Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) is a unique ATP-binding cassette (ABC) ion channel mutated in patients with cystic fibrosis. The most common mutation, deletion of phenylalanine 508 (ΔF508) and many other disease-associated mutations occur in the nucleotide binding domains (NBD) and the cytoplasmic loops (CL) of the membrane-spanning domains (MSD). A recently constructed computational model of the CFTR three-dimensional structure, supported by experimental data (Serohijos, A. W., Hegedus, T., Aleksandrov, A. A., He, L., Cui, L., Dokholyan, N. V., and Riordan, J. R. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 3256–3261) revealed that several of these mutations including ΔF508 disrupted interfaces between these domains. Here we have used cysteine cross-linking experiments to verify all NBD/CL interfaces predicted by the structural model and observed that their cross-linking has a variety of different effects on channel gating. The interdomain contacts comprise aromatic clusters important for stabilization of the interfaces and also involve the Q-loops and X-loops that are in close proximity to the ATP binding sites. Cross-linking of all domain-swapping contacts between NBDs and MSD cytoplasmic loops in opposite halves of the protein rapidly and reversibly arrest single channel gating while those in the same halves have lesser impact. These results reinforce the idea that mediation of regulatory signals between cytoplasmic- and membrane-integrated domains of the CFTR channel apparently relies on an array of precise but highly dynamic interdomain structural joints.
The cystic fibrosis transmembrane conductance regulator (CFTR)3, the mutation of which causes cystic fibrosis (CF), belongs to the superfamily of ATP-binding cassette (ABC) proteins but functions as an ion channel rather than an active transporter. The chloride channel activity is crucial for maintaining salt and fluid homeostasis in epithelial tissues (1). In patients with CFTR mutations that compromise its maturation or channel activity, the airway surface liquid volume is diminished, impeding mucociliary clearance (2, 3). The absence of functional CFTR also impairs submucosal gland secretion (4).
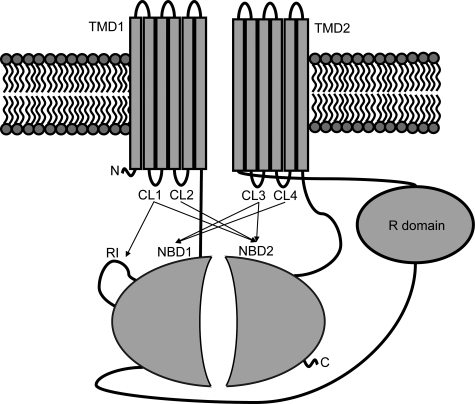
Like many other ABC family proteins, CFTR (also known as ABCC7) contains two membrane-spanning domains (MSDs) and two nucleotide-binding domains (NBDs), with an additional unique R domain (Fig. 1). While many ABC proteins are multisubunit proteins composed of two identical NBDs and MSDs, CFTR is a single polypeptide containing two distinct NBDs and MSDs. The proper folding of the individual domains and the interactions between these domains during or after protein synthesis are essential for CFTR assembly, a process that is inefficient with the majority of CFTR being degraded at the endoplasmic reticulum by the proteasome (5, 6). The most prevalent CF-causing mutation is the deletion of a phenylalanine at position 508 (ΔF508). Recent studies suggest that the folding kinetics of NBD1 and the interdomain interactions between MSDs and the NBDs are disrupted by this mutation (7–9), although the crystal structures of isolated wild-type and mutant NBD1 show no major alteration in its overall three-dimensional structure (10).
FIGURE 1.
CFTR scheme. CFTR is composed of two nucleotide binding domains (NBD1 and NBD2), two membrane-spanning domains (MSD1 and MSD2), and a regulatory region (R domain). Indicated by arrows are the confirmed interactions between the NBDs and the CLs of the MSDs.
