for growth on selective medium (left) and the ability to grow on high-osmolarity medium containing 1.5 M sorbitol (right).
STE11ΔSAM was found to be unable to complement STE11 function in the activation of the HOG pathway, because a ssk2Δ ssk22Δ ste11Δ strain transformed with STE11ΔSAM was still osmosensitive; however, STE11ΔSAM could partially activate the mating pheromone pathway in the same strain (Figure 3B and Table 3), so the STE11ΔSAM allele is not simply nonfunctional. The STE11ΔEE construct gave the opposite result: STE11ΔEE was able to activate the HOG pathway just as well as the wild-type STE11 in a ssk2Δ ssk22Δ ste11Δ strain but was not able to fully complement the mating defect of the strain (Figure 3B and Table 3). Finally, expression of the most extensive N-terminal deletion STE11ΔSAMEE was able to activate the HOG pathway but was completely nonfunctional in the mating pheromone pathway (Figure 3B and Table 3). These results suggest that the SAM domain-containing region of Ste11p is required for the activation of the HOG pathway and is somewhat dispensable for the mating pheromone pathway, whereas the EE region is involved in the mating pathway and is dispensable in the HOG pathway.
Table 3.
Differential roles of the Ste11p N terminus in the HOG and mating response pathways
|
ssk2Δ ssk22Δ ste11Δ
|
ssk2Δ ssk22Δ ste11Δ ste50Δ
|
|||||||
|---|---|---|---|---|---|---|---|---|
| OsmRa | Mating (%)b | FUS1::LacZc | Arrest (halo)d | OsmRa | Mating (%)b | Fus1::LacZc | Arrest (halo)d | |
| Vector | <1 × 10−4 | <0.1 | < × 10−4 | <0.1 | ||||
| STE11WT | ++ | 11.3 | 156 | Clear | 4.3 | 29 | Fill-in | |
| STE11ΔSAM | 3.0 | 30 | Fill-in | 4.2 | 36 | Fill-in | ||
| STE11ΔEE | ++ | 0.1 | 5 | ++ | <1 × 10−4 | ND | ||
| STE11ΔSAMEE | + | <1 × 10−4 | <0.1 | + | <1 × 10−4 | ND | ||
The hyperosmolarity-resistant phenotype of yeast cells transformed with different STE11 alleles was scored for growth (+) on selective media with 1.5 M sorbitol for 4 d at 30°C. + indicates hyperosmolarity-resistant phenotype but with noticeably slower growth than wild type (++) on high-osmolarity media.
Mating was performed by mixing exponentially growing cells bearing different alleles of STE11 with fivefold excess of DC17 (his1) and incubating for 4 h at 30°C (MATERIALS AND METHODS). Mating efficiency was defined as (the number of diploid cells)/(the number of the experimental haploid cells) × 100. Data represent the mean of two to three independent experiments.
β-galactosidase activity was determined with yeast cells indicated transformed with different alleles of STE11 and FUS1::LacZ::URA3 plasmid, treated with 1 μM of α-factor for 1.5 h, and the activity was expressed as Miller units (see MATERIALS AND METHODS). Data represent the mean value of two to three independent experiments (the estimate of the error for each value was < 25% of the mean value).
Growth inhibition was assessed by spotting 5 μg of α-factor on media agar plate embedded with experimental cells bearing different alleles of STE11 and scored after 24 h of incubation at 30°C (see MATERIALS AND METHODS).
Deletions of the Ste11p N Terminus Can Bypass the Need for Ste50p in the HOG Pathway
We next determined whether the STE11ΔSAMEE mutant was able to bypass the requirement for Ste50p in the activation of the HOG pathway. A quadruple mutation strain (ssk2Δ ssk22Δ ste11Δ ste50Δ) was created and transformed with the various STE11 constructs. This strain by itself was osmosensitive and unable to grow on medium supplemented with 1.5 M sorbitol. Both the STE11ΔEE and STE11ΔSAMEE constructs were able to confer an osmoresistant phenotype (Figure 3C and Table 3), suggesting that the function provided by these mutant alleles was Ste50p-independent, and thus there was no essential requirement for Ste50p downstream of Ste11p in the HOG pathway. In contrast, wild-type Ste11p was unable to activate the HOG pathway in the quadruple mutation strain (Figure 3C and Table 3), suggesting an essential role of Ste50p in modulating the activity of the wild-type Ste11p.
In contrast to the situation with the osmotic pathway, deletion of N-terminal domains did not bypass the need for Ste50p in the pheromone response pathway. In fact, the EE deletion, which allowed weak mating in the triple mutant, was completely unable to restore mating to the quadruple mutant strain (ssk2Δ ssk22Δ ste11Δ ste50Δ) (Table 3). The deleted region is involved in the association of Ste11p with Ste5p (see below). Therefore mutations in Ste11p that disrupt the normal association with Ste5p do not totally abolish mating, but the residual mating ability is now Ste50p dependent.
