Abstract
The extreme variation in gene content among phylogenetically related microorganisms suggests that gene acquisition, expansion, and loss are important evolutionary forces for adaptation to new environments. Accordingly, phylogenetically disparate organisms that share a habitat may converge in gene content as they adapt to confront shared challenges. This response should be especially pronounced for functional genes that are important for survival in a particular habitat. We illustrate this principle by showing that the repertoires of two different types of carbohydrate-active enzymes, glycoside hydrolases and glycosyltransferases, have converged in bacteria and archaea that live in the human gut and that this convergence is largely due to horizontal gene transfer rather than gene family expansion. We also identify gut microbes that may have more similar dietary niches in the human gut than would be expected based on phylogeny. The techniques used to obtain these results should be broadly applicable to understanding the functional genes and evolutionary processes important for adaptation in many environments and useful for interpreting the large number of reference microbial genome sequences being generated for the International Human Microbiome Project.
Keywords: comparative genomics, glycoside hydrolases, glycosyltransferases, gut ecosystem, microbial genome evolution
Closely related microorganisms can differ radically in genome content, particularly if they are adapted to different habitats (1). For instance, in Escherichia coli, a species in which closely related strains can have very different lifestyles, only 2,241 of the 9,433 genes observed in 32 sequenced strains were present in all genomes (2). This variation suggests that gene content plasticity may play a key role in adaptation to new environments. Evolutionary processes that affect gene content include new gene acquisition via horizontal gene transfer (HGT), gene family expansion from duplication, and gene loss from deletion (3). HGT can increase fitness by allowing microbes to acquire useful functions from other microbes that live in their environment, such as antibiotic resistance (4). Gene duplication generates paralogs that can diverge and acquire new functions (5). Gene loss occurs when genes do not provide a selective advantage, or when they are deleterious for a particular lifestyle (6).
The identification of gene families that have recently expanded by duplication or were acquired by HGT in a particular genome can reveal functions important for an organism's lifestyle (7, 8). Gene families that have independently expanded in diverse lineages that live in the same habitat may have functions that are important for shared challenges within that habitat, rather than for a distinctive niche. Methods that (i) show when functionally important genes converge in response to shared habitat and the evolutionary processes that cause this convergence and (ii) can evaluate many such functional gene groups using information from many genomes will become increasingly important as more sequenced genomes become available (e.g., through the International Human Microbiome Project; ref. 9).
In this report, we show convergence in functional gene repertoires by determining whether these gene families cluster genomes together significantly better than a 16S rRNA phylogeny. Methods for comparing genomes based on gene content have been extensively explored for a different application, namely inferring phylogenetic relationships among genomes: these methods can either examine the presence/absence patterns of specific gene families or take the degree of sequence similarity into account (10). Despite the potentially confounding factors of HGT and gene duplication and loss, the overwhelming conclusion has been that these phylogenomic methods yield genome trees that are remarkably consistent with 16S rRNA phylogeny and that gene content plasticity creates random noise that does not obscure shared phylogenetic history (10, 11). Although these phylogenomic approaches can inform development of clustering methods that seek to detect convergence of functional genes, they are not directly applicable because they are optimized to disregard, rather than to detect, genome plasticity by correcting for HGT, duplication, and parallel gene loss, [e.g., by excluding gene families with paralogs or that have phylogenies suggestive of HGT (10, 12, 13)]. Methods to cluster genomes to detect for convergence of functional gene repertoires have generally focused on the presence or absence of gene families alone, and not the degree of similarity between genes or the number of representatives in a particular gene family. For instance, Ren and Paulsen (14) used hierarchical clustering of the “presence” or “absence” profiles of homologues in a set of fully sequenced genomes (phylogenetic profiling) to explore the evolution of membrane transport content. The resulting clusters correlated with both evolutionary history and lifestyle: the obligate intracellular pathogens/symbionts, the soil/plant associated microbes, and a collection of autotrophs formed clusters despite phylogenetic differences (14).