Control of CFTR channel activity is modulated by the phosphorylation of the R domain by protein kinase A, which allows the regulation of gating by ATP binding at the NBD1/NBD2 interface. Stable binding of ATP at NBD1 and binding and hydrolysis of ATP at NBD2, together with R domain phosphorylation, may alter allosteric interactions between these domains and impact the channel gating cycle (11–13). Although considerable progress has been made toward understanding the integrated control of CFTR channel gating by phosphorylation and ATP binding/hydrolysis (12), details at the level of interactions of specific secondary and tertiary structural elements remain to be elucidated and require knowledge of the three-dimensional structure.
As yet, no high resolution crystal structure of any eukaryotic ABC proteins is available. However, the crystal structures of several bacterial ABC transporter proteins have been recently solved (14–18). Of particular interest is the structure of the bacterial exporter Sav1866, which shows its NBDs in close contact with both MSDs, a configuration that is unlike those of the importers (BtuCD, ModBC, HI1470/71 and the Escherichia coli maltose transporter), which show each of their MSD in contact solely with one NBD. Although CFTR functions as a channel, it belongs to the exporter subclass of the ABC family. In our recent work, a molecular model of CFTR was constructed based on its homology to Sav1866. From the model, we predicted and confirmed experimentally the interdomain interactions between CL2 (MSD1) and NBD2 and between CL4 (MSD2) and NBD1 (19). The later interaction between NBD1 and CL4 is most crucial to CFTR biogenesis and assembly because of the known sensitivity of CFTR conformational maturation to many disease-associated mutations in CL4 as well as NBD1 (20). Single channel activity measurements also show that both interfaces are important for the regulation of channel gating as cross-linking of Cys on either side of these interfaces arrests channel gating. This observation, corroborated by rate equilibrium-free energy relationship (REFER) analysis of the single channel kinetics, suggest that these interfaces act as connecting joints between MSDs and NBDs, thus coordinating movement on either side of the contact (19).
Many disease-causing mutations also occur in other cytoplasmic loops such as CL1 and CL3, compromising CFTR maturation and channel function (21, 22), which suggests that these regions of CFTR may also play an important role in interdomain interactions and channel regulation. Further identification and analysis of the CL/NBD interfaces should help understanding of how these mutations disrupt interdomain interactions during CFTR biosynthesis and how the signals of ATP binding and hydrolysis at NBDs are transmitted to MSDs.
EXPERIMENTAL PROCEDURES
Antibodies—Mouse monoclonal CFTR antibodies to an N-terminal fragment (mAb 13-4, IgG1κ), NBD2 (mAb 596, IgG2b), and R domain (mAb 450, IgG1κ) were generated as described (9). Goat anti-mouse IgG-IR800, IgG1-IR800, and IgG2b-IR680 were from LiCor Corp.
Construction and Expression of Mutants—Cysteine (Cys) was introduced into the Cys-less CFTR construct in pcDNA3 vector by the Stratagene Quick Exchange protocol as described (19). A stop codon (TAA) was introduced at residue Glu-1172 to produce the ΔNBD2 construct, 1172X (9). 5′-GGACCCCAGCGCCCGAGAGACCATGGAAGGTAAACCTACCAAGTCAACC-3′ and its reverse primer were used to make an NBD2-containing construct (residues 1172–1480). Point mutations and PCR-generated DNA fragments were confirmed by automated DNA sequencing (UNC-CH Genome Analysis Facility).
Human embryonic kidney 293 (HEK) cells were transiently transfected using Effectene transfection reagent (Qiagen) according to the manufacturer's instructions. To promote maturation of Cys-less CFTR variants, 24 h after transfection, HEK cells were incubated at 27 °C for 48 h before the cross-linking experiment. For stable expression, constructs were cotransfected with pNUT plasmid into baby hamster kidney (BHK-21) cells, which were selected and maintained in methotrexate-containing medium (23). To phosphorylate CFTR, cells were incubated for 10 min with 10 μm forskolin, 100 μm DiBu-cAMP, and 1 mm 3-isobutyl-1-methylxanthine before harvesting. This stimulation mixture was also present during the cross-linking reaction.