Deletion of the SAM Domain-containing Region of Ste11p Acts Like Deletion of Ste50p
Intriguingly, the phenotypic consequences of loss of Ste50p function and loss of the Ste50 interaction domain on Ste11p (SAM domain-containing region) are identical (Figure 3 and Table 3). Cells containing either the ste50Δ allele or the STE11ΔSAM allele were osmotically sensitive, showed moderate levels of mating and FUS1 induction compared with wild-type, and responded to pheromone treatment with a transient arrest (Figure 4 and Table 3). In cells with the STE11ΔSAM allele, the presence or absence of Ste50p was not important (Table 3). These results suggest that the role of Ste50p in this activation requires physical interaction with the Ste11p N terminus. In the HOG pathway activation, this interaction is made unnecessary by deletion of the EE region or by the STE11–1 mutation.
Figure 4.
Ste11p is regulated by both Ste50p and Ste5p through their respective interactions with its SAM domain-containing region and the EE region. (A) The ste50Δ and the deletion of Ste50p binding domain of Ste11p had the same effect on the pheromone-induced cell cycle arrest. Yeast strain YCW340 (MATa ssk2Δ ssk22Δ ste11Δ) (left) and YCW555 (MATa ssk2Δ ssk22Δ ste11Δ ste50Δ) (right) were transformed with plasmids indicated or the cloning vector and examined for α-factor–induced growth inhibition by halo assays (MATERIALS AND METHODS). Approximately 1 × 106 exponentially growing cells were embedded in 5 ml melted agar and spread on prewarmed plates, and 5 μg of α-factor in 50% methanol were spotted on each plate. The halos were scored after 24 h incubation at 30°C. (B) The ste50Δ and the deletion of the Ste50p binding domain of Ste11p had the same effect on the pheromone-induced FUS1 expression. Strains used in A were cotransformed with plasmid pSB234 (FUS1::LacZ::URA3). Exponentially growing cells were treated with 3 μM of α-factor, and samples were taken at the time intervals indicated. β-galactosidase activity was assayed as described in MATERIALS AND METHODS and expressed in Miller units. Data represent the mean of two to three independent experiments.
Dominant Interference by Wild-type Ste11p in the Absence of Ste50p in the HOG Pathway
We further tested the ability of the STE11ΔSAMEE construct to bypass the HOG pathway requirement of both Ste11p and Ste50p in the absence of Ssk2p and Ssk22p. As noted, STE11ΔSAMEE activated the HOG pathway in strains with either the ssk2Δ ssk22Δ ste11Δ triple mutant or in the ssk2Δ ssk22Δ ste11Δ ste50Δ quadruple mutant; however, this allele of STE11 could not activate the HOG pathway in a ssk2Δ ssk22Δ ste50Δ triple mutant (Figure 3D). Therefore the presence of the wild-type Ste11p interferes with the ability of STE11ΔSAMEE to activate the HOG pathway in the absence of Ste50p. This dominant negative effect of Ste11p did not require an intact kinase domain because a catalytically inactive mutant of Ste11p generated the same phenotype when introduced into the quadruple mutant strain along with the STE11ΔSAMEE construct (our unpublished results).
Effect of N-Terminal Deletions of Ste11p on Pheromone-mediated Cell Cycle Arrest and FUS1 Induction
Activation of the pheromone response pathway causes cell cycle arrest and induction of gene expression. We used halo assays to monitor the effect of the N-terminal deletions of Ste11p in cells with and without Ste50p on mating pheromone-mediated cell cycle arrest. The STE11ΔEE and STE11ΔSAMEE constructs did not make detectable halos in either the triple (ssk2Δ ssk22Δ ste11Δ) or quadruple (ssk2Δ ssk22Δ ste11Δ ste50Δ) mutant strains (Figure 4A). Wild-type STE11 gave the expected clear halos in the triple mutant and fuzzy halos in the quadruple mutant, although the halo diameters are the same (Figure 4A). This indicates that Ste50p is required for the maintenance or propagation of the pheromone-mediated signals required for cell cycle arrest, but as suggested previously (Xu et al., 1996), it does not affect the initial sensitivity of the strain. As noted, the STE11ΔSAM construct gave identical fuzzy halos in both the triple and quadruple mutant strains (Figure 4A).
We next examined the ability of various mutants of STE11 to allow pheromone-induced transcriptional activation of FUS1 expression in the presence or absence of STE50 using β-galactosidase as a reporter. As shown in Figure 4B, wild-type cells showed a typical FUS1-LacZ induction profile, whereas cells with STE11ΔEE or STE11ΔSAMEE showed no induction of FUS1-LacZ. Interestingly, cells with ste50Δ, STE11ΔSAM, or ste50Δ together with STE11ΔSAM showed weak FUS1-LacZ induction compared with wild type, suggesting that the function of Ste50p is exerted through the interaction with the SAM domain-containing region of Ste11p. Deletion of this region of Ste11p made the presence or absence of Ste50p irrelevant.