The methods we describe below account for similarity between gene sequences and family number to show convergence. This approach is illustrated using two classes of carbohydrate active enzymes in human gut-associated microbes, glycoside hydrolases (GH) and glycosyltransferases (GT). GH catalyze the hydrolysis of glycosidic bonds between sugar resides or between a carbohydrate and non-carbohydrate moiety and are important for the degradation of complex plant polysaccharides in the gut (7). GT catalyze the transfer of sugars from activated donor molecules to specific acceptors and are important for the formation of surface structures recognized by the host's immune system (15) and enabling microbes to colonize the gut (16). Previous comparisons of the sequenced genomes of several human gut-derived Bacteroidetes have revealed plasticity in their repertoires of both GTs and GHs; GTs were significantly enriched in horizontally transferred genes and GHs were among the most markedly expanded paralogous groups (17, 18). We applied clustering methods to genome sequences from (i) 36 human gut-derived microbes representing four bacterial phyla (the Firmicutes and Bacteroidetes, whose members predominate in the distal intestine, as well as the Actinobacteria and Proteobacteria), and two members of Archaea, plus (ii) 31 phylogenetically related organisms that do not live in the gut, and show that clusters based on GH and GT gene repertoires group gut-associated genomes together better than does a 16S rRNA phylogeny.
Results
Methods for Clustering Isolates Based on Functional Gene Repertoires.
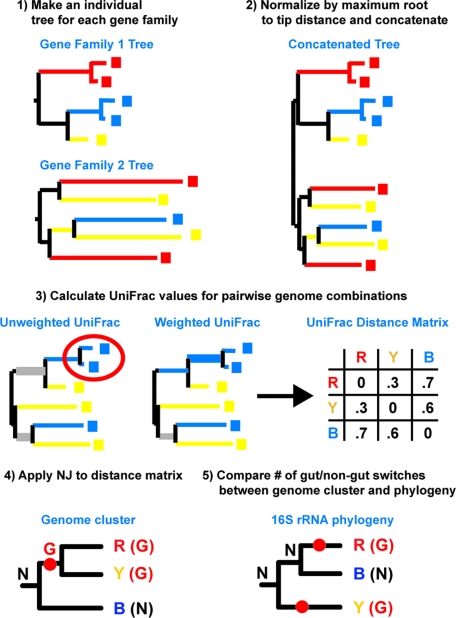
The four methods for gene content-based clustering of genomes described here (Table 1) are optimized to detect genome plasticity due to different evolutionary forces. The first two methods are adapted from the unweighted and weighted UniFrac method that we described for comparing microbial communities (19–21). Unweighted UniFrac measures the distance between two collections of sequences as the percent of branch length in a phylogenetic tree that leads to one collection or the other but not both (Fig. 1). Sequence collections with phylogenetically related sequences will have lower UniFrac values than those with phylogenetically distinct sequences. Likewise, weighted UniFrac accounts for phylogenetic relatedness between sequences and also for the number of times a sequence is observed (19, 20) (Fig. 1).
Table 1.
The four different techniques for making genomic distance matrices
| UniFrac | Counts | |
|---|---|---|
| Unweighted | Accounts for presence/absence of gene family members and their phylogenetic relationship. Sensitive to gene acquisition and loss. | Accounts only for presence/absence of gene family members. Sensitive to gene acquisition and loss. |
| Weighted | Accounts for relative abundance of gene family members and their phylogenetic relationship. Sensitive to gene acquisition, loss, and duplication. | Accounts only for relative abundance of gene family members. Sensitive to gene acquisition, loss, and duplication. |
Fig. 1.
Methodologic approaches. Schematic of genomic UniFrac that can be applied to a “forest” of trees. 1) First we use NJ to generate a phylogenetic tree for each gene family. In this example, we are comparing three genomes that are colored red, blue, and yellow using two gene families. 2) Trees are joined by addition to the same root with a branch length of zero. The trees are first normalized by dividing the branches by the maximum root to tip distance to correct for differential rates of evolution in the different gene families. 3) Pairwise UniFrac distances are calculated between all possible combinations of genomes using both unweighted and weighted UniFrac. For each pair, all sequences not from either genome are first removed. Unweighted UniFrac distances are the fraction of branch length that leads to one genome or the other (yellow and blue branches) but not both (gray branches). Paralogous genes (circled in red) do not heavily affect the results because they introduce little unique branch length. Weighted UniFrac weights each branch by the differential representation of its descendants in the two genomes (represented by line thickness; gray branches carry no weight). The blue genome will look more different from the yellow because of the paralogs. 4) The final genome cluster is made by applying the NJ algorithm to the UniFrac distance matrix. It takes only one switch between Non-gut (N) and Gut (G) to describe the distribution of states on the tree. 5) Determining if genome clusters group gut genomes together better than phylogeny. If the ancestral state was N, it would require more changes to explain the distribution of states in the 16S rRNA phylogenetic tree than in the genome cluster, suggesting convergence.