Isolation of Membrane Vesicles and Single-Channel Measurements—Membrane vesicles were isolated from BHK or HEK cells expressing variants of Cys-less CFTR as described previously (19). To phosphorylate CFTR, membranes were treated with 100 units/ml PKA (Promega) in the presence of 2 mm ATP and 2 mm MgCl2 for 15 min at room temperature, sonicated briefly, and incubated for another 15 min to achieve complete phosphorylation. Phosphorylation was confirmed by the diminution of the signal detected by the phosphorylation-sensitive mAb 450. Single-channel recordings were collected as previously described (19, 24).
Cross-linking in Whole Cells and Membrane Vesicles—Disulfide cross-linking in cells with bifunctional methanethiosulfonate (MTS, Toronto Research Chemicals) cross-linkers with spacer arms ranging from 3.9 to 24.7 Å was performed as described (19). To cross-link CFTR in vesicles, membranes (1 mg/ml total proteins) were incubated with 20 μm MTS reagent for 15 min at room temperature. The cross-linking reaction was stopped with Laemmli sample buffers with or without DTT. Proteins were resolved with 7.5% SDS-PAGE and CFTR detected with mAb 596 and secondary goat anti-mouse IgG-IR800 using the Odyssey infrared scanner (LiCor Corp.). For dual antibody labeling, isotype-specific secondary antibodies labeled with different infrared (IR) dyes were used. Specifically, goat-anti-mouse IgG1-IR800 and IgG2b-IR680 were used to detect mAb 450 and mAb 596, respectively.
Limited Trypsin Digestion—For limited trypsin digestion, BHK membranes were resuspended at 1 mg of protein/ml in a buffer containing 40 mm Tris-HCl, pH 7.4, 2 mm MgCl2, and 0.1 mm EGTA. Membrane proteins were first treated with 20 μm M8M cross-linker for 15 min at room temperature and centrifuged to remove cross-linker before trypsin digestion. Membranes were incubated on ice with 240 and 480 μg/ml of TPCK-treated trypsin for 15 min, and digestion was stopped with excess soybean trypsin inhibitor. CFTR tryptic fragments separated on 4–20% gradient gels (Bio-Rad) were detected by Western blots probed with mAb 13-4.
RESULTS
Aromatic Clusters Mediate Domain-swapping Interactions—Many disease-associated CFTR mutations occur in cytoplasmic loops, compromising CFTR maturation and channel activity (20–22). Determination of how these loops interact with other domains may help the understanding of how these mutations disrupt CFTR biosynthesis and channel activity. We have constructed a three-dimensional model of CFTR based on Sav1866 and shown that there are close contacts between CL2 and NBD2, and between CL4 and NBD1 (19) (Fig. 1). This configuration supported the so-called “domain-swapping” where each MSD interacts with the NBD in the opposite half of the molecule as was earlier observed in Sav1866 (14) and also in MsbA (25).
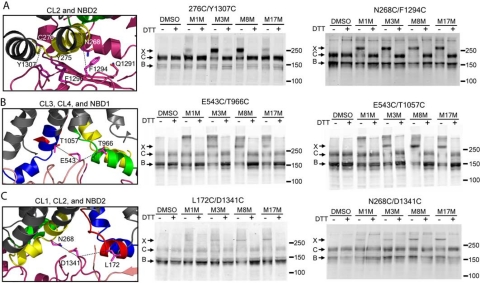
A striking feature of the CL4/NBD1 interface is the interaction of Phe-508 with a cluster of aromatic residues (Phe-1068, Tyr-1073, and Phe-1074) in CL4, which may serve to stabilize this crucial interface (19). The structural model predicts an analogous aromatic cluster in the interface between CL2 (Tyr-275) and NBD2 (Phe-1294, Phe-1296, and Tyr-1307). To verify these specific interactions, we performed cross-linking experiments in HEK cells expressing a Cys-less CFTR construct containing the Cys pair 276C and Y1307C (Fig. 2A). The 276 and 1307 sites could be cross-linked using the MTS reagents M3M and M8M, but to a lesser extent with M1M and M17M, suggesting that the two residues are in relatively close contact. These cross-linking results confirm the existence of an analogous aromatic cluster between CL2 and NBD2.