Physical Interaction of Ste11p with Ste50p and Ste5p through Its Noncatalytic N-regulatory Domain
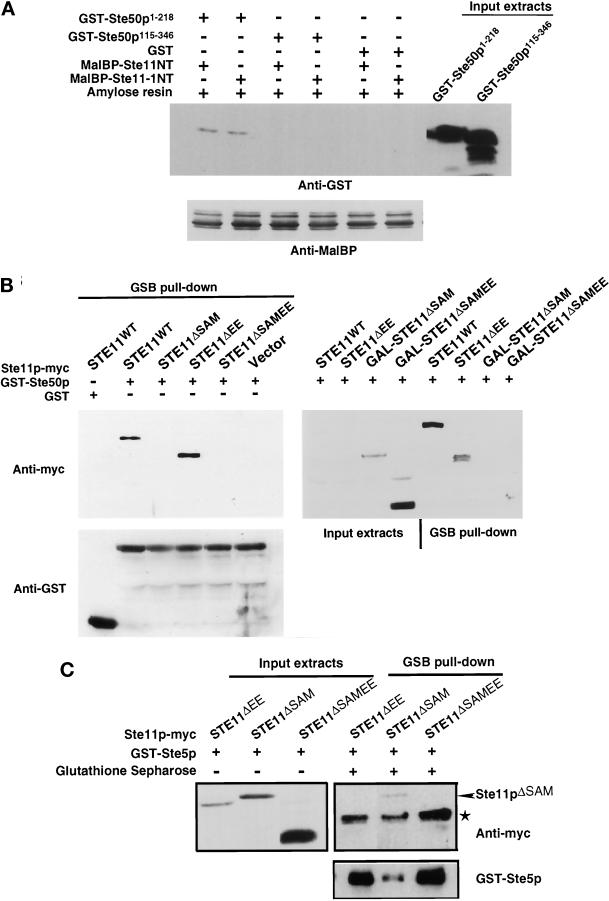
To determine biochemically the regions of Ste50p that interact with Ste11p, we expressed in E. coli the N-terminal part (aa 1–386) of Ste11p as a fusion protein with MalBP and expressed different parts of Ste50p as GST fusions proteins. Lysates containing MalBP-Ste11pNT were mixed with lysates containing GST fusions of various Ste50p fragments. MalBP-Ste11pNT was then isolated with amylose resin, subjected to SDS-PAGE, and the presence or absence of association of the different GST-Ste50p fusions was detected with anti-GST antibody (Figure 5A). The region of Ste50p that was required for the interaction with the N terminus of Ste11p resided in the first 115 aa of the N terminus of Ste50p. In contrast to previous data using the two-hybrid system (Xu et al., 1996), we observed that the N terminus of the constitutively active allele of STE11, STE11–1, was able to bind the N terminus of Ste50p in a manner identical to that of wild-type Ste11p (Figure 5A).
Figure 5.
Physical interaction of the N-terminal regulatory domain of Ste11p with Ste50p and Ste5p. (A) The SAM domain-containing region of Ste50p is required for the interaction with the N-terminal domain of Ste11p in vitro. Bacterial extracts containing GST alone or Ste50p fragments as GST fusions were mixed with bacterial extracts containing bacterially expressed N terminus of Ste11p or Ste11–1p as MalBP fusion proteins. Amylose agarose beads were added to the mixtures, incubated, washed extensively, and subjected to SDS-PAGE, and then immunoblot analysis was performed with anti-GST or anti-MalBP polyclonal antibodies and visualized with the ECL detection system (see MATERIALS AND METHODS). The input MalBP are shown in the bottom panel, and the input Ste50p fragments as GST fusion proteins are shown on the right of the top panel. The input GST is not shown. (B) The SAM domain-containing region of Ste11p is required for the interaction with Ste50p in vivo. Yeast strain YCW555 (MATa ssk2Δ ssk22Δ ste11Δ ste50Δ) was transformed with STE11 constructs (tagged with c-myc epitope) or the cloning vector as well as with either the GST-Ste50p expression construct (pVL57) or the GST vector. Exponentially growing cells (∼5 × 108 cells) in selective medium were shifted to selective medium or to selective medium with 3% galactose (to induce expression of GAL promoter-driven Ste11p constructs) for 3 h at 30°C. All media were supplemented with 0.5 mM CuSO4 to induce GST-Ste50 expression under the CUP1 promoter. Cells were then harvested, and cell extracts were prepared as described in MATERIALS AND METHODS. The cell extracts were either analyzed by Western blotting (input extracts) or incubated with glutathione-Sepharose beads at 4°C for 2 h, and the beads were then washed and subjected to Western blotting analyses (GSB pull-down) with either anti-myc (9E10) monoclonal antibody (to detect myc-tagged Ste11p constructs, top panels) or anti-GST polyclonal antibodies (to detect GST and GST-Ste50p, bottom panel) and visualized with the ECL detection system. Although all Ste11p constructs were not detectable by Western blotting using the crude extracts when they were expressed from their own promoter, GST-Ste50p was able to pull down specifically Ste11pWT and Ste11pΔEE. Neither Ste11pΔSAM nor Ste11pΔSAMEE were able to be pulled down by GST-Ste50p, although these proteins were overexpressed and readily detectable in the crude extracts by Western blotting. (C) The EE region of the N-terminal regulatory domain of Ste11p is required for the association of Ste5p with Ste11p. Yeast strain YCW340 (MATa ssk2Δ ssk22Δ ste11Δ) was transformed with STE11 constructs (tagged with c-myc epitope) and a GST-Ste5p expression construct (pVL23). Exponentially growing cells (∼5 × 108 cells) in selective medium with 3% raffinose were shifted to selective medium with 3% galactose (to induce expression of GAL promoter-driven Ste11p constructs) and supplemented with 0.5 mM CuSO4 (to induce GST-Ste5 expression under the CUP1 promoter) for 3 h at 30°C. The experiment was then performed as described in B. * indicates a nonspecific immunoreactivity.