A major difference in using UniFrac to cluster genomes, as opposed to microbial community comparisons based on 16S rRNA genes, is that phylogenetic trees from multiple gene families must be evaluated, rather than a single phylogenetic tree from one gene. For instance, the GH and GT sequence collections in the Carbohydrate-Active Enzymes database (CAZy) (22) currently consist of members of 112 and 91 distinct protein families, respectively. To integrate these data, we can create a separate phylogenetic tree for each gene family and concatenate these trees to a single root (Fig. 1). Protein families can differ dramatically in the rate of evolution of their sequences, and “fast-evolving” sequences will produce trees with longer branches than “slow-evolving” sequences (23). Since this rate heterogeneity can bias the UniFrac results toward relationships found primarily in the fast-evolving protein families, we normalize the trees before concatenation by dividing the branch lengths by the maximum root-to-tip distance (Fig. 1).
The other two methods that we used to create distance matrices between genomes are weighted and unweighted count methods (Table 1). In the weighted count approach, the difference between two genomes is the sum of the absolute value of the difference in gene counts for each gene family. The unweighted count approach is the same, except the gene families are scored with 0 or 1 depending on whether the gene families are present or absent in each genome: this approach is related to the phylogenetic profiling method used by Ren and Paulsen (14). The distance matrices from the unweighted and weighted UniFac and count measures were used to create genome clusters by applying the neighbor-joining (NJ) algorithm (24). We chose NJ, rather than clustering methods that produce an ultrametric tree (in which all tips are equidistant from the root) such as UPGMA (25), because it allows for variation in the rates of evolutionary change. The rate of change of gene content varies substantially among organisms, especially as they adapt to different environments (1), although models describing these changes are still in their infancy (preventing likelihood or Bayesian approaches to cluster building). For comparison, however, we also applied UPGMA to the distance matrices.
The four clustering techniques differ in the degree to which they are sensitive to underlying processes of genome evolution (Table 1). Thus, comparing the results can suggest which processes, such as HGT or parallel gene duplication/loss, underlie the adaptation. For instance, the unweighted UniFrac test should be particularly sensitive to convergence due to HGT, because HGT will cause the input tree to have more closely related sequences than expected from phylogeny. It will also be sensitive to gene loss, because genomes that have members of the same gene family will have additional shared branch length. However, unweighted UniFrac will not be sensitive to parallel expansion of families by duplication, because it is a qualitative measure, and the addition of identical or near identical sequences will have little to no effect on the results (20) (Fig. 1). In contrast, weighted UniFrac accounts for the number of representatives a gene family has in a genome and should be sensitive to gene family expansion as well as HGT (Fig. 1). If there is parallel expansion of each organism's original copy of a gene (as opposed to a horizontally acquired copy), genome clustering by lifestyle should be best observed with the weighted counts method. Like unweighted UniFrac, the unweighted counts method is sensitive to gene acquisition and loss, but we expect that unweighted UniFrac would be more sensitive to HGT because highly similar genes from a recent HGT event will share much more branch length than genes whose original version of the gene is present in both genomes.
Selecting Gut and Related Non-Gut Microbes.