FIGURE 2.
Contact interfaces of CLs with opposite NBDs. HEK 293 cells transiently transfected with Cys-less CFTR containing Cys pairs introduced at different interfaces between cytoplasmic loops and NBDs were incubated with 200 μm MTS reagents of different spacer arm lengths. Cell lysates in SDS-PAGE sample buffer with or without DTT were subjected to Western blot analysis using CFTR antibody mAb 596. Cross-linked proteins migrate above 250 kDa. A, 276C/Y1307C and N268C/F1292C at the CL2/NBD2 interface involving an aromatic cluster and the Q-loop. B, T966C/E543C at the CL3/NBD1 interface and T1057C/E543C at the CL4/NBD1 interface at the X-loop of NBD1. C, L172C/D1341C at the CL1/NBD2 interface and N268C/D1341C at the CL2/NBD2 interface at the X-loop of NBD2. The three bands, X, C, and B, represent the cross-linked, the mature complex glycosylated, and the immature core glycosylated CFTR, respectively.
Q- and X-Loops of Both NBDs Also Participate in CL2/NBD2 and CL4/NBD1 Interactions—Conformational changes due to ATP binding and hydrolysis at the NBD1/NBD2 interface may be transmitted to the MSDs through the CL/NBD interface contacts. Thus it is important to determine whether or not these interfaces are spatially proximal to the ATP binding sites. In agreement with what is predicted by our model, we were able to cross-link residue pairs W496C/T1064C, M498C/L1065C (19), which proves that indeed CL4 also interacts with NBD1 through the so called Q-loop (Q493). The Q-loop connects the canonical α-helical subdomain containing the ABC signature with the core subdomain containing Walker A and Walker B motifs (26–28). In the histidine permease structure, the side chain of the Q-loop glutamine (Q100) contacts the γ-phosphate of ATP via a water molecule (28). Analogous to the interaction of CL4 with the Q-loop of NBD1, our model also predicts a close contact between CL2 and the Q-loop (Q1291) of NBD2. The successful cross-linking of the residue pairs N268C (CL2) and F1294C (NBD2) by the MTS reagents of various spacer arm lengths confirmed this contact (Fig. 2A). The interaction of the Q-loops with the CLs observed in the Sav1866 crystal structure and identified by cross-linking in CFTR is also observed in ABC importers such as BtuCD, ModBC, HI1470/1, and the maltose transporter (15–18). However, one feature that is unique to the Sav1866 structure is that its so-called X-loop (469TEVGERG) in the NBD interacts with both CL1 and CL2. The X-loop sequence is conserved in exporters but not in importers (14). Moreover, X-loops are in close proximity to the ABC signature motifs, making them also good candidates for the transmission of signals of ATP binding/hydrolysis at the NBDs to the MSDs. Sequence alignment of CFTR with Sav1866 indicates that in CFTR, Glu-543 (NBD1), and Asp-1341 (NBD2) correspond to Glu-473 of the Sav1866 X-loop. Cys pair cross-linking experiments showed that indeed E543C could be cross-linked with both T966C (CL3) and T1057C (CL4, Fig. 2B), while D1341C was in close contact with both L172C (CL1) and N268C (CL2, Fig. 2C).