It has been shown that the N terminus of Ste11p also interacts with Ste5p (Choi et al., 1994; Marcus et al., 1994; Printen and Sprague, 1994). We examined various N-terminal deletion mutants of Ste11p for their ability to associate with Ste50p and Ste5p in vivo. Ste50p or Ste5p GST fusion proteins were coexpressed in yeast with various deletion mutants of Ste11p tagged with the c-myc epitope. Cell extracts of the double transformants were incubated with glutathione-Sepharose beads, washed, subjected to SDS-PAGE, and assayed for the presence of c-myc–tagged Ste11p by immunoblot analysis. As shown in Figure 5B, although wild-type Ste11p and Ste11pΔEE were coprecipitated by GST-Ste50p, Ste11pΔSAM and Ste11pΔSAMEE were not. The association of various Ste11p mutants with GST-Ste50p was specific because Ste11p did not coprecipitate with GST alone. The level of Ste11p expressed from its own promoter was too low to be detected by Western blotting with crude cell extracts. To rule out the possibility that the failure to detect association of GST-Ste50p with Ste11pΔSAM and Ste11pΔSAMEE was due to lower level expression of these proteins, Ste11pΔSAM and Ste11pΔSAMEE were overexpressed and used for the coprecipitation assays. As shown in the right panel of Figure 5B, although the over-expressed Ste11pΔSAM and Ste11pΔSAMEE were readily detectable in the crude extracts by Western blotting, neither of these proteins was able to coprecipitate with GST-Ste50p. Thus, the N-terminal SAM domain-containing region (aa 26–129) of Ste11p was required for in vivo association with Ste50p. Ste11pΔEE and Ste11pΔSAMEE failed to be coprecipitated by GST-Ste5p, whereas Ste11pΔSAM was precipitated. Therefore, a distinct N-terminal region (133–335) of Ste11p was required for in vivo association with Ste5p (Figure 5C). These results indicate that the N-terminal regulatory region of Ste11p can associate with both Ste50p and Ste5p and thus might be regulated differentially by these two proteins. These experiments did not determine, however, whether Ste11p can associate with Ste5p and Ste50p simultaneously or whether the interactions are mutually exclusive.
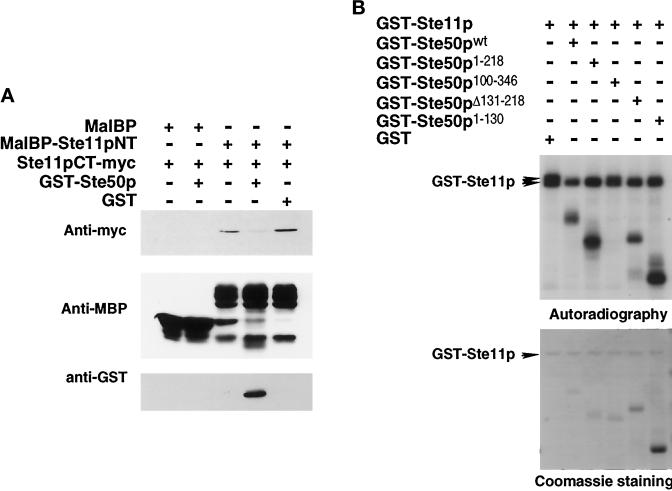
Ste50p Displaces the N-Terminal Regulatory Domain of Ste11p from Its Interaction with the C-Terminal Catalytic Domain of Ste11p
The N terminus of Ste11p as a MalBP fusion protein and the C terminus of Ste11p tagged with c-myc epitope as a GST fusion were expressed in E. coli, purified, and used in an in vitro binding assay. We found that the N-terminal portion of Ste11p interacted with the C-terminal portion of Ste11p. This interaction was specific because the C-terminal portion of Ste11p was unable to interact with MalBP alone. We asked if Ste50p, through its ability to interact with the N-terminal SAM domain-containing region of Ste11p, could influence the interaction between the N- and C-terminal portions of Ste11p. Purified MalBP-Ste11NT (aa 1–386) immobilized on amylose agarose beads was incubated with the same amount of purified GST-Ste11CT–myc (aa 385–738) in the presence of or absence of purified GST-Ste50p, washed, subjected to SDS-PAGE, and assayed for the presence of c-myc immunoreactivity by immunoblot analysis. As shown in Figure 6A, the MalBP-Ste11NT interacted with GST-Ste11CT–myc, and this interaction was effectively inhibited by GST-Ste50p, which under these conditions bound to MalBP-Ste11pNT. Thus, binding of GST-Ste50p to MalBP-Ste11pNT was responsible for the displacement of MalBP-Ste11pNT from interacting with GST-Ste11CT–myc.
Figure 6.