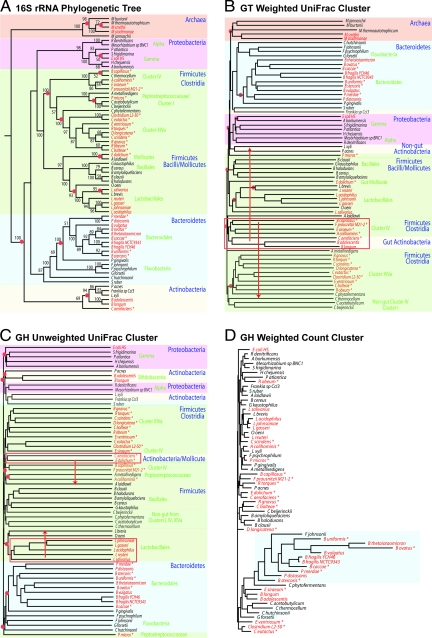
Our analysis included 67 microbial genomes: of the 36 obtained from the human gut, 21 were part of an ongoing project to sequence 100 cultured representatives of major phylogenetic lineages in the normal distal human intestinal microbiota [Human Gut Microbiome Initiative HGMI; http://genome.wustl.edu/sub_genome_group.cgi?GROUP = 3&SUB_GROUP = 4], and 15 came from other projects [supporting information (SI) Tables S1 and S2]. To identify phylogenetically related non-gut isolates with sequenced genomes, we made a NJ 16S rRNA phylogenetic tree of organisms in the HGMI genome sequencing pipeline and completed microbial genomes deposited in GenBank as of March 19, 2008, and culled sequenced isolates that were related to the 36 gut isolates (Fig. 2A). To determine the environmental distribution of these relatives and to verify that they were not found in the guts of any vertebrate hosts, we used metadata about the isolates deposited in NCBI (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi), publications describing the isolate, and the titles of culture-independent environmental surveys that deposited near identical (>98% sequence identity) 16S rRNA sequences in GenBank's ENV database (see SI Methods and Table S2).
Fig. 2.
Clustering of the 67 gut and non-gut associated microbes included in this study. (A) 16S rRNA-based phylogenetic tree that is the majority rule consensus of 1,000 bootstrapped NJ trees (see SI Methods). Gut microorganisms are highlighted in red. Sequenced genomes from the HGMI are marked with an asterisk. Red dots denote the 13 internal nodes where Fitch parsimony counted a gut/non-gut switch (see Fig. 1). Higher-level taxonomic categories are noted with both text and shading. (B) The GT weighted UniFrac cluster. Higher-level taxonomic categories are shaded as in A and gut organisms are colored red. The red box highlights interdivision clustering between gut Actinobacteria and Firmicutes. Red arrows show where related, non-gut organisms (non-gut Actinobacteria and C. thermocellum) cluster instead. (C) The GH unweighted UniFrac cluster. Shading and text colors are as described for B. The red boxes and arrows highlight habitat related clustering (the gut Mollicute E. dolichum clusters with a gut Actinobacteria instead of with its relative Acholeplasma laidlawii and the non-gut Lactobacillus brevis clusters with O. oeni instead of its relative from the gut L. salivarus). (D) The GH weighted count cluster. Gut organisms are in red text and members of the Bacteroidetes are highlighted in blue.
Clustering Isolates Based on GH and GT Gene Repertoires: Results.
Table S3 and Table S4 summarize CAZy annotations for the GH and GT family members. 2,370 genes from 29 GT families and 3,537 genes from 84 GH families were present in the included genomes. Each of the four clustering methods was used to group the genomes based on these data. To quantify the recovery of phylogenetic relationships, we calculated the fraction of nodes in the phylogenetic tree that were shared by the GT/GH-based “genome cluster” (26). We assessed whether gut genome clustering was significant by determining the fraction of the time that the number of Fitch parsimony changes was lower when the gut and non-gut states were assigned randomly. The 16S rRNA phylogeny itself clusters gut genomes together better than chance expectation; a minimum of 13 changes between gut and non-gut are required to explain the distribution of these states on the 16S rRNA tree (Fig. 2A, red dots). This number is less than chance (22 changes for the same tree with random state assignments), because organisms that live in the gut tend to be phylogenetically related. Since a significant result with this test could be due to the recovery of phylogenetic relationships alone, we also compared the genome clusters with the phylogeny by measuring the fraction of bootstrapped 16S rRNA trees that had less than or equal parsimony counts than the genome cluster. Functional groups that have few informative characters (i.e., few gene families or “fast-evolving” gene families with few informative characters) may cluster genomes by habitat no better than phylogeny, even if some convergence has occurred, due to lack of statistical power. However, a significant result provides strong evidence for convergence because it must recapture both phylogenetically-related gut clustering and habitat-related deviations from phylogeny.
