Abstract
Although many stimuli activate extracellular signal-regulated kinases 1 and 2 (ERK1/2), the kinetics and compartmentalization of ERK1/2 signals are stimulus-dependent and dictate physiological consequences. ERKs can be inactivated by dual specificity phosphatases (DUSPs), notably the MAPK phosphatases (MKPs) and atypical DUSPs, that can both dephosphorylate and scaffold ERK1/2. Using a cell imaging model (based on knockdown of endogenous ERKs and add-back of wild-type or mutated ERK2-GFP reporters), we explored possible effects of DUSPs on responses to transient or sustained ERK2 activators (epidermal growth factor and phorbol 12,13-dibutyrate, respectively). For both stimuli, a D319N mutation (which impairs DUSP binding) increased ERK2 activity and reduced nuclear accumulation. These stimuli also increased mRNA levels for eight DUSPs. In a short inhibitory RNA screen, 12 of 16 DUSPs influenced ERK2 responses. These effects were evident among nuclear inducible MKP, cytoplasmic ERK MKP, JNK/p38 MKP, and atypical DUSP subtypes and, with the exception of the nuclear inducible MKPs, were paralleled by corresponding changes in Egr-1 luciferase activation. Simultaneous removal of all JNK/p38 MKPs or nuclear inducible MKPs revealed them as positive and negative regulators of ERK2 signaling, respectively. The effects of JNK/p38 MKP short inhibitory RNAs were not dependent on protein neosynthesis but were reversed in the presence of JNK and p38 kinase inhibitors, indicating DUSP-mediated cross-talk between MAPK pathways. Overall, our data reveal that a large number of DUSPs influence ERK2 signaling. Together with the known tissue-specific expression of DUSPs and the importance of ERK1/2 in cell regulation, our data support the potential value of DUSPs as targets for drug therapy.
The extracellular signal-regulated kinase 1/2 (ERK1/2)2 pathway forms a major part of the mitogen-activated protein kinase (MAPK) network and is activated by a diverse array of extracellular cues (1–3). Activated ERK1/2 can phosphorylate a growing list of substrate proteins in the nucleus and cytoplasm and represents a focal point of integration in cellular responses (1–3). The specificity of biological outcome from ERK1/2 stimuli is apparently achieved through tight control of the duration, magnitude, and localization of ERK1/2 signals (2, 3).
Activation of ERK1/2 commonly causes its translocation from the cytoplasm to the nucleus, which is necessary for the expression of many immediate early gene products such as c-Fos, c-Jun, and early growth response gene-1 product (Egr-1) (4–8). In fibroblasts and epithelial cells, sustained ERK1/2 activity causes expression and stabilization of immediate early gene products, culminating in G1/S transition (6–9). This does not occur in cells where nuclear localization of ERK1/2 is prevented (10). In contrast, transient ERK1/2 signals similarly cause the transcription of immediate early genes, but this is not sustained, and the protein products are rapidly degraded (6–8). Thus, both the signal duration and localization of ERK1/2 determine cell fate.
The dual specificity phosphatases (DUSPs) are an important family of proteins that influence spatiotemporal aspects of ERK1/2 signaling. Two major groups in this family are the MAPK phosphatases (MKPs) and the highly related atypical DUSPs (11, 12). The MKP group consists of 10 proteins that can remove activating Thr and Tyr phosphate groups from ERK1/2 and/or the other major MAPKs, c-Jun N-terminal kinase (JNK) and p38, and thus act in direct opposition to activating signals from upstream kinases (13, 14). The MKPs are characterized by their variable N-terminal MAPK binding region that governs substrate specificity and stability of interaction (14–18). This region includes the docking (D)-domain motif and can determine whether DUSPs remain associated with ERK1/2 following dephosphorylation (16–19). The MKPs are further divided into subgroups according to subcellular localization and substrate specificity. DUSP1, -2, -4, and -5 constitute the nuclear inducible MKPs, all of which are able to dephosphorylate ERK1/2 and, with the exception of DUSP5, can also dephosphorylate JNK and/or p38 (18, 20–22). DUSP6, -7, and -9 preferentially dephosphorylate ERK1/2, are not restricted to the nucleus, and are termed the cytoplasmic ERK MKPs (23–25). DUSP8, -10, and -16 have greater activity toward JNK and/or p38 and are known as the JNK/p38 MKPs (26–28). In contrast, the atypical DUSPs are smaller, lack obvious MAPK targeting motifs, and dephosphorylate a diverse group of substrates (11, 12). However, members of this group are known to act directly on MAPK family members. Notably, DUSP3 dephosphorylates ERK1/2 (29, 30), whereas DUSP18 and -22 dephosphorylate JNK or its upstream activators (31–33). A recent genome-wide phosphatase screen revealed that a number of DUSPs are essential for cell survival (34). These include DUSPs that can directly dephosphorylate ERKs, highlighting the potential value of DUSP targeting as a means to control ERK1/2 activity and cell fate (35–38).
We recently began to explore spatiotemporal aspects of ERK2 activation using a high content imaging-based model in which endogenous ERKs are knocked down with siRNAs and a GFP fusion protein reporter is added back with recombinant adenovirus (Ad) expressing wild-type (WT) ERK2-GFP or a similar construct mutated to prevent D-domain binding. We showed that ERK2 dephosphorylation and trafficking are coordinately regulated by DUSP1, -2, and -4 in a stimulus-specific manner and that whereas DUSP2 and -4 dephosphorylate and scaffold ERK2 in the nucleus, DUSP1 dephosphorylates ERK2 and releases it for return to the cytoplasm (19). Here we have used this model to explore possible effects of other DUSP family members on ERK responses to transient or sustained ERK activators (EGF and PDBu, respectively). We show that a surprisingly large number of these enzymes (12 of 16 in an siRNA screen) are able to shape ERK2 responses. These include members of each DUSP subgroup. The effects of the siRNAs were stimulus-specific and were mostly inhibitory. For most DUSPs, the reduction in ERK2 activity and/or nuclear localization was paralleled by decreases in ERK-dependent transcription, but this was not the case for the nuclear inducible MKPs. When all four members of the nuclear inducible MKP subgroup were knocked out simultaneously, levels of ERK2-GFP in the nucleus were decreased, whereas both active nuclear ERK2 levels and ERK-dependent transcription were greatly increased. In contrast, targeting of JNK/p38 MKPs reduced ERK2 activation, ERK2-GFP nuclear accumulation, and ERK-dependent transcription. These siRNA effects were prevented by pharmacological JNK or p38 kinase inhibitors, indicating the JNK/p38 MKP siRNA effects were mediated by JNK/p38 activation. These data indicate opposing collective functional roles of the nuclear inducible MKPs and JNK/p38 MKPs during ERK2 regulation.
EXPERIMENTAL PROCEDURES
Engineering of Plasmids and Viruses—Adenoviral (Ad) shuttle vectors of WT, Y261A, and D319N ERK2-GFP in pacAd5 CMV K-N pA were constructed as described (19). A 1.2-kb SalI fragment of the murine egr-1 promoter from an Egr-1-Luc vector (39) was subcloned into an XhoI digest of pAd5-Luc2 (made initially by subcloning an SspI-BamHI fragment of pGL4.17 into pacAd5 K-N pA). Viruses were generated from shuttle vectors as described (40). Briefly, 4.5 μg of shuttle vectors were digested alongside 1.5 μg of pacAd5 9.2–100 sub360 backbone vector (donated by Prof. Beverly Davidson, University of Iowa, Iowa City) with PacI. Cut shuttle and backbone vectors were then mixed and transfected into low passage HEK293 cells using Superfect (Qiagen, Crawley, UK). Cells were left to allow recombination between shuttle and backbone vectors. Verification of recombination was performed by restriction digest and sequence analysis, and Ad vectors were grown to high titer and purified according to standard protocols (41). The Ad CMV β-galactosidase vector was a gift from Prof. James Uney (University of Bristol, UK).