Ste50p blocks the N-terminal regulatory domain of Ste11p from binding to its C-terminal catalytic domain and is itself an in vitro substrate for Ste11p kinase. (A) Ste50p prevents the N-terminal regulatory domain of Ste11p from binding to the C-terminal catalytic domain of Ste11p. The myc-tagged C-terminal domain of Ste11p (Ste11CT-myc) and Ste50p were expressed as GST fusion proteins and purified from E. coli. The N-terminal domain of Ste11p was expressed in E. coli as an MalBP fusion protein, purified, and immobilized on amylose agarose beads. The beads were then incubated with ∼1 μg of purified myc-tagged C-terminal domain of Ste11p for 30 min at 4°C in the presence of ∼1 μg of either GST-Ste50p or GST alone. The beads were then washed, resuspended into sample loading buffer, and subjected to SDS-PAGE. Immunoblot analyses were then performed with anti-myc (9E10) monoclonal antibody or anti-GST polyclonal antibodies or anti-MalBP polyclonal antibodies using the ECL detection system. (B) Ste50p influences Ste11p kinase activity and is itself a substrate for Ste11p kinase in vitro. GST-Ste11p, various GST-Ste50p constructs, and GST were expressed and purified from yeast strain YCW555 (ssk2Δ ssk22Δ ste11Δ ste50Δ). In vitro kinase assays were performed with ∼250 ng of GST-Ste11p with 250 ng GST or GST-Ste11p in the presence of various GST-Ste50p. Kinase reactions were stopped and subjected to SDS-PAGE, followed by autoradiography (top panel) and Coomassie staining (bottom panel). GST is not shown.
Ste50p Is an In Vitro Substrate of the Ste11p Kinase
We next tested whether Ste50p is an in vitro substrate for Ste11p kinase. To do this, GST-Ste11p, GST-Ste50p, and GST fusions to various deletion constructs of Ste50p were purified from yeast strain YCW 555 (ste11Δ ste50Δ ssk2Δ ssk22Δ), and in vitro kinase assays were performed. As shown in Figure 6B, purified GST-Ste11p showed kinase activity as judged by autophosphorylation. Interestingly, autophosphorylation in the presence of GST alone resulted in the appearance of two distinct forms of GST-Ste11p of different electrophoretic mobilities. Addition of GST-Ste50p prevented the formation of the slower migrating form of Ste11p, and the GST-Ste50p itself was phosphorylated. Similar results were obtained with constructs containing the N terminus of Ste50p (either amino acid residues 1–130 or 1–218) as well as a construct containing an internal deletion (deletion of amino acid residues 131–218 inclusive) of Ste50p. These three constructs were in vitro substrates of Ste11p kinase, and their presence in the kinase reaction prevents the formation of the slower migrating form of Ste11p. In contrast, a GST-Ste50p construct lacking the N-terminal 115 amino acid residues was not an in vitro substrate for Ste11p kinase, and its presence was unable to prevent the formation of the slower migrating form of Ste11p. The ability of GST-Ste50p constructs to prevent the formation of the slower migrating form of Ste11p was not likely due to substrate competition, because the amount of phosphate incorporated into GST-Ste11p from the kinase reaction with the substrate Ste50p construct was very similar to that with the nonsubstrate Ste50p construct (our unpublished results).
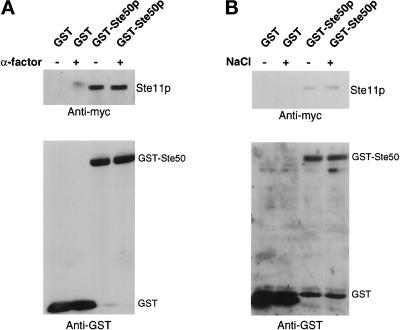
The Association of Ste50p with Ste11p Is Constitutive In Vivo
To determine whether the association of Ste50p with Ste11p is an environmental signal-regulated process, we performed pull-down assays with differentially tagged Ste50p and Ste11p. Yeast cells coexpressing c-myc epitope-tagged Ste11p constructs with either GST-Ste50p as a fusion protein or with GST as negative control were treated with or without stimuli (3 μM α-mating factor or 0.4 M NaCl high extracellular osmolarity). The GST fusion protein or GST alone was recovered with glutathione-Sepharose beads, and the presence of c-myc–tagged Ste11p was determined. As shown in Figure 7, Ste11p was specifically associated with GST-Ste50p in vivo. The amount of Ste11p associated with GST-Ste50p in vivo was the same in cells either treated with the stimuli or untreated. Thus, the association of Ste11p with Ste50p in vivo appeared to be constitutive and independent of pheromone treatment and extracellular hyperosmolarity stress.
Figure 7.
In vivo association of Ste50 and Ste11p is constitutive. Yeast strain YCW555 (MATa ssk2Δ ssk22Δ ste11Δ ste50Δ) was transformed with myc-tagged Ste11p expression construct (pCW199) and either a GST-Ste50p expression construct (pVL57) or a GST expression vector. Exponentially growing cells (3 × 109 cells) were induced with 0.5 mM CuSO4 (for the expression of GST-Ste50p or GST) for 2 h and then treated either with 3 μM α-factor for 1 h (A) or with 0.4 M NaCl for 10 min (B) before cells were harvested. Cell extracts were prepared and incubated with glutathione-Sepharose beads for 2 h at 4°C. The beads were washed, resuspended into sample buffer, and subjected to SDS-PAGE. Immunoblotting analyses were then performed with either anti-myc (9E10) monoclonal antibody or anti-GST polyclonal antibodies and detected with the ECL detection system.