The UniFrac methods outperformed the count methods in both the clustering of gut genomes and the recovery of phylogenetic relationships (Table 2). For the GTs, both the unweighted and weighted UniFrac methods clustered gut organisms significantly better than phylogeny (P ≤ 0.001), with 10 and 9 versus 13 parsimony changes. The unweighted and weighted UniFrac methods also recovered 48% and 43% of the nodes in the 16S rRNA tree respectively. In contrast, neither the unweighted nor the weighted count method clustered gut genomes significantly better than phylogeny (12 and 15 parsimony changes respectively), and they only recovered 5–11% of the nodes in the 16S rRNA tree (Table 2). The UPGMA results were similar to the NJ results but generally did not cluster gut genomes as well (Table S5).
Table 2.
Comparison of GT and GH clustering results to the 16S rRNA phylogeny
| Cluster type | Fraction shared nodes | Cluster parsimony count | 16S rRNA parsimony count | Probability better than phylogeny | Probability better than chance |
|---|---|---|---|---|---|
| GT UniFrac UW | 0.48 | 10 | 13 | ≤0.001 | ≤0.001 |
| GT UniFrac W | 0.43 | 9 | 13 | ≤0.001 | ≤0.001 |
| GT Count UW | 0.11 | 12 | 13 | 0.20 | ≤0.001 |
| GT Count W | 0.05 | 15 | 13 | 1.0 | 0.002 |
| GH UniFrac UW | 0.42 | 8 | 11 | ≤0.001 | ≤0.001 |
| GH UniFrac W | 0.33 | 10 | 11 | 0.18 | ≤0.001 |
| GH Count UW | 0.10 | 12 | 11 | 0.98 | ≤0.001 |
| GH Count W | 0.12 | 14 | 11 | 1.0 | 0.001 |
UW, unweighted; W, weighted.
Although some of the weighted UniFrac clustering was consistent with phylogeny (e.g., the clustering of the gut Bacteroidetes), there are remarkable examples where gut organisms cluster together despite major differences in phylogeny (Fig. 2B). For instance, diverse gut Actinobacteria, including two species in the genus Bifidobacteria and the distantly related Colinsella aerofaciens, cluster together rather than with the three non-gut Actinobacteria that occupy an intermediate position in the 16S rRNA tree (compare A and B in Fig. 2). The gut Actinobacteria cluster with Firmicutes related to members of Clostridia Clusters IV, which are all gut organisms, rather than with the non-gut Actinobacteria, which are located in completely different parts of the tree (Fig. 2B, red arrow). Additionally, the one non-gut species representing Clostridium Cluster IV, Clostridium thermocellum, groups with non-gut Firmicutes related to Clostridium Cluster I (Fig. 2B; red arrow). Also, the predominant gut archaeon Methanobrevibacter smithii clusters with another gut archaeon Methanosphaera stadtmanae, rather than with the non-gut Methanobacterium thermoautotrophicum as it does in the 16S rRNA tree (Fig. 2 A and B).
Comparison of Clustering Methods Suggests that the Convergence of GT Genes in Gut Bacteria Is Due to HGT and Parallel Gene Loss.
The GT weighted and unweighted UniFrac clusters were very similar; the unweighted UniFrac cluster had one more gut/non-gut parsimony change (Table 2) but the same clustering patterns between phylogenetically disparate gut organisms noted for weighted UniFrac (data not shown). This indicates that the presence/absence of phylogenetically similar GTs alone was largely sufficient to observe the pattern but that it may have been slightly enhanced by parallel gene family expansion. Weighted UniFrac clustered gut genomes slightly better, even though the introduced noise caused a smaller fraction of nodes to be shared with the 16S rRNA tree (Table 2). However, parallel gene expansion was not a major cause of the GT convergence: the weighted count cluster grouped gut organisms together the worst of all of the methods used and substantially worse than the 16S rRNA tree, requiring 15 gut/non-gut changes. Although the unweighted count method is sensitive to the same factors as unweighted UniFrac (HGT and parallel gene loss), it was clearly not as powerful as the unweighted UniFrac method for detecting GT convergence since the gut clustering was not significantly better than phylogeny.