Cell Culture and Transfection—HeLa cells were cultured in 10% FCS-supplemented Dulbecco's modified Eagle's medium (DMEM). For 96-well plate experiments, cells were transfected with 1 nm nontargeting control siRNAs or siRNAs targeted to noncoding regions of ERK1/2 as described (19, 42). For DUSP siRNA transfection, 10 nm SMARTpool or nontargeting control siRNA mixtures (Dharmacon, Cramlington, UK) were included in transfections. Sixteen hours after siRNA transfection, cells were transduced with 2 × 106 plaque-forming units/ml Ad WT or D319N ERK2-GFP vector in DMEM with 10% FCS. For luciferase assays, Ad Egr-1-luciferase and Ad CMV β-galactosidase vectors were included at 1 × 106 plaque-forming units/ml. The Ad-containing medium was removed after 4–6 h and replaced with fresh DMEM supplemented with 0.1% FCS. The cells were then maintained for 16–24 h in culture prior to stimulation with EGF (Calbiochem) or PDBu (Sigma). In inhibitor studies, cells were pretreated for 30 min with 10 μm SP600125 (Ascent Scientific, Weston-super-Mare, UK), 20 μm SB203580 (Calbiochem), or 30 μm cycloheximide (Sigma). Expression levels of GFP-tagged fusions were compared by Western blotting (Ref. 9, see also supplemental Fig. 1) as well as comparison of mean cell fluorescence in microscopy assays (as demonstrated in Fig. 2C).
FIGURE 2.
Influence of D-domains on the potency of ERK2 signaling. Cells transfected with ERK1/2 siRNAs were transduced with Ad wild-type (WT) or D319N-mutated ERK2-GFP and analyzed for activation, localization, and transcriptional activation as follows. A–C, cells were stimulated in 96-well plates with the indicated concentrations of EGF for 5 min and stained before image acquisition and analysis (as described in Fig. 1) for the calculation of whole-cell ppERK2 intensity (A), the ERK2-GFP N:C ratio (B), and whole-cell ERK2-GFP (N+C) levels (C). D, cells were additionally transduced with Ad Egr-1 luciferase and Ad CMVβ-galactosidasevectorsbeforestimulationwithindicatedconcentrationsof EGF for 6 h and prior to lysis and luciferase assay (as described in Fig. 1) for the assessment of egr-1 promoter activity. Data shown were obtained from three separate experiments, each with duplicate wells (mean± S.E., n = 3). * = p < 0.05 and ** = p < 0.01, comparing WT with D319N conditions, according to two-way ANOVA and Bonferroni post hoc tests.
Western Blotting—HeLa cells were simultaneously plated and transfected in 6-well plates (2.5 × 105 cells/well) with 1 nm ERK1/2 siRNAs and 10 nm control or SMARTpool siRNAs prior to Ad transduction as above. Following treatment noted in figure legends, cells were lysed as described (19, 41), prior to Western blotting. Total and ppERK1/2 were detected using polyclonal rabbit anti-total ERK1/2 and rabbit anti-ppERK1/2 (1:1000; Cell Signaling Technology, Hitchin, UK), respectively. Loading controls were assayed by staining parallel blots with mouse anti-α-tubulin (Sigma).
Quantitative PCR—HeLa cells were simultaneously plated and transfected in 6-well plates (2.5 × 105 cells/well) with 1 nm ERK1/2 siRNAs and 10 nm control or SMARTpool siRNAs prior to Ad transduction as described above. Cells were either kept in 10% FCS DMEM or kept in reduced serum media prior to stimulation with 10 nm EGF or 1 μm PDBu. Extraction of total RNA was performed using an RNeasy kit according to the manufacturer's instructions (Qiagen). Contaminating genomic DNA was removed from columns using an additional DNase (Qiagen) digestion step. Complementary DNA was then prepared for 1 μg of each total RNA sample using a cloned avian myeloblastosis virus first-strand synthesis kit according to the manufacturer's instructions (Invitrogen). cDNAs were then quantified relative to expression of human GTPase-activating protein using the following primers: human GTPase-activating protein, 5′-GGG AAG GTG AAG GTC GGA GT-3′ and 5′-GAG TTA AAA GCA GCC CTG GTG A-3′; DUSP1, 5′-CAA CGA GGC CAT TGA CTT CAT AG-3′ and 5′-CAA ACA CCC TTC CTC CAG CA-3′; DUSP2, 5′-AAA ACC AGC CGC TCC GAC-3′ and 5′-CCA GGA ACA GGT AGG GCA AG-3′; DUSP3, 5′-GCG CTT ACT TTG AAA GGG CTG-3′ and 5′-TGC CGC ATC ATG AGG TAG G-3′; DUSP4, 5′-CTG GTT CAT GGA AGC CAT AGA GT-3′ and 5′-CGC CCA CGG CAG TCC-3′; DUSP5, 5′-CCG CGG GTC TAC TTC CTC A-3′ and 5′-GGG TTT TAC ATC CAC GCA ACA-3′; DUSP6, 5′-CTG CCG GGC GTT CTA CCT-3′ and 5′-CCA GCC AAG CAA TGT ACC AAG-3′; DUSP7, 5′-GTG CTC GGC CTG CTC CT-3′ and 5′-GAA GAG CTG TCC ACG TTG GTC-3′; DUSP8, 5′-GCA TCC TGC CTC ACC TCT ACC-3′ and 5′-CCA TTT TGC GTC ATC AGA TCC-3′; DUSP9, 5′-CTG CTG CAG AAG CTG CGA-3′ and 5′-CCT GGA ATC TGC TGA AGC CT-3′; DUSP10, 5′-GCC AGC CAC TGA CAG CAA C-3′ and 5′-TCC CAC ACT GGT GAG CTT CC-3′; DUSP11, 5′-AAG ACT ATC TCC CAG TTG GAC AGC-3′ and 5′-GGA AAA GCA TTC TTC TGG AGC A-3′; DUSP12, 5′-TGG AAT CTG CTT TGT TGG GAG-3′ and 5′-GAA GGA ACC CAA CTT GGC ACT-3′; DUSP13, 5′-CAC ACT GAA CCA TAT CGA TGA GG-3′ and 5′-AGC TGG ATC AGC TTG CTC TTG-3′; DUSP14, 5′-GAT CCG GAC CCA GGC AG-3′ and 5′-GGC GGC AAG ACC AGA GTG-3′; DUSP15, 5′-CTA TCC ATG AGT CAC CCC AGC-3′ and 5′-GTG CTT TTT GAT GGG TAC CTC AG-3′; DUSP16, 5′-TCA CTG TAC TTC TGG GTA AAC TGG AG-3′ and 5′-AAG GCT GAG AAA TGC AGG TAG G-3′; DUSP18, 5′-CTC TCC CGA AGA ACC TTG CC-3′ and 5′-GTC AGC AGT CAG CGA AGC AC-3′; DUSP19, 5′-TGC AGG ACC TTA GCT CGG AC-3′ and 5′-TGT ATC CAA ATC ATG AGC AGC ATC-3′; DUSP21, 5′-GTC CAG CAA TCG CAT CAC C-3′ and 5′-CCC TCG AAG AAT ACG TTG ACC A-3′; and DUSP22, 5′-CGC TAG CGT TCG CCT TCA-3′ and 5′-GCT CAA TTG TTC CGC GTC TC-3′. PCR primers were mixed with 50 ng of reverse transcription-PCR template and SYBR green PCR master mix (Applied Biosystems, Warrington, UK), and the comparative CT method was used to detect relative expression curves on an ABI PRISM 7500 detection system (Applied Biosystems).
Semi-automated Image Acquisition and Analysis—Cells were transfected with siRNA, transduced with Ad vectors, and plated as described above on Costar plain black-wall 96-well plates (Corning Glass). Following treatment with EGF or PDBu (and/or inhibitors), cells were washed in ice-cold phosphate-buffered saline before fixation and staining for ppERK1/2 and imaging as described (19). Image acquisition in each well was performed on an IN Cell Analyzer 1000 microscope, using a ×10 objective (GE Healthcare). Analysis of ppERK1/2 staining and localization was performed using the Dual Area Object Analysis algorithm in the IN Cell Analyzer work station (IN Cell Investigator, GE Healthcare) using DAPI and ppERK1/2 images. ERK2-GFP localization and ppERK2 staining were simultaneously analyzed using the Multitarget Analysis algorithm (IN Cell Investigator, GE Healthcare) using ERK2-GFP, ppERK2, and DAPI images (ERK2-GFP and DAPI images were used to define whole-cell and nuclear regions, respectively). Single cells expressing superphysiological levels of ERK2-GFP were excluded from analysis (∼20% of cells) using appropriate gating parameters to prevent misleading localization data (19). 300–500 cells per field were typically analyzed, and up to four fields per well were captured in experiments performed in duplicate or quadruplicate, meaning that in each experiment data were normally derived from at least 1000 individual cells per time point. Imaging data are reported as ppERK2 intensity (mean fluorescence intensity per cell) or as a ratio of nuclear to cytoplasmic intensity (N:C ratio) of either ERK2-GFP or ppERK2 signal.