DISCUSSION
Ste50p, originally identified as an ORF in the yeast genome project and shown to be involved in the mating pheromone response pathway (Rad et al., 1992), also plays an essential role in the Sho1p-Ste11p–mediated activation of the HOG pathway in S. cerevisiae. In the absence of the two-component osmoregulation branch, Ste11p (MAPKKK) kinase activity is essential for the survival of cells challenged with hyperosmotic stress (Posas and Saito, 1997). Under such conditions, Ste50p is also essential, because deletion of either STE11 (ssk2Δ ssk22Δ ste11Δ) or STE50 (ssk2Δ ssk22Δ ste50Δ) creates identical hyperosmosensitive phenotypes (O’Rourke and Herskowitz, 1998; Posas et al., 1998). The fact that constitutively active versions of the Ste11p kinase bypass the requirement of Ste50p suggests a role for Ste50p in the modulation of Ste11p activity. Thus Ste50p appears to be a structural and functional analogue of S. pombe Ste4p, which is required for the activation of Byr2p MAPKKK (Tu et al., 1997).
The N Terminus of Ste11p Is Regulatory
Ste11p, in common with many other members of the MAPKKK family, has a large N-terminal regulatory domain that is believed to play a negative regulatory role, because its deletion creates a constitutively active kinase (Wang et al., 1991; Cairns et al., 1992; Posas and Saito, 1997). We have therefore investigated the involvement of this domain and the proteins that interact with it in the pheromone response and osmoregulation pathways. As detailed below, the N terminus of Ste11p is a multifunctional domain. The region encompassing amino acid residues 26–129 contains sequence similarity to a motif termed a SAM domain found in a number of signaling proteins (Ponting, 1995; Schultz et al., 1997). This region is required for the physical interaction of Ste11p with the Ste50p protein, and this interaction plays a positive role in Ste11p activity. The next 203 amino acid residues (133–335) contain no recognized sequence motif but perform at least two functions. One of these is to direct the interaction between Ste11p and Ste5p and is thus required for proper pheromone response; the other is to provide negative regulation of the kinase, because its deletion leads to a Ste50p-independent activation of the HOG pathway.
Ste50p and Ste11p Interaction Requires Their N-Terminal Regions
The N terminus of Ste11p, which contains a region of sequence similarity to SAM domains, is required for physical association of Ste11p and Ste50p, and deletion of this region of the kinase creates a phenotype identical to that of deletion of Ste50p. As noted for STE50 deletion mutants, cells containing Ste11p lacking the SAM domain-containing region are weakly defective in mating, have a modified cell cycle arrest in response to pheromone, and are unable to fully induce pheromone-responsive genes. Such cells are also sensitive to osmotic stress in the absence of the two-component branch of the osmotic response network. Cells that contain the mutant STE11 allele are no longer influenced by the presence or absence of Ste50p; the phenotype of the STE50 deletion, the Ste11p SAM domain-containing region deletion, and the double mutant are all identical. This implies that the role of Ste50p in the regulation of the Ste11p kinase function depends on its association with the N terminus of Ste11p.
We have investigated the consequences of Ste50p binding to the Ste11p kinase. This binding appears to relieve the action of a negative regulatory domain adjacent to the SAM domain-containing region of the kinase. Deletion of the EE region of the Ste11p N terminus (aa 133–335) eliminates the requirement of both Ste50p and the SAM domain-containing region of Ste11p in activating the HOG pathway. The STE11ΔSAMEE allele of Ste11p confers resistance to hyperosmotic stress in either a STE50 or ste50Δ mutant background, and the STE11ΔEE allele also bypasses the need for Ste50p function in the osmotic stress pathway. The STE11–1 allele (Stevenson et al., 1992) can also bypass the requirement for Ste50p in the activation of the HOG pathway. This allele has a substitution of proline for serine at amino acid residue position 300 (P300S) within the EE region, consistent with the idea that this region plays a negative regulatory role in Ste11p function that requires Ste50p binding to relieve it.
The N terminus of Ste50p, like the N terminus of Ste11p, has a region of sequence similarity to the SAM domain motif. Deletions of this region of Ste50p block the association between Ste11p and Ste50p as assayed by resin binding experiments, so the link between these proteins appears to involve the interaction of two SAM domain-containing regions. Two-hybrid analysis also shows the involvement of these regions in Ste11p/Ste50p association (Posas et al., 1998; M. Rad, personal communication); however, the requirement for Ste50p in Ste11p function does not simply involve the interaction of the SAM domain-containing regions of the two proteins. Although deletion of the internal region of Ste50p does not influence either function or binding to Ste11p, deletions of the Ste50p C terminus that do not block interaction with Ste11p have a dramatic effect on Ste50p function. Some of these deletions create a dominant negative phenotype when overexpressed from the CUP1 promoter; cells lacking the two-component osmosensing branch of the HOG pathway become osmosensitive even in the presence of a normal Ste50p and Ste11p. This result implies that the Ste50p C terminus plays a critical role in cellular response to hyperosmotic stress.