Unweighted UniFrac also Detects Significant Convergence of GH Gene Repertoires in Gut Genomes.
Unweighted UniFrac of the GH gene families also clustered gut organisms together significantly better than phylogeny (P ≤ 0.001), with 8 versus 11 parsimony changes (note that the number of parsimony changes in the 16S rRNA tree is lower for the GHs than for the GTs; this is because the GH analysis excludes the Archaea since neither of the gut Archaea had any annotated GHs). Weighted UniFrac performed worse than unweighted UniFrac, with 10 parsimony changes needed to explain the distribution. Although this is still less than in the 16S rRNA phylogeny, the result is not significant when accounting for the uncertainty in the topology of the 16S rRNA tree (Table 2). Unweighted UniFrac again recaptured phylogenetic relationships better than weighted UniFrac (42% vs. 33% of the 16S rRNA tree recovered). The recovery of phylogeny was slightly worse for GHs than for GTs, indicating that GH genes generally have a less strong phylogenetic signal, either due to fewer phylogenetically informative positions in the aligned sequences of families or a greater tendency for confounding factors such as gene loss or gain. As with the GTs, the unweighted and weighted count methods did not recover phylogenetic relationships as well as the UniFrac methods, sharing 10% and 12% of the nodes with the phylogeny respectively. Although both count methods clustered gut genomes worse than phylogeny, all of the clusters were better than expected by chance alone (Table 2), which may indicate a role for gut-related clustering for the count clusters since they recapture phylogeny so poorly.
Unweighted UniFrac clustered phylogenetically disparate gut organisms, although the deviations from phylogeny are not the same as for the GTs (Fig. 2C). For instance, with the GHs, Lactobacillus salivarus clusters with other gut Lactobacilli rather than with the non-gut Lactobacillus brevis, which instead clusters with the non-gut Oenococcus oeni. Similarly, the gut Mollicute Eubacterium dolichum and a gut Actinobacteria (Collinsella aerofaciens) cluster together rather than with their relatives, indicating that these phylogenetically disparate organisms may occupy more similar niches in terms of carbohydrate utilization than would be predicted by phylogeny alone. Non-gut members of three of the main groups of Clostridia in the analysis (related to Clusters I, IV, and XIVa) also cluster together rather than with their gut relatives. This may be related to the shared environment of these species, which are all prevalent in soil (Table S2).
Before performing these analyses, we expected that the weighted clustering methods might cluster gut genomes better based on GHs because a substantial expansion of GH families had been observed in gut Bacteroidetes (7, 17). However, since there are no non-gut isolates interspersed among the gut Bacteroidetes in the 16S rRNA tree, improved gut genome clustering could only be observed if a gut-associated member of a different phylum experienced the same type of gene expansion. The comparatively poor performance of both weighted clustering techniques indicates that this is not the case, and that different phyla do not undergo expansion of the same gene families. The expansion of GHs in gut genomes, however, is evident from the long branches among gut Bacteroidetes in the weighted count cluster (Fig. 2D). Interestingly, the clustering pattern reveals that two soil organisms may have similar expansions of their GH repertoires as the gut Bacteroidetes: Flavobacterium johnsoniae, a Bacteroidetes species characterized by the ability to use a wide variety of naturally occurring complex glycans, such as chitin, cellulose, and lignin (27, 28), and the Firmicute Clostridium phytofermentans, which performs anaerobic fermentation of cellulose to acetate and ethanol (29). The fact that this change was not detected by unweighted or weighted UniFrac indicates that HGT did not play a role in this association, but rather that soil and gut microbes independently expanded their own versions of genes within the same families to address similar challenges in different environments.