Luciferase Assays—Cells were transfected with siRNA, transduced with Ad vectors, and plated as described above on Costar plain black-wall 96-well plates (Corning Glass), but including Ad Egr-1-luciferase and Ad CMV β-galactosidase reporter vectors. Following treatment with EGF or PDBu (and/or inhibitors), cells were washed in ice-cold phosphate-buffered saline, lysed, and assessed for luciferase activity by chemical luminescence following the addition of luciferin substrate (Promega, Southampton, UK). β-Galactosidase activity was used to correct luciferase activity for transduction efficiency, as measured following the addition of chlorophenol red β-d-galactopyranoside substrate (Roche Applied Science).
RESULTS AND DISCUSSION
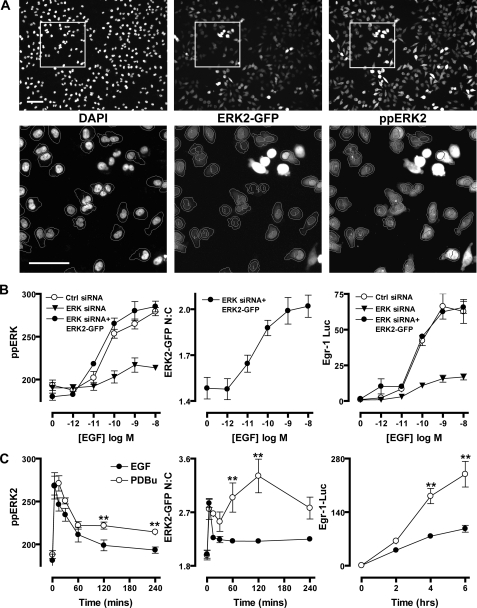
Comparison of EGF and Protein Kinase C-induced ERK2 Responses—To examine the spatial and temporal aspects of ERK2 regulation, we have used siRNAs targeted to noncoding regions to knock down endogenous ERK1/2 and recombinant Ad to restore ERK2 expression with fusion proteins of WT or the phosphatase-resistant D319N ERK2-GFP. When combined with antibody staining for dually phosphorylated, active ERK1/2 (ppERK1/2) and a DAPI stain to identify nuclear regions, this system allows direct monitoring of both total and phosphorylated forms of ERK2-GFP in nuclear and cytoplasmic compartments (Fig. 1A). The key readouts from this assay are as follows: 1) whole-cell ppERK2 intensity, which reflects ERK2 activation state irrespective of scaffolding or localization; 2) ppERK2 nuclear to cytoplasmic (N:C) ratio, which indicates changes in the compartmentalization and scaffolding of active ERK2; and 3) N:C distribution of ERK2-GFP, a readout for changes in ERK2 distribution irrespective of activation state. Combining these with an Egr-1 luciferase reporter assay (as a downstream readout for ERK1/2-dependent transcriptional activation), we initially determined the effects of ERK1/2 siRNA transfection and transduction with Ad ERK2-GFP on responses to EGF and the protein kinase C-activating phorbol ester, PDBu. In cells transfected with control siRNAs, 5 min of stimulation with EGF caused robust and dose-dependent increases of ppERK1/2 staining, and 6 h of stimulation caused induction of Egr-1 transcription. Potencies (log EC50 values) were identical for both end points (-10.3, see Fig. 1B, left and right panels). Transfection with ERK1/2 siRNAs had no effect on cell number or the expression of α-tubulin in microscopy or Western blotting assays (supplemental Fig. 1) (19), but ERK1/2 expression and maximal effect of EGF on ppERK1/2 and Egr-1 luciferase were reduced by >85% (Fig. 1B, left and right panels, and supplemental Fig. 1). Subsequent transduction with Ad ERK2-GFP restored ERK2 expression levels (as judged by Western blotting for ERK1/2, see supplemental Fig. 1). It also restored the whole-cell ppERK2 and Egr-1 luciferase responses, so that maximal responses to EGF and log EC50 values were indistinguishable between cells receiving control siRNAs and those receiving ERK1/2 siRNAs and Ad ERK2-GFP together. Monitoring of the ERK2-GFP nuclear cytoplasmic (N:C) ratio revealed that increases in ERK2-GFP nuclear distribution paralleled ppERK2 responses in the same cells (Fig. 1B, left and middle panels). Similar profiles were seen in parallel experiments with PDBu, but with a log EC50 of -6.3 (not shown).
FIGURE 1.
High content imaging methods for studying ERK1/2 regulation. A, cells were transfected with ERK1/2 siRNAs and transduced with Ad ERK2-GFP. Following treatment, cells were stained, and images were acquired for DAPI, ERK2-GFP, and ppERK2 stains, as described under “Experimental Procedures.” Top panels show whole fields of acquired images, and bottom panels show blow-up images of areas denoted by white squares. Outlines on lower panels denote the segmentation of individual cells according to DAPI and ERK2-GFP intensity using Multitarget Analysis software. Cells without outlines indicate cells excluded from analysis either for expressing superphysiological levels of ERK2-GFP or failing to meet other criteria needed for accurate segmentation. Bars, 100 μm. B, cells were transfected with control (Ctrl) siRNAs, ERK1/2 siRNAs, or ERK1/2 siRNAs as well as Ad ERK2-GFP as indicated. Cells were stimulated for 5 min with indicated concentrations of EGF before fixation, ppERK1/2 staining, image acquisition, and analysis as described in A to assess whole-cell levels of ERK1/2 phosphorylation (ppERK, left panel) and nucleocytoplasmic distribution of ERK2-GFP (ERK2-GFP N:C, middle panel). For Egr-1 luciferase assays, Ad Egr-1 luciferase and Ad CMV β-galactosidase vectors were also added to cells before stimulation with EGF for 6 h and assay of luciferase activity (Egr-1 Luc), as described under “Experimental Procedures,” and are expressed as fold change compared with unstimulated conditions. C, cells were transfected with ERK1/2 siRNAs and transduced with Ad ERK2-GFP (with or without Ad Egr-1 luciferase and Ad CMV β-galactosidase), prior to stimulation with 10 nm EGF or 1 μm PDBu, as indicated, in time course studies. Cells were fixed and assessed for ppERK2 levels (left panel) and ERK2-GFP N:C localization (middle panel) simultaneously, or lysed and assayed for luciferase activity (right panel) as described in B. Data shown are from four separate experiments (mean ± S.E., n = 4). ** = p < 0.01, comparing PDBu and EGF-stimulated cells using two-way ANOVA and Bonferroni post hoc tests.
We next used the knockdown and add-back system to define time courses of EGF and PDBu treatment. EGF caused a rapid and transient increase in whole-cell ppERK2 levels, which was paralleled by a transient relocalization of ERK2-GFP to the nucleus in the same cells (Fig. 1C, left and middle panels). EGF also mediated strong activation of the Egr-1 luciferase reporter, peaking at approximately a 100-fold induction over basal levels after 4–6 h of stimulation (Fig. 1C, right panel). PDBu caused a comparably rapid ppERK2 response to that of EGF (maximum at 5–15 min) with a subsequent reduction to ∼40% of peak values for the remainder of the experiment (Fig. 1C, left panel). PDBu also caused a similar rapid increase in N:C ERK2-GFP ratio to EGF, but rather than paralleling the activation profile, ERK2-GFP nuclear localization was sustained, peaking at 120 min (Fig. 1C, middle panel). This echoes previous findings that transient ERK2 activation does not mediate sustained nuclear accumulation of ERK2 (19, 43). In contrast, PDBu causes sustained ERK2 activation as well as sustained nuclear localization of dephosphorylated ERK2 (19) (Fig. 3C, Fig. 5, and Fig. 7B). The more sustained ppERK2 response to PDBu is associated with more pronounced activation of Egr-1 luciferase. These responses reached ∼240-fold induction after 6 h of stimulation, almost 2.5 times higher that that induced by EGF (Fig. 1C, right panel). Together, these data reveal that the knockdown, add-back, and imaging-based model recapitulates key features of ERK1/2 signaling seen with more conventional models (e.g. Western blotting (6–9)). These include relative potencies, duration of responses, and effects on nucleocytoplasmic distribution in cells stimulated with EGF and PDBu. The parallel effects on Egr-1-dependent transcription also demonstrate that the transient and sustained ERK1/2 activation modes are interpreted by the cell at the level of immediate early gene transcription (6–9).