Differential Regulation of Ste11p in the Pheromone Response and HOG Pathways
The regulation of the pheromone response pathway appears somewhat more complex than that of the hyperosmotic stress response pathway. First, deletion of the Ste11p SAM domain-containing region, or deletion of Ste50p, does not totally eliminate pheromone responsiveness, although it does eliminate the response to hyperosmotic stress. Second, the mating defect caused by deletion of STE50 can be bypassed by simple overexpression of wild-type Ste11p, although this overexpression does not bypass the need for Ste50p in the HOG pathway. These observations imply that the role of Ste50p in modulating the activity of Ste11p in the mating pathway is not as critical as was found for the HOG pathway. This result may be because the absolute level of Ste11p function needed for the mating pathway is less than that required for the HOG pathway. Alternatively, it may reflect the involvement of other regulators, such as Ste5p (see below), that can at least partially activate the mating pathway in the absence of Ste50p function. A third difference in the regulation of the mating response and the HOG pathway is that deletion of the negative regulatory EE region of Ste11p does not activate the pheromone response pathway, yet it does activate the HOG pathway. In fact, deletion of this region greatly reduces the response to pheromone signaling. This apparently positive involvement of the EE region in Ste11p function in mating can be explained by the requirement for this region in the Ste11p/Ste5p interaction. The STE11–1 allele of Ste11p, which relieves the requirement of Ste50p in the HOG pathway, also bypasses the need for Ste50p in the mating pathway but fails to do so in a ste5Δ mutant (Stevenson et al., 1992). This observation emphasizes the dual role of the EE region in both binding Ste5p and repressing Ste11p function. Although deletion of Ste5p leads to complete sterility, deletion of the Ste5p binding region of Ste11p only reduces mating. The residual mating is dependent on the association of Ste11p with Ste50p, because deletion of either the Ste11p SAM domain-containing region or the Ste50p protein reduces mating to zero (Table 3). The observation that the STE11ΔSAMEE construct is sterile appears inconsistent with a previous report (Cairns et al., 1992) that identified a constitutively active Ste11p created by deleting its N terminus; however, this previous work used an overexpressed version of the mutant Ste11p, whereas the current work involved the expression of wild-type levels of the protein.
Comparison of MAPKKK Regulation
It appears that Ste50p association with Ste11p is constitutive, suggesting that simple binding, although required, is not sufficient for the function of Ste50p in regulating Ste11p function. Other yet unidentified factor(s) may play a role. Consistent with this notion, the C-terminally truncated mutant of Ste50p can bind Ste11p, yet has a dominant interference phenotype in the activation of the HOG pathway in ssk2Δ ssk22Δ cells (Figure 2B) and shows a worse-than-null phenotype as judged by pheromone-induced morphogenesis (Figure 2A). This result indicates the importance of the C-terminal region of Ste50p in the activation of Ste11p, because the binding of Ste50p lacking the C-terminal domain to Ste11p is nonproductive. In addition, S. pombe homologues of Ste50p and Ste11p (Ste4p and Byr2p, respectively) associate with each other through their N-terminal SAM domain-containing regions, yet the regions of greatest sequence similarity between Ste50p and Ste4p reside in their respective C termini, suggesting a possible conserved function for this region as well. As is found with Ste50p, the Ste4p C terminus is required for proper function (Tu et al., 1997).
The theme of protein–protein interactions influencing kinase function is a common one. The serine/threonine protein kinase Raf-1 is one of the most extensively studied members of the MAPKKK family, and its activation is a complex multistep process that requires the proper interaction of its regulatory/autoinhibitory N-terminal domains with many regulatory proteins, including Ras, 14-3-3, and KSR (Therrien et al., 1996; Morrison and Cutler, 1997; Tzivion et al., 1998). The activation of mammalian PAK requires the binding of GTP-bound forms of either the Cdc42p or Rac1p small G-proteins (Manser et al., 1994). In S. pombe both Ras1p and Ste4p interact with and are required for the proper activation of Byr2p (MAPKKK) (Barr et al., 1996; Tu et al., 1997). In the current context, one attractive possibility is that Ste50p modulates Ste11p kinase function by creating structural changes in Ste11p that relieve the inhibitory function within the EE region and create a conformation that is favorable for further modulation by other factors, including autophosphorylation. Many signaling protein kinases, including Raf-1, PKC, and MEKK1 as well as Byr2p of S. pombe have regulatory domains that are autoinhibitory, because removal of these domains leads to constitutive activation of the kinases (Fanger et al., 1997 and refs therein; Morrison and Cutler, 1997; Newton, 1997; Tu et al., 1997). Disruption of such an autoinhibitory complex could be a step leading to kinase activation. Our in vitro resin binding assay showed both that the N and C termini of Ste11p can associate and that this association is disrupted by the addition of Ste50p. In addition, this in vitro interaction modulates autophosphorylation of Ste11p. These results imply that Ste50p binding to Ste11p could modify its conformation/phosphorylation state and thus potentially its function. The observation that both the basal and pheromone-induced transcriptional activation signals are lower in strains with ste50Δ than wild type is consistent with this notion. It is worth noting, however, that Ste50p associates with Ste11p constitutively in vivo, suggesting that Ste50p is necessary for but not sufficient for maximal activation of Ste11p in response to extracellular stimuli. Overall, given the remarkable similarities of structure and function between Ste50p and Ste11p on one hand, and Ste4p and Byr2p on the other, and the conserved nature of the MAP kinase module, it is possible that Ste50p/Ste4p-like molecules will play roles in MAPKKK activation in higher cells.