Discussion
Comparison of genome clusters from the four methods described here reveal that diverse gut microbes had a convergence of carbohydrate-active genes and that HGT and parallel gene loss, as opposed to parallel duplication, are likely to be important for this convergence. This is consistent with previous studies showing that gene duplication has a minor impact on gene content variation between related genomes compared to HGT and gene loss (30). The unweighted count method did not perform as well for GHs and GTs as it did in a previous study of membrane transport proteins (14), indicating that these gene families are not as stably inherited vertically. Therefore, it is notable that the UniFrac methods were still sufficiently powerful to detect convergence and recover phylogenetic relationships.
The four clustering methods are better suited for detecting the convergence of functional gene repertoires than the techniques that have been described for phylogeny. For instance, clustering genomes with unweighted UniFrac is similar to the superalignment (12) and supertree (13) phylogenomic methods: it clusters genomes based on phylogenetic information from multiple protein families, but is influenced more by parallel gene loss and can either account or correct for paralogy by using the weighted and unweighted versions respectively. The unweighted UniFrac method can also be further explored for phylogenomics as it has some interesting advantages for this application: it is insensitive to paralogs and allows “missing” genes. Therefore, it can use most of the information in a set of genomes, and build individual gene trees using parameters that are specific for each gene family, allowing normalization of evolutionary rates.
The UniFrac methods outperformed the count methods, both for detecting convergence and for recovering phylogenetic relationships between genomes. Thus, accounting for the relatedness between gene family members in genome comparisons utilizes more of the data and has increased power. However, the count methods can occasionally detect relationships that the influence of sequence relatedness may obscure, such as parallel expansion of non-horizontally acquired genes as shown here for GHs in gut and soil Bacteroidetes.
In this analysis, we specifically looked for convergence in gene families with functions already believed to be important in the gut. The significant clustering of gut microbes based on these functions supports the notion that the level of gene family convergence can predict functions important for adaptation to shared challenges for a given lifestyle. Convergence should not be as strong, however, for functions that are important only for a particular niche within a habitat for which there is strong competition. This work paves the way for genome-wide adaptation of the techniques to provide an unbiased look at which of all of the types of functional genes converge the most in gut genomes. As the UniFrac approaches require the computationally intensive steps of sequence alignment and phylogenetic reconstruction, successful genome-wide application to the growing set of available genomes will require further exploration of how fast, approximate methods impact the results.
The degree of gene repertoire convergence and correlation with 16S rRNA phylogeny also indicates how informative surveys of 16S rRNA gene sequences are for describing specific functions in human gut samples. It will be interesting to apply these techniques to other features of carbohydrate metabolism (e.g., nutrient sensors, carbohydrate binding proteins, transporters, fermentation pathways), for a more detailed look at how predictive 16S rRNA sequences are likely to be for important functions in the gut. This will aid in the interpretation of the large 16S rRNA datasets being generated in the International Human Microbiome Project and other efforts that seek to understand how the gut microbiota changes with diet, age, and disease.
Finally, the power of comparative genome analysis for understanding how microbes adapt to the gut habitat is greatly affected by the availability of genome sequences for phylogenetically related microorganisms that do not inhabit the gut. Therefore, the International Human Microbiome Project should profit from sponsoring efforts to sequence non-gut organisms that (i) intersperse deep branching lineages for which only genome sequences from gut dwellers are available, and (ii) live in a diverse set of habitats so that clustering of gut organisms does not appear as an artifact of convergence in organisms from another heavily represented environment.
Methods
Implementation of the Genome Clustering Methods.
For each GH and GT family with ≥3 sequences, a multiple sequence alignment was made using MUSCLE (31). To exclude low-quality alignment regions, positions at which >25% of the sequences had a gap were removed. NJ trees were made from these alignments using ClustalW (32). Each tree was normalized by dividing the branches by the maximum root-to-tip distance and concatenated to a single tree root. The NJ algorithm and UPGMA were applied to the distance matrices from all four methods to produce the genome clusters. All computations were performed with the Python programming language using PyCogent (33) and the UniFrac Python API (19).
Supplementary Material
Acknowledgments.
We are grateful to Janaki Guruge, Ruth Ley, Priya Sudarsanam, Brian Muegge, and Michael Mahowald for their help at various stages of this project. This work was supported by grants from the NIH (DK78669, DK30292, DK70977), NSF (OISE-0812861), and the Crohn's and Colitis Foundation of America. M.H. was supported by the NIH Molecular Biophysics Training Program (T32GM065103) and an NSF EAPSI fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807339105/DCSupplemental.