FIGURE 3.
Enhancement of ERK2 signaling by D319N mutation of ERK2-GFP. A–C, cells transfected with ERK1/2 siRNAs were transduced with Ad wild-type (WT) or D319N-mutated ERK2-GFP as indicated prior to stimulation with 10 nm EGF (A) or 1 μm PDBu (B and C) for the times indicated. Cells were stained before image acquisition and analysis (as described in Fig. 1) for the calculation of whole-cell ppERK2 intensity (A and B, left panels), ppERK2 N:C ratio (A and B, middle-left panels), and the ERK2-GFP N:C ratio (A and B, middle-right panels). Cells were additionally transduced with Ad Egr-1 luciferase and Ad CMV β-galactosidase vectors before stimulation to assess Egr-1 induction by luciferase assay as described in Fig. 1 (A and B, right panels). Data shown were obtained from three separate experiments, each with duplicate wells (mean ± S.E., n = 3). * = p < 0.05 and ** = p < 0.01, comparing WT with D319N conditions, according to two-way ANOVA and Bonferroni post hoc tests. C, representative cropped images, collected under conditions described in A and B showing differences in ERK2-GFP distribution (top panels) and ppERK2 levels (bottom panels) following stimulation with 1 μm PDBu as indicated. Bar, 50 μm.
FIGURE 5.
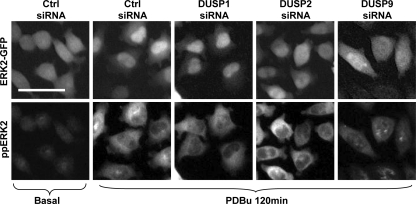
DUSP siRNAs affect spatiotemporal ERK2-GFP regulation. Cells were transfected in 96-well plates with 1 nm ERK1/2 siRNAs and 10 nm control (Ctrl) or siRNA SMARTpools targeting individual DUSPs (as indicated) before addition of Ad ERK2-GFP. Cells were stimulated with 1 μm PDBu as indicated, prior to staining and imaging (as described under Fig. 1). Representative cropped images are shown for each condition from ERK2-GFP (top panels) and ppERK2 images (bottom panels). Bar, 50 μm.
FIGURE 7.
Contribution of DUSP subfamilies to ERK2 regulation. A, cells were transfected with ERK1/2 siRNAs and either 40 nm control siRNAs (Ctrl si), 40 nm nuclear inducible MKP siRNAs (Nuc Ind MKP si, 10 nm each of DUSP1, -2, -4, and -5 siRNAs), 40 nm cytoplasmic ERK MKP siRNAs (Cyt ERK MKP si, 10 nm each of DUSP6, -7, -9 and ctrl siRNAs), or 40 nm JNK/p38 siRNAs (JNK/p38 MKP si, 10 nm each of DUSP10 and -16 siRNAs with 20 nm ctrl siRNAs) as indicated. Cells were transduced with Ad ERK2-GFP prior to stimulation with 1 μm PDBu for the times indicated. Cells were stained before image acquisition and analysis (as described in Fig. 1) for the calculation of whole-cell ppERK2 intensity (top panels), ppERK2 N:C ratio (middle panels), and the ERK2-GFP N:C ratio (lower panels). B, representative cropped images for control siRNA and nuclear inducible MKP siRNA conditions (as indicated) collected as described (A, left panels) and showing differences in ERK2-GFP (top panels) and ppERK2 (bottom panels) distribution following stimulation with 1 μm PDBu as indicated. Bar, 50 μm. C, cells treated as described in A were additionally transduced with Ad Egr-1 luciferase and Ad CMV β-galactosidase vectors before stimulation with PDBu for times indicated to assess Egr-1 induction by luciferase assay (as described in Fig. 1). Data shown in A and C were obtained from four separate experiments, each with duplicate wells (mean± S.E., n = 4). * = p < 0.05 and **= p < 0.01, comparing control siRNA to test conditions, according to two-way ANOVA and Bonferroni post hoc tests.
Effects of D-domain Interference on ERK2 Activation, Trafficking, and Egr-1 Transcription—The D319N mutation within the common docking domain of ERK2 is analogous to the sevenmaker gain-of-function mutation in Drosophila (44), which perturbs the ability of ERK2 to associate with D-domain binding partners without affecting intrinsic kinase activity (15, 42). These partners include all MKPs (1, 14, 16), making the D319N variant a useful tool to study the influence of DUSP interaction on ERK2 responses. We first assessed the dose dependence of EGF and PDBu effects under conditions of ERK1/2 knockdown and reconstitution with WT or D319N ERK2-GFP. We found that D319N mutation did not affect the potency (log EC50 values) of EGF or PDBu effects on ppERK2, ERK2-GFP N:C ratio, and Egr-1 luciferase activity (Fig. 2, A, B, and D, and data not shown). However, maximal ERK2-GFP N:C and Egr-1 luciferase responses were significantly altered. As shown (Fig. 2B), EGF-mediated changes in ERK2-GFP N:C ratio at 5 min of stimulation were reduced by up to 50% by D319N mutation, whereas whole-cell ERK2-GFP intensity (N+C) values remained unchanged (indicating comparable whole-cell expression levels, Fig. 2C), but Egr-1 luciferase responses were increased ∼2.5-fold (Fig. 2D). In contrast, inhibition of D-domain interactions had no effect on 5-min ppERK2 responses to EGF (Fig. 2A). Similar trends were observed in cells stimulated with PDBu (not shown).
We characterized these differences further in time course studies using maximally stimulating concentrations of EGF or PDBu. Early peak levels of ppERK2 responses were unaltered by D319N mutation, but responses to both stimuli were greatly increased at later time points (Fig. 3, A and B, left panels, and C). Comparing the distribution of the ppERK2 signal intensity in the nucleus and cytoplasm (ppERK2 N:C) revealed that prolongation of the whole-cell ppERK2 signal by D319N mutation is associated with an increased proportion of ppERK2 in the nucleus (Fig. 3, A and B, middle left panels, and C). In contrast, the ERK2-GFP N:C ratios from the same cells show that D319N actually reduces the total amount of ERK2-GFP in the nucleus (Fig. 3, A and B, middle right panels, and C). The D319N mutation also increased effects of both stimuli on Egr-1 transcription, almost doubling responses at 4–6 h (Fig. 3, A and B, right panels). Taken together, these data show that D319N-mediated inhibition of phosphatase binding does not increase the sensitivity of ERK2-GFP to low concentrations of stimuli (see also Refs. 15, 19) but increases ERK2-dependent transcriptional responses by prolonging its activation and increasing the proportion of active ERK2 in the nucleus. The corresponding decrease in ERK2-GFP N:C ratio is consistent with the fact that, in these cells, high levels of nuclear accumulation are because of D-domain-dependent scaffolding and signal termination by nuclear inducible MKPs (19).
Regulation of DUSP Transcription and ERK2 Responses—Because D319N mutations inhibit ERK2 association with MKP family DUSPs (14, 16, 19), the data above are indicative of a major role for them in shaping ERK2 responses. Many ERK1/2 stimuli induce the transcription of nuclear inducible MKPs to act in negative feedback loops, but the involvement of other DUSPs has not been extensively explored in this context. We first assessed expression and knockdown of DUSP1–16, -18, -19, -21, and -22 using qPCR and siRNAs. Of these 20 transcripts, we found that DUSP13, -15, and -21 mRNAs were not detectable in these cells and that DUSP8 mRNA could not be reduced by more than 30% following siRNA transfection (not shown), so these enzymes were excluded from further analysis. The qPCR revealed that a large number of the remaining DUSP transcripts (9 of 16) were increased by EGF and/or PDBu (Fig. 4A). PDBu had a greater effect than EGF on both the number of transcripts regulated (six compared with three) and, in some cases, the magnitude of transcription (Fig. 4A). Thus, although transcriptional regulation appeared characteristic of the nuclear inducible MKPs, it also occurred among the cytoplasmic ERK MKPs (DUSP6), the JNK/p38 MKPs (DUSP10), and the atypical DUSPs (DUSP14) (Fig. 4A). ERK2 activity and signal duration clearly plays a major role in these transcriptional responses, as evidenced by the sensitivity to ERK1/2 knock-down (inhibiting effects on DUSP1 and -2 transcription) and D319N ERK2-GFP expression (enhancing transcription of DUSP2, -5, and -6) (Fig. 4A).