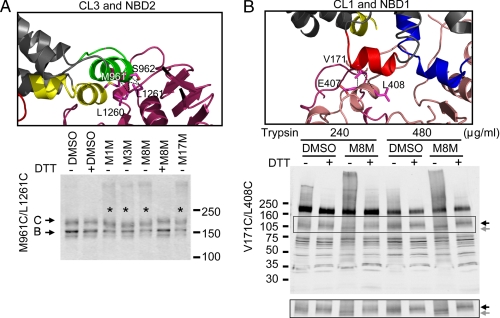
CL/NBD Interfaces within Each Half of CFTR—So far, we have only confirmed the cross-linking between MSDs with their opposite NBDs through CLs, i.e. CL1 and CL2 with NBD2, and CL3 and CL4 with NBD1. However, contact interfaces of the N-terminal cytoplasmic loops in each MSD (CL1 and CL3) with NBDs of the same half of the molecule have not been established in any eukaryotic ABC proteins. According to our Sav1866-based CFTR model, CL1 and CL3 should contact both NBDs, as shown in the scheme in Fig. 1. To confirm these contacts in the case of CL3 and NBD2, we designed several Cys pairs from CL3 (M961C and S962C) and NBD2 (L1260C and L1261C) (Fig. 3A). Cross-linking experiments were carried out in HEK cells overexpressing Cys-less CFTR with these Cys pairs. In constructs containing the Cys pairs M961C/L1261C (Fig. 3A), M961C/L1260C, and S962C/L1261C (supplemental Fig. S1A), MTS reagent treatment produced a slightly, albeit distinguishably, faster moving band, which could be reversed by DTT. We hypothesized that this different mobility shift pattern may be due to the smaller number of amino acids between these cross-linked Cys pairs (∼300 amino acids) than those between CLs and opposite NBDs (∼400 to ∼1200 amino acids), which produce a clear cross-linked band that migrates much slower (for example Fig. 2). A faster moving band after cross-linking was also observed when cross-linking T123C (CL1) with S428C (NBD) of the multidrug ABC transporter BmrA (29), which is separated by ∼300 amino acids between the cross-linked cysteines. Cross-linking of Cys pairs between CL3 and NBD2 was also confirmed with co-expression of constructs of Cys-less ΔNBD2 CFTR containing M961C together with the Cys-less NBD2 fragment containing L1261C in HEK cells (supplemental Fig. S1, B and C).
FIGURE 3.
Contact interfaces of CLs with NBDs within each half of CFTR. A, cross-linking at interfaces between CL3 and NBD2. Transiently transfected HEK cells were incubated with 200 μm MTS reagents, and cross-linking was detected as described under “Experimental Procedures.” The cross-linked proteins that moved faster than the mature C band with a Cys pair introduced at Met-961 and Leu-1261 are marked with *. B, cross-linking at interface between CL1 and NBD1. Membrane vesicles prepared from BHK cells overexpressing Cys-less CFTR containing the Cys pair V171C and L408C were treated with 20 μm MTS cross-linker M8M before limited trypsin digestion with concentrations indicated in the figures. Partially digested fragments were resolved with 4–20% SDS-PAGE and Western blotting with mAb 13-4. The band highlighted in the rectangle is shown separately at the bottom. The gray arrow denotes the cross-linked fragment that moved faster than its uncross-linked counterpart (black arrow).
Baker et al. (30) reported NMR data indicating that CL1 binds to NBD1 in a phosphorylation-dependent mechanism, which may be mediated by a short sequence in NBD1 termed the regulatory insertion (RI). The absence of the RI in the crystal structure of the human NBD1 and the elevated b-factor in the mouse NBD1 structure (10, 26) indicate that this region is highly dynamic, potentially adopting multiple conformations in solution. In the construction of the whole CFTR model (19), we modeled the RI conformation such that it points toward the solution, as suggested by one of the crystal structures of the human NBD1 with Phe-508 deleted (10). However, another possible conformation that may be adopted by the RI is suggested by the crystal structure of the mouse NBD1 (26), which shows its RI “flipped” toward the NBD1/NBD2 interface and CL1 (Fig. 3B).