References
- 1.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willenbrock H, Hallin PF, Wassenaar TM, Ussery DW. Characterization of probiotic Escherichia coli isolates with a novel pan-genome microarray. Genome Biol. 2007;8:R267. doi: 10.1186/gb-2007-8-12-r267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraser-Liggett CM. Insights on biology and evolution from microbial genome sequencing. Genome Res. 2005;15:1603–1610. doi: 10.1101/gr.3724205. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 5.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 6.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 7.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 8.White O, et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snel B, Huynen MA, Dutilh BE. Genome trees and the nature of genome evolution. Annu Rev Microbiol. 2005;59:191–209. doi: 10.1146/annurev.micro.59.030804.121233. [DOI] [PubMed] [Google Scholar]
- 11.Dutilh BE, Huynen MA, Bruno WJ, Snel B. The consistent phylogenetic signal in genome trees revealed by reducing the impact of noise. J Mol Evol. 2004;58:527–539. doi: 10.1007/s00239-003-2575-6. [DOI] [PubMed] [Google Scholar]
- 12.Brown JR, Douady CJ, Italia MJ, Marshall WE, Stanhope MJ. Universal trees based on large combined protein sequence data sets. Nat Genet. 2001;28:281–285. doi: 10.1038/90129. [DOI] [PubMed] [Google Scholar]
- 13.Daubin V, Gouy M, Perriere G. A phylogenomic approach to bacterial phylogeny: Evidence of a core of genes sharing a common history. Genome Res. 2002;12:1080–1090. doi: 10.1101/gr.187002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren Q, Paulsen IT. Large-scale comparative genomic analyses of cytoplasmic membrane transport systems in prokaryotes. J Mol Microbiol Biotechnol. 2007;12:165–179. doi: 10.1159/000099639. [DOI] [PubMed] [Google Scholar]
- 15.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 16.Walter J, Schwab C, Loach DM, Ganzle MG, Tannock GW. Glucosyltransferase A (GtfA) and inulosucrase (Inu) of Lactobacillus reuteri TMW1106 contribute to cell aggregation, in vitro biofilm formation, and colonization of the mouse gastrointestinal tract. Microbiology. 2008;154:72–80. doi: 10.1099/mic.0.2007/010637-0. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 18.Xu J, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:1574–1586. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozupone C, Hamady M, Knight R. UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371–385. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coutinho PM, Henrissat B. Carbohydrate-active enzymes: An integrated database approach. In: Gilbert HJ, Davies G, Henrissat B, Svensson B, editors. Recent Advances in Carbohydrate Bioengineering. Cambridge: Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 23.Pupko T, Huchon D, Cao Y, Okada N, Hasegawa M. Combining multiple data sets in a likelihood analysis: Which models are the best? Mol Biol Evol. 2002;19:2294–2307. doi: 10.1093/oxfordjournals.molbev.a004053. [DOI] [PubMed] [Google Scholar]
- 24.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein J. Inferring Phylogenies. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 26.Penny D, Hendy MD. The use of tree comparison metrics. Syst Zool. 1985;34:75–82. [Google Scholar]
- 27.Larkin JM. Nonphotosynthetic, nonfruiting gliding bacteria. In: Staley JT, Bryant MP, Pfenning N, Holt JG, editors. Bergey's Manual of Systematic Bacteriology. Baltimore: Williams and Wilkins; 1989. pp. 2010–2138. [Google Scholar]
- 28.Stanier RY. Studies on non-fruiting myxobacteria I Cytophaga Johnsonae, N Sp, a chitin-decomposing myxobacterium. J Bacteriol. 1947;53:297–315. [PMC free article] [PubMed] [Google Scholar]
- 29.Warnick TA, Methe BA, Leschine SB. Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil. Int J Syst Evol Microbiol. 2002;52:1155–1160. doi: 10.1099/00207713-52-4-1155. [DOI] [PubMed] [Google Scholar]
- 30.Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:807–814. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knight R, et al. PyCogent: A toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.