FIGURE 4.
Stimulus and ERK dependence of DUSP transcription and effects of DUSP siRNAs on ERK2 signaling. A, cells were transfected in 6-well plates with control siRNAs (Ctrl) or ERK1/2 siRNAs and transduced with Ad wild-type (WT) or D319N-mutated ERK2-GFP as indicated. Cells were either left unstimulated (Basal) or treated with 10 nm EGF (left panel) or 1 μm PDBu (right panel) for 120 min. Total RNA isolates were analyzed for relative levels of DUSP2 mRNA using qPCR protocols described under “Experimental Procedures.” Data shown are average values from three independent experiments represented as fold change from basal levels and presented as a heat map. DUSPs are grouped according to sequence similarity and substrate specificity, and data included are values found to differ significantly from control (basal) conditions using one-way ANOVA and Dunnett's post hoc test, accepting p < 0.05 as significant. B, cells were transfected in 96-well plates with 1 nm ERK1/2 siRNAs and 10 nm control (Ctrl) or siRNA SMARTpools targeting individual DUSPs (as indicated) before addition of Ad ERK2-GFP. Cells were stimulated with 1 μm PDBu or 10 nm EGF as indicated, prior to staining and imaging (as described in Fig. 1). Data are expressed in the heat map as the extent of difference above or below control values for each condition and time point for ppERK2 intensity (left panel) and ERK2-GFP N:C ratio (right panel) from four separate experiments performed in duplicate. Targets are again grouped according to sequence similarity and substrate specificity. Statistical analysis was performed using one-way ANOVA and Dunnett's post hoc test, accepting p < 0.05 as significant. Nonsignificant changes are shown as white blocks for both experiments.
To investigate possible roles of these DUSPs, we reduced the mRNA levels of each DUSP through siRNA targeting. Assessment of mRNA levels 48 h after siRNA transfection revealed at least 70% reduction in mRNA levels for each of the 16 genes tested (supplemental Table 1). These DUSP knockdown conditions were used in conjunction with ERK1/2 siRNAs and Ad WT ERK2-GFP to assess DUSP regulation of ERK2. None of the siRNAs significantly altered whole-cell ERK2-GFP expression levels, cell number, or the proportion of apoptotic cells (supplemental Table 1) arguing against nonspecific cytotoxic effects. Forty eight hours after transfection, cells were either left untreated (basal) or stimulated with EGF (5 and 120 min) or PDBu (15 and 120 min) prior to assessment of ppERK2 staining and ERK2-GFP localization. As shown (Fig. 4B), we found 12 phosphatases that had a significant effect on PDBu and/or EGF-stimulated ERK2 phosphorylation and/or compartmentalization (Fig. 4B). None of the siRNAs had a measurable effect on whole-cell ppERK2 levels in unstimulated cells (Fig. 4B, left panel), but siRNAs to DUSP3, -5, and -10 significantly reduced the ERK2-GFP N:C ratio, and DUSP2 and -7 siRNAs increased ERK2-GFP nuclear localization under basal conditions (Fig. 4B, right panel). siRNAs to DUSP3, -9, -10, and -16 all reduced ppERK2 and/or ERK2-GFP responses to both EGF and PDBu (Fig. 4B). No siRNAs increased ppERK2 responses to EGF or PDBu, but siRNAs to DUSP5, -7, -12, -14, and -18 specifically inhibited the PDBu-mediated ppERK2 response (particularly at later time points) without altering the EGF response at either time point (Fig. 4B). As expected (19), siRNAs to DUSP2 and -4 reduced the ERK2-GFP N:C ratio in cells stimulated with PDBu without measurably altering the ppERK2 response nor any aspect of the EGF response (Fig. 4B). DUSP1 knockdown was the sole condition that increased the ERK2-GFP N:C ratio in stimulated cells. DUSP3 and -9 siRNAs were the only treatments to significantly reduce the effect of both EGF and PDBu on ppERK2 levels and ERK2-GFP N:C ratio (Fig. 4B). The striking findings from these experiments are as follows. First, many DUSPs from both the MKP and atypical groups play specific roles in shaping ERK2 responses, with little overlap or redundancy of function (illustrated in representative images in Fig. 5). Second, the unifying theme of action across each subgroup of DUSPs was that the knockdowns inhibited ligand effects on ppERK2 and/or ERK2-GFP N:C ratio. However, the nuclear inducible MKPs stood out as the only group that had pronounced stimulus-specific effects on ERK2-GFP N:C ratio without an overall effect on phosphorylation state.
Effects of DUSP Knockdown on Stimulus-specific Induction of Egr-1—As most DUSP knockdowns reduced ppERK2 responses and/or ERK2-GFP N:C ratio, we focused on a selection of DUSPs showing a range of effects on ERK2-GFP activation and trafficking to see if they had corresponding effects on Egr-1 transcription. We compared EGF and PDBu-mediated Egr-1 induction in the presence and absence of siRNAs to DUSP1–3, -9, -10, and -16 (Fig. 6). Knockdown of DUSP3, -9, -10, and -16 reduced PDBu effects on Egr-1 luciferase, paralleling their ability to reduce the PDBu effect on ppERK2 and/or ERK2-GFP N:C response. Similar effects were seen in EGF-stimulated cells, suggesting that the effects of these DUSPs were not stimulus-specific (Fig. 6). For each of these DUSPs, effects of knockdown on ERK2 activity were predictive of functional outcome: a reduction in ppERK2 response led to a reduction in transcriptional activation. In contrast, knockdown of the nuclear inducible MKPs, DUSP1 and -2, did not affect ppERK2 responses but increased and decreased ERK2-GFP N:C localization, respectively (Fig. 6). They also had stimulus-specific effects on Egr-1 transcription. DUSP2 knockdown caused a 40% increase in PDBu-stimulated Egr-1 luciferase (without altering the EGF-mediated response), and DUSP1 knockdown had no effect on the levels of Egr-1 luciferase activity under any condition (Fig. 6).
FIGURE 6.
Comparison of ERK2-GFP activation, localization, and regulation of Egr-1 following DUSP knockdown. Cells were transfected with 1 nm ERK1/2 siRNAs and 10 nm control (Ctrl) or siRNA SMARTpools targeting individual DUSPs (as indicated) before addition of Ad ERK2-GFP. Top and middle panels, cells were either left unstimulated (basal) or treated with 1 μm PDBu or 10 nm EGF for 120 min as indicated, prior to staining and imaging (as described in Fig. 1). Bottom panel, cells were additionally transduced with Ad Egr-1 luciferase and Ad CMV β-galactosidase vectors before stimulation with 1 μm PDBu or 10 nm EGF for 6 h as indicated to assess Egr-1 induction by luciferase assay (as described in Fig. 1). Data are expressed as average ppERK2 values (top panel), ERK2-GFP N:C ratio (middle panel), and fold change in Egr-1 luciferase activity compared with unstimulated control siRNA-transfected cells (bottom panel) and were obtained from three separate experiments, each with duplicate observations (mean ± S.E., n = 3). * = p < 0.05 and ** = p < 0.01, comparing DUSP and control siRNA conditions for each stimulus, according to one-way ANOVA and Dunnett's post hoc test.