ACKNOWLEDGMENTS
We thank G. F. Sprague for plasmid pSL1654, M. Peter for plasmid pRD56, and H. Saito, F. Posas, G. Janson, and M. R. Rad for strains, for plasmids, and for communication of results before publication. We also thank A. Nantel, B. Klebl, J. Sherk, and other members of the Thomas lab for helpful discussions, J. Ash, L. Johnson, and D. Dignard for excellent technical assistance, and V. Lytvyn for valuable technical assistance during the initial phase of the work. The National Research Council of Canada publication number for this work is 42914.
REFERENCES
- Banuett F. Signaling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr MM, Tu H, Van Aelst L, Wigler M. Identification of Ste4 as a potential regulator of Byr2 in the sexual response pathway of Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:5597–5603. doi: 10.1128/mcb.16.10.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguslawski G, Polazzi JO. Complete nucleotide sequence of a gene conferring polymyxin B resistance on yeast: similarity of the predicted polypeptide to protein kinases. Proc Natl Acad Sci USA. 1987;84:5848–5852. doi: 10.1073/pnas.84.16.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Ramer SW, Kornberg RD. Order of action of components in the yeast pheromone response pathway revealed with a dominant allele of the STE11 kinase and the multiple phosphorylation of the STE7 kinase. Genes Dev. 1992;6:1305–1318. doi: 10.1101/gad.6.7.1305. [DOI] [PubMed] [Google Scholar]
- Choi K-Y, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- Fanger GR, Gerwins P, Widmann C, Jarpe MB, Johnson GL. MEKKs, GCKs, MLKs, PAKs, TAKs, and tpls: upstream regulators of the c-Jun amino-terminal kinases? Curr Opin Genet Dev. 1997;7:67–74. doi: 10.1016/s0959-437x(97)80111-6. [DOI] [PubMed] [Google Scholar]
- Feng Y, Song LY, Kincaid E, Mahanty SK, Elion EA. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Leberer E, Dignard D, Hougan L, Thomas DY, Whiteway M. Dominant-negative mutants of a yeast G-protein β subunit identify two functional regions involved in pheromone signaling. EMBO J. 1992;11:4805–4813. doi: 10.1002/j.1460-2075.1992.tb05586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall JE, Thomas DY. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Manser E, Leung T, Salihuddin H, Zhao Z, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- Marcus S, Polverino A, Barr M, Wigler M. Complexes between STE5 and components of the pheromone mitogen-activated protein kinase module. Proc Natl Acad Sci USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK, Cutler RE. The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- Newton AC. Regulation of protein kinase C. Curr Opin Cell Biol. 1997;9:161–167. doi: 10.1016/s0955-0674(97)80058-0. [DOI] [PubMed] [Google Scholar]
- O’Rourke SM, Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. SAM: a novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 1995;4:1928–1930. doi: 10.1002/pro.5560040927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- Posas F, Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Witten EA, Saito H. Requirement of STE50 for the osmostress induced activation of the STE11 MAPKKK in the HOG pathway. Mol Cell Biol. 1998;18:5788–5796. doi: 10.1128/mcb.18.10.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Printen JA, Sprague G., Jr Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad MR, Xu G, Hollenberg CP. STE50, a novel gene required for activation of conjugation at an early step in mating in Saccharomyces cerevisiae. Mol Gen Genet. 1992;236:145–154. doi: 10.1007/BF00279653. [DOI] [PubMed] [Google Scholar]
- Rhodes N, Connell L, Errede B. STE11 is a protein kinase required for cell-type-specific transcription and signal transduction in yeast. Genes Dev. 1990;4:1862–1874. doi: 10.1101/gad.4.11.1862. [DOI] [PubMed] [Google Scholar]
- Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- Saiki RJ, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Schultz J, Ponting CP, Hofmann K, Bork P. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 1997;6:249–253. doi: 10.1002/pro.5560060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson BJ, Rhodes N, Errede B, Sprague G., Jr Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- Therrien M, Michaud NR, Rubin GM, Morrison DK. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- Tu H, Barr M, Dong DL, Wigler M. Multiple regulatory domains on the Byr2 protein kinase. Mol Cell Biol. 1997;17:5876–5887. doi: 10.1128/mcb.17.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G, Luo Z, Avruch J. A dimeric 14–3-3 protein is an essential cofactor for Raf kinase activity. Nature. 1998;394:88–92. doi: 10.1038/27938. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu H-P, Riggs M, Rodgers L, Wigler M. byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol Cell Biol. 1991;11:3554–3563. doi: 10.1128/mcb.11.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway MS, Wu C, Leeuw T, Clark K, Fourest A, Thomas DY, Leberer E. Association of the yeast pheromone response G protein βγ subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- Wu C, Lytvyn V, Thomas DY, Leberer E. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J Biol Chem. 1997;272:30623–30626. doi: 10.1074/jbc.272.49.30623. [DOI] [PubMed] [Google Scholar]
- Wu C, Whiteway M, Thomas DY, Leberer E. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J Biol Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- Xu G, Jansen G, Thomas DY, Hollenberg CP, Ramezani Rad M. Ste50p sustains mating pheromone-induced signal transduction in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1996;20:773–783. doi: 10.1111/j.1365-2958.1996.tb02516.x. [DOI] [PubMed] [Google Scholar]