Functional Profiling of DUSP Family Effects on ERK Signaling—The data outlined above reveal the stimulus-specific shaping of ERK2 responses by members of each DUSP class. They also demonstrate the importance of assaying localization as well as phosphorylation state when screening for changes in ERK2 regulation. This appears to be especially pertinent when dealing with the nuclear inducible MKPs, presumably because they can both dephosphorylate and scaffold ERK2 (18, 19) and because knockdown of one can lead to compensatory changes in another. For example, we have found that knockdown of DUSP1 increases PDBu-stimulated DUSP2 expression, just as knockdown of DUSP2 increases PDBu-stimulated DUSP1 expression (19). In light of such compensation we found that function of these proteins could best be revealed by simultaneous knockdown of multiple nuclear inducible DUSPs (19). By extending this we have combined siRNAs to knock down all nuclear inducible MKPs (DUSP1, -2, -4, and -5), all cytoplasmic ERK MKPs (DUSP6, -7, and -9), or both JNK/p38 MKPs (DUSP10 and -16) before determining PDBu effects. As shown (Fig. 7, A and C, left panels, and representative images in B), knockdown of the nuclear inducible MKPs did not alter whole-cell ppERK2 responses but caused a marked prolongation of PDBu effects on the ppERK2 N:C ratio while reducing its effects on the ERK2-GFP N:C ratio and greatly enhancing its effect on transcriptional activation of Egr-1. These data demonstrate the importance of the nuclear inducible MKPs (collectively) as inhibitors of sustained ERK signaling by virtue of their ability to inactivate and scaffold ERK within the nucleus. In contrast, siRNAs targeting the cytoplasmic ERK MKPs did not alter PDBu effects on whole-cell ppERK2 responses or on the ppERK2 N:C ratio and had only a modest inhibitory effect on ERK2 distribution (reducing the PDBu effect on ERK2-GFP N:C ratio at 240 min only). They also failed to alter PDBu-stimulated Egr-1 luciferase activity, arguing against a major role for cytoplasmic ERK MKPs in shaping of sustained ERK signaling in this model. Knockdown of the JNK/p38 MKPs also failed to alter PDBu effects on ppERK2 distribution but it did reduce the PDBu effect on the whole-cell ppERK2 levels and ERK2-GFP N:C ratio (significant reductions of both at 60–120 min) and also inhibited the PDBu effect on Egr-1 luciferase activity. Thus, in this model the JNK/p38 MKPs act (together) as positive regulators of ERK2 signaling, supporting Egr-1 luciferase activity by enhancing ERK2 phosphorylation. This is in sharp contrast to the nuclear inducible MKPs that act (collectively) as negative regulators of ERK2 signaling, primarily by reducing the proportion of active ERK2 within the nucleus (Fig. 7).
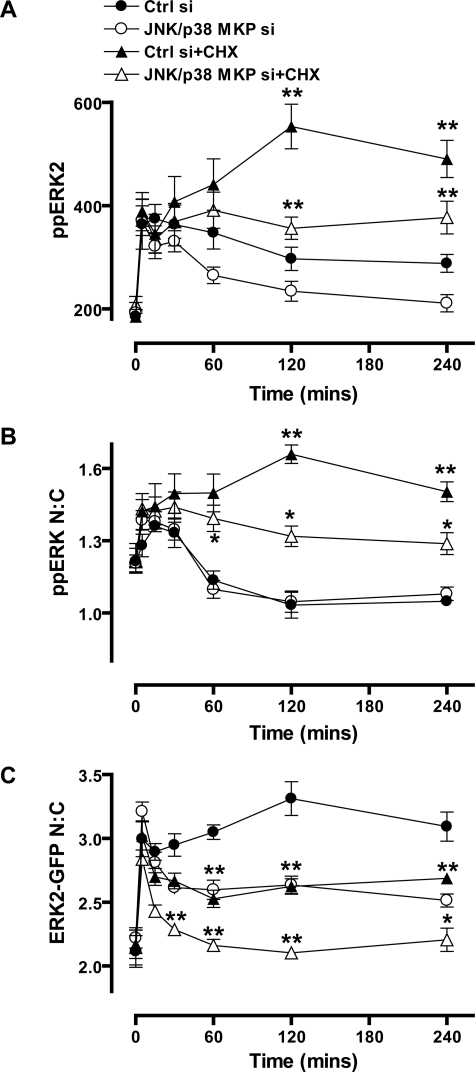
Relevance of Protein Neosynthesis and JNK or p38 MAPKs for JNK/p38 MKP Effects on ERK2 Signaling—We next explored mechanisms of JNK/p38 MKP action, focusing first on relevance of protein neosynthesis. We have previously shown that the sustained effects of PDBu on whole-cell ppERK2 levels and ERK2-GFP N:C ratio and its more transient effect on ppERK2 N:C ratio are all increased and/or prolonged by the protein synthesis inhibitor, CHX. In contrast, when EGF is used to elicit transient ERK activation, CHX has little or no effect (19) (supplemental Fig. 2). These effects are thought to reflect prevention of nuclear inducible MKP neosynthesis as evidenced by the close parallels between effects of CHX and knockdown of nuclear inducible MKPs as well as with effects of the D319N mutation that inhibits ERK2 binding to nuclear inducible MKPs (Fig. 3). To determine the relevance of protein synthesis for JNK/p38 effects, we repeated the JNK/p38 siRNA experiments (shown in Fig. 7) in the presence and absence of CHX. As shown (Fig. 8) CHX caused the expected prolongation of PDBu effects on ppERK2 levels and nuclear localization of ppERK2, whereas inhibiting its effects on ERK2-GFP N:C localization. JNK/p38 siRNAs reduced PDBu effects on whole-cell ppERK2 levels and on ERK2-GFP distribution but did not alter its effects on ppERK2 N:C ratio (Fig. 8). JNK/p38 siRNAs also caused a pronounced inhibition of PDBu effects on each of these imaging end points in the presence of CHX (Fig. 8). The fact that effects of JNK/p38 knockdown are maintained or increased by CHX argues that shaping of ERK2 responses by JNK/p38 MKPs is not dependent upon protein neosynthesis despite the fact that PDBu increased DUSP10 transcription (Fig. 4A) This is in direct contrast to the nuclear inducible MKPs, for which neosynthesis is clearly necessary (13, 14, 18, 19, 43, 45).
FIGURE 8.
Effects of protein synthesis inhibition on JNK/p38 MKP regulation of ERK2. A–C, cells were transfected with 1 nm ERK1/2 siRNAs and either 40 nm control siRNAs (Ctrl si) or 40 nm JNK/p38 siRNAs (JNK/p38 MKP si, 10 nm each of DUSP10 and -16 siRNAs with 20 nm ctrl siRNAs) as indicated. Cells were transduced with Ad ERK2-GFP prior to treatment with 30 μm cycloheximide (CHX) as indicated for 30 min prior to stimulation with 1 μm PDBu for the times indicated. Cells were stained before image acquisition and analysis (as described in Fig. 1) for the calculation of whole-cell ppERK2 intensity (A), ppERK2 N:C ratio (B), and the ERK2-GFP N:C ratio (C). Data shown in all panels were obtained from four separate experiments, each with duplicate wells (mean ± S.E., n = 4). * = p < 0.05 and ** = p < 0.01, comparing control and JNK/p38 siRNA conditions to those with CHX, according to two-way ANOVA and Bonferroni post hoc tests.
Changes in JNK/p38 activity can have profound effects on the ERK MAPK pathway (46), and a recent study demonstrated that impairment of ERK1/2 activation in DUSP2-/- mice was because of increased JNK activity (45). The observation that JNK/p38 MKPs influence ERK2 signaling in this model implies that PDBu effects on ERK2 may be influenced by cross-talk from concomitantly activated p38 and JNK MAPKs. To address this possibility we determined effects of pharmacological inhibition of JNK (using SP600125) and p38 (using SB203580) on ERK2 signaling and also determined the effects of the JNK/p38 MKP siRNAs in the presence of these inhibitors. We found that when used alone, these inhibitors did not measurably influence responses to PDBu (Fig. 9). As expected, knockdown of the JNK/p38 MKPs inhibited PDBu effects on whole-cell ppERK2 levels, ERK2-GFP N:C ratio, and Egr-1 luciferase responses without altering its effects on ppERK2 N:C ratios (Fig. 7 and Fig. 9). However, the knockdown effects on Egr-1 activation and ERK2-GFP nuclear localization were reversed by pharmacological inhibition of JNK or p38 activity; Fig. 9 shows data for 120 min of stimulation with PDBu, and similar effects were seen with 30 or 60 min of stimulation (not shown). Thus, the observed effects of JNK/p38 MKP siRNAs are, at least partially, dependent upon activation of JNK and/or p38 MAPKs. These data therefore identify the JNK/p38 MAPKs as modulators of ERK2 signaling and as the pertinent targets for JNK/p38 MKPs in this respect.
FIGURE 9.
Effects of JNK and p38 kinase inhibition on JNK/p38 MKP regulation of ERK2. Cells were transfected with 1 nm ERK1/2 siRNAs and either 40 nm control siRNAs (Ctrl si) or 40 nm JNK/p38 siRNAs (JNK/p38 MKP si, 10 nm each of DUSP10 and -16 siRNAs with 20 nm ctrl siRNAs) as indicated. Cells were transduced with Ad ERK2-GFP prior to treatment with 10 μm SP600125 (SP) or 20 μm SB203580 (SB) as indicated for 30 min prior to stimulation with 1 μm PDBu for 120 min. Cells were stained before image acquisition and analysis (as described in Fig. 1) for the calculation of whole-cell ppERK2 intensity (middle panel) and the ERK2-GFP N:C ratio (lower panel). Cells in the upper panel were treated as described above but were additionally transduced with Ad Egr-1 luciferase and Ad CMV β-galactosidase vectors before stimulation with 1 μm PDBu for 6 h prior to assessment of Egr-1 induction by luciferase assay (as described in Fig. 1). Data shown were obtained from three separate experiments, each with triplicate wells (mean ± S.E., n = 3). * = p < 0.05 and ** = p < 0.01, comparing JNK/p38 siRNA conditions with and without SP600125 (SP) or SB203580 (SB), according to one-way ANOVA and Dunnett's post hoc tests.
Summary—Multiple DUSPs clearly contribute to the regulation of ERK1/2, and at least 12 DUSPs can dephosphorylate ERK1/2 in vitro (13, 14). However, the prediction of DUSP regulation of ERK1/2 in cells is complicated by a number of factors. First, they can often dephosphorylate multiple MAPK substrates (13, 14). Second, their transcription can be rapidly regulated in a stimulus-specific manner. Third, their expression is highly cell type-specific (Fig. 4A) (13, 14, 19, 45). These properties make them exciting therapeutic targets, and a number of DUSP inhibitors have been discovered in recent years (35–38). It is important to recognize, however, that DUSPs may influence many interconnecting signaling and feedback pathways so their loss or inhibition can have positive or negative effects on ERK1/2 signaling (13, 14, 19, 45). This underlines the need to explore DUSP function within intact systems, but we know of no previous work in which mammalian DUSPs have been systematically knocked down to assess their influence on ERK1/2 signaling. Here, we have done so and have determined the effects of siRNA-mediated knockdown of individual DUSPs on multiple aspects of ERK2 regulation. Our data reveal that a surprisingly large number of DUSPs influence ERK2 signaling in this model. These include members of the nuclear inducible MKPs (DUSP1, -2, -4, and -5). They also include MKPs of the cytoplasmic ERK1/2-selective group (DUSP7 and -9), the JNK/p38-selective group (DUSP10 and -16), as well as four of the atypical DUSPs.
Although a screen of this nature cannot reveal mechanisms, it did reveal a remarkable degree of specificity. The functional profiles of DUSP knockdowns were almost all distinct when the stimulation conditions (stimulus and time) and end points (ppERK2 levels, ERK2-GFP N:C ratio, and Egr-1 transcription) were taken into consideration (Figs. 4, 5, 6). The other obvious finding is that the effects of DUSP knockdown (with the exception of the nuclear inducible MKPs) were all negative; DUSP knockdown caused a decrease in ppERK2 response and a corresponding decrease in Egr-1 transcription (Fig. 6). This is in stark contrast to the experiments we performed expressing a D319N variant of ERK2-GFP to inhibit interactions with phosphatases, which had a pronounced enhancing effect on ERK2 activity (Fig. 2 and Fig. 3). In this comparison it is important to note that this mutation will only affect the binding of phosphatases that contain D-domains, and will only affect association with ERK2, whereas the catalytic activity of the DUSPs and their effect on other MAPKs will be left intact. In contrast, the siRNA knockdowns represent the impact of selective phosphatase inhibition. These data may therefore prove useful in the design and use of isotype-specific DUSP inhibitors and inhibitors that reduce DUSP-ERK1/2 interaction, as the outcomes can clearly be very different. This study also sounds a cautionary note in screening assays for ERK1/2 regulation. By monitoring two end points (ppERK2 and ERK2-GFP N:C) with two stimuli (EGF and PDBu) and two time points (for each stimulus), we have found that 12 of the 16 DUSPs tested influence ERK2 signaling. Had we performed a single end point assay screening for effects on ERK2-GFP N:C ratio after 15 min of stimulation with PDBu, we would have obtained only one hit, and if we had just screened for effects on the acute ERK2-GFP N:C response to EGF, we would have obtained none.
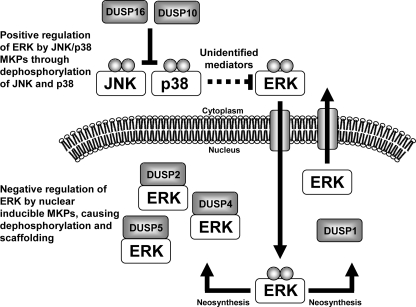
Following up the screen with more mechanistic studies, we have found that the nuclear inducible MKPs act (collectively) as negative regulators of ERK2 signaling, whereas the JNK/p38 MKPs act (together) as positive regulators (summarized in Fig. 10). Interestingly, they do so by distinct mechanisms as the nuclear inducible MKPs inhibit ERK-dependent transcription by reducing the proportion of ppERK2 in the nucleus (but without altering the whole-cell level of active kinase), whereas the JNK/p38 MKPs support ERK-dependent transcription by increasing whole-cell ppERK2 levels. Moreover, the JNK/p38 MKPs appear to shape PDBu-mediated ERK2 signaling by acting upon JNK and p38 MAPKs.
FIGURE 10.
Model of ERK2 regulation by nuclear inducible and JNK/p38 MKPs. ERK2 activation and translocation to the nucleus causes neosynthesis of the nuclear inducible MKPs (DUSP1, -2, -4, and -5) that collectively mediate both dephosphorylation and scaffolding of ERK2 in the nucleus. Previous studies have revealed that all nuclear inducible MKPs can contribute to ERK2 dephosphorylation, but only DUSP2, -4, and -5 stably associate with ERK2, whereas DUSP1 inactivates and releases ERK2 for reactivation in the cytosol (18, 19). This presumably facilitates sustained ERK2 signals in the face of persistent upstream stimuli. In contrast, we find that the JNK/p38 MKPs (DUSP10 and -16) have a positive role in ERK2 regulation by inactivating JNK and p38 kinases. The proteins that mediate negative cross-talk between the JNK, p38, and ERK2 pathways have not been identified in this system but have been defined in others (46).
Together, our findings support the notion that DUSPs represent nonredundant regulators of ERK2 signal regulation in response to external stimuli. The degree of specificity revealed in this study support the use of multiple end point microscopy-based assays to resolve effects of DUSP regulation of ERK1/2. Most importantly, the specificity revealed in this investigation may well prove important in the exploitation of DUSPs as therapeutic targets.
Supplementary Material
This work was supported by Wellcome Trust Project and Equipment Grants 062918, 076557, and 078407. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; EGF, epidermal growth factor; MKP, MAPK phosphatase; DUSP, dual specificity phosphatase; JNK, c-Jun N-terminal kinase; siRNA, short inhibitory RNA; GFP, (enhanced) green fluorescent protein; PDBu, phorbol 12,13-dibutyrate; CHX, cycloheximide; Ad, adenovirus; qPCR, quantitative PCR; DAPI, 4′,6-diamidino-2-phenylindole; N:C, nuclear:cytoplasmic; N+C, nuclear+cytoplasmic; ANOVA, analysis of variance; WT, wild type; DMEM, Dulbecco's modified Eagle's medium; FCS, fetal calf serum; D-domain, docking domain; CMV, cytomegalovirus.
References
- 1.Caunt, C. J., Finch, A. R., Sedgley, K. R., and McArdle, C. A. (2006) Trends Endocrinol. Metab. 17 276-283 [DOI] [PubMed] [Google Scholar]
- 2.Ebisuya, M., Kondoh, K., and Nishida, E. (2005) J. Cell Sci. 118 2997-3002 [DOI] [PubMed] [Google Scholar]
- 3.Murphy, L. O., and Blenis, J. (2006) Trends Biochem. Sci. 31 268-275 [DOI] [PubMed] [Google Scholar]
- 4.Chen, R. H., Sarnecki, C., and Blenis, J. (1992) Mol. Cell. Biol. 12 915-927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenormand, P., Sardet, C., Pages, G., L'Allemain, G., Brunet, A., and Pouyssegur, J. (1993) J. Cell Biol. 122 1079-1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook, S. J., Aziz, N., and McMahon, M. (1999) Mol. Cell. Biol. 19 330-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy, L. O., Smith, S., Chen, R. H., Fingar, D. C., and Blenis, J. (2002) Nat. Cell Biol. 4 556-564 [DOI] [PubMed] [Google Scholar]
- 8.Murphy, L. O., MacKeigan, J. P., and Blenis, J. (2004) Mol. Cell. Biol. 24 144-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto, T., Ebisuya, M., Ashida, F., Okamoto, K., Yonehara, S., and Nishida, E. (2006) Curr. Biol. 16 1171-1182 [DOI] [PubMed] [Google Scholar]
- 10.Brunet, A., Roux, D., Lenormand, P., Dowd, S., Keyse, S., and Pouyssegur, J. (1999) EMBO J. 18 664-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alonso, A., Sasin, J., Bottini, N., Friedberg, I., Friedberg, I., Osterman, A., Godzik, A., Hunter, T., Dixon, J., and Mustelin, T. (2004) Cell 117 699-711 [DOI] [PubMed] [Google Scholar]
- 12.Tonks, N. K. (2006) Nat. Rev. Mol. Cell Biol. 7 833-846 [DOI] [PubMed] [Google Scholar]
- 13.Jeffrey, K. L., Camps, M., Rommel, C., and Mackay, C. R. (2007) Nat. Rev. Drug Discov. 6 391-403 [DOI] [PubMed] [Google Scholar]
- 14.Owens, D. M., and Keyse, S. M. (2007) Oncogene 26 3203-3213 [DOI] [PubMed] [Google Scholar]
- 15.Bott, C. M., Thorneycroft, S. G., and Marshall, C. J. (1994) FEBS Lett. 352 201-205 [DOI] [PubMed] [Google Scholar]
- 16.Tanoue, T., Adachi, M., Moriguchi, T., and Nishida, E. (2000) Nat. Cell Biol. 2 110-116 [DOI] [PubMed] [Google Scholar]
- 17.Tanoue, T., Yamamoto, T., and Nishida, E. (2002) J. Biol. Chem. 277 22942-22949 [DOI] [PubMed] [Google Scholar]
- 18.Mandl, M., Slack, D. N., and Keyse, S. M. (2005) Mol. Cell. Biol. 25 1830-1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caunt, C. J., Rivers, C. A., Conway-Campbell, B. L., Norman, M. R., and McArdle, C. A. (2008) J. Biol. Chem. 283 6241-6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alessi, D. R., Smythe, C., and Keyse, S. M. (1993) Oncogene 8 2015-2020 [PubMed] [Google Scholar]
- 21.Rohan, P. J., Davis, P., Moskaluk, C. A., Kearns, M., Krutzsch, H., Siebenlist, U., and Kelly, K. (1993) Science 259 1763-1766 [DOI] [PubMed] [Google Scholar]
- 22.Chu, Y., Solski, P. A., Khosravi-Far, R., Der, C. J., and Kelly, K. (1996) J. Biol. Chem. 271 6497-6501 [DOI] [PubMed] [Google Scholar]
- 23.Groom, L. A., Sneddon, A. A., Alessi, D. R., Dowd, S., and Keyse, S. M. (1996) EMBO J. 15 3621-3632 [PMC free article] [PubMed] [Google Scholar]
- 24.Muda, M., Boschert, U., Smith, A., Antonsson, B., Gillieron, C., Chabert, C., Camps, M., Martinou, I., Ashworth, A., and Arkinstall, S. (1997) J. Biol. Chem. 272 5141-5151 [DOI] [PubMed] [Google Scholar]
- 25.Dowd, S., Sneddon, A. A., and Keyse, S. M. (1998) J. Cell Sci. 111 3389-3399 [DOI] [PubMed] [Google Scholar]
- 26.Muda, M., Theodosiou, A., Rodrigues, N., Boschert, U., Camps, M., Gillieron, C., Davies, K., Ashworth, A., and Arkinstall, S. (1996) J. Biol. Chem. 271 27205-27208 [DOI] [PubMed] [Google Scholar]
- 27.Tanoue, T., Yamamoto, T., Maeda, R., and Nishida, E. (2001) J. Biol. Chem. 276 26629-26639 [DOI] [PubMed] [Google Scholar]
- 28.Theodosiou, A., Smith, A., Gillieron, C., Arkinstall, S., and Ashworth, A. (1999) Oncogene 18 6981-6988 [DOI] [PubMed] [Google Scholar]
- 29.Rahmouni, S., Cerignoli, F., Alonso, A., Tsutji, T., Henkens, R., Zhu, C., Louis-dit-Sully, C., Moutschen, M., Jiang, W., and Mustelin, T. (2006) Nat. Cell Biol. 8 524-531 [DOI] [PubMed] [Google Scholar]
- 30.Todd, J. L., Tanner, K. G., and Denu, J. M. (1999) J. Biol. Chem. 274 13271-13280 [DOI] [PubMed] [Google Scholar]
- 31.Aoyama, K., Nagata, M., Oshima, K., Matsuda, T., and Aoki, N. (2001) J. Biol. Chem. 276 27575-27583 [DOI] [PubMed] [Google Scholar]
- 32.Chen, A. J., Zhou, G., Juan, T., Colicos, S. M., Cannon, J. P., Cabriera-Hansen, M., Meyer, C. F., Jurecic, R., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Fletcher, F., Tan, T. H., and Belmont, J. W. (2002) J. Biol. Chem. 277 36592-36601 [DOI] [PubMed] [Google Scholar]
- 33.Wu, Q., Huang, S., Sun, Y., Gu, S., Lu, F., Dai, J., Yin, G., Sun, L., Zheng, D., Dou, C., Feng, C., Ji, C., Xie, Y., and Mao, Y. (2006) Front. Biosci. 11 2714-2724 [DOI] [PubMed] [Google Scholar]
- 34.MacKeigan, J. P., Murphy, L. O., and Blenis, J. (2005) Nat. Cell Biol. 7 591-600 [DOI] [PubMed] [Google Scholar]
- 35.Lazo, J. S., Nunes, R., Skoko, J. J., Queiroz de Oliveira, P. E., Vogt, A., and Wipf, P. (2006) Bioorg. Med. Chem. 14 5643-5650 [DOI] [PubMed] [Google Scholar]
- 36.Usui, T., Kojima, S., Kidokoro, S., Ueda, K., Osada, H., and Sodeoka, M. (2001) Chem. Biol. 8 1209-1220 [DOI] [PubMed] [Google Scholar]
- 37.Vogt, A., Cooley, K. A., Brisson, M., Tarpley, M. G., Wipf, P., and Lazo, J. S. (2003) Chem. Biol. 10 733-742 [DOI] [PubMed] [Google Scholar]
- 38.Vogt, A., Tamewitz, A., Skoko, J., Sikorski, R. P., Giuliano, K. A., and Lazo, J. S. (2005) J. Biol. Chem. 280 19078-19086 [DOI] [PubMed] [Google Scholar]
- 39.Cohen, D. M., Gullans, S. R., and Chin, W. W. (1996) J. Biol. Chem. 271 12903-12908 [DOI] [PubMed] [Google Scholar]
- 40.Anderson, R. D., Haskell, R. E., Xia, H., Roessler, B. J., and Davidson, B. L. (2000) Gene Ther. 7 1034-1038 [DOI] [PubMed] [Google Scholar]
- 41.Caunt, C. J., Finch, A. R., Sedgley, K. R., Oakely, L., Luttrell, L. M., and McArdle, C. A. (2006) J. Biol. Chem. 281 2701-2710 [DOI] [PubMed] [Google Scholar]
- 42.Dimitri, C. A., Dowdle, W., MacKeigan, J. P., Blenis, J., and Murphy, L. O. (2005) Curr. Biol. 15 1319-1324 [DOI] [PubMed] [Google Scholar]
- 43.Volmat, V., Camps, M., Arkinstall, S., Pouyssegur, J., and Lenormand, P. (2001) J. Cell Sci. 114 3433-3443 [DOI] [PubMed] [Google Scholar]
- 44.Brunner, D., Oellers, N., Szabad, J., Biggs, W. H., III, Zipursky, S. L., and Hafen, E. (1994) Cell 76 875-888 [DOI] [PubMed] [Google Scholar]
- 45.Jeffrey, K. L., Brummer, T., Rolph, M. S., Liu, S. M., Callejas, N. A., Grumont, R. J., Gillieron, C., Mackay, F., Grey, S., Camps, M., Rommel, C., Gerondakis, S. D., and Mackay, C. R. (2006) Nat. Immunol. 7 274-283 [DOI] [PubMed] [Google Scholar]
- 46.Shen, Y. H., Godlewski, J., Zhu, J., Sathyanarayana, P., Leaner, V., Birrer, M. J., Rana, A., and Tzivion, G. (2003) J. Biol. Chem. 278 26715-26721 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.