Based on the latter model, we attempted to detect cross-linking of the residue pairs V171C/E407C and V171C/L408C in the whole protein, but did not observe any indication of cross-linking of the mature band (supplemental Fig. S2A). Because the number of amino acids between CL1 and NBD1 (∼240 amino acids) is even smaller than that between CL3 and NBD2 (∼300 amino acids), we speculated that any mobility shift caused by cross-linking of CL1 and NBD1, if it existed, may not be detectable with the resolution of SDS-PAGE. We also speculated that the mobility shift between CL1 and NBD1 might be detected if the molecular fragments were smaller. To test this hypothesis, membrane vesicles were prepared from BHK cells overexpressing Cys-less CFTR containing V171C and L408C. To obtain cross-linked smaller fragments of CFTR, membrane vesicles were subjected to limited trypsin digestion after M8M treatment. The digested proteins were then resolved with gradient SDS-PAGE, and the CFTR fragments were detected by Western blot using the N-terminal CFTR antibody mAb 13-4. As shown in Fig. 3B, limited trypsin digestion produced a band of ∼120 kDa in control membrane vesicles (highlighted in the rectangle, indicated by the black arrow). This band was unique to the Cys-less CFTR and was not previously observed in wild-type CFTR (9). M8M treatment caused the band to move faster (apparent molecular mass ∼110 kDa, indicated by the gray arrow), which could be reversed by DTT. We found similar results for the Cys pair V171C and E407C (supplemental Fig. 2B), but not with a Cys pair introduced at Gln-958 and Leu-1261 (supplemental Fig. S2C), confirming that the faster moving CFTR fragment was indeed due to the cross-linking at the CL1/NBD1 interface. Contrary to what was found by NMR analysis using a synthetic CL1 peptide and purified NBD1 (30), our cross-linking experiments using a functional full-length CFTR, fully integrated in the membrane, detected a CL1/NBD1 interaction which was independent of PKA phosphorylation (supplemental Fig. S2B). No cross-linking was detected when Cys pairs were introduced at L172C/E543C, T966C/D1341C, V171C/L1261C, or M961C/L408C, which are not predicted to be in association in the structural model (supplemental Fig. S3).
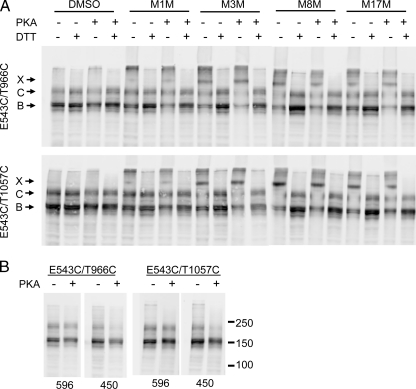
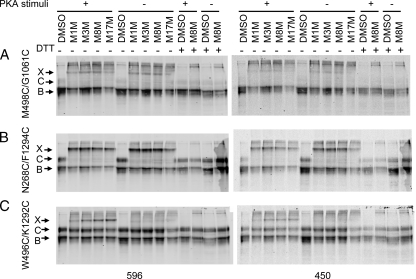
PKA Phosphorylation Does Not Affect CL/NBD Cross-linking—Positioned at a crucial contact point between MSDs and NBDs, CLs are good structural candidates for transmitting the conformational signals initiated by ATP binding/hydrolysis to the MSDs to regulate channel gating. Previously, we have found that the interfaces at CL2/NBD2 and CL4/NBD1 are not strongly influenced by PKA stimuli or by the binding of AMP-PNP, or by trapping of ADP-vanadate at the NBDs (19). In the present study, we also determined whether our newly detected CL and NBD interactions were affected by PKA phosphorylation and ATP binding. As shown in Fig. 2, B and C, of particular interest are the interactions of the NBD1 X-loop (Glu-543) with both CL3 and CL4 and of the NBD2 X-loop (Asp-1341) with CL1 and CL2. That the X-loops are in close proximity with the ABC signature motif and with both CLs of opposite MSDs suggest that the signals from ATP binding/hydrolysis at NBDs may be transmitted to MSDs through these interfaces. To determine whether the interfacial interaction of Glu-543 with CL3 and CL4 was indeed affected by PKA phosphorylation, membrane vesicles from HEK cells overexpressing Cys-less CFTR with Cys pairs E543C/T966C and E543C/T1057C were pretreated with PKA in the presence of ATP and Mg2+ before cross-linking with various MTS reagents. As shown in Fig. 4A, similar to the experiments with whole cells in membranes not treated with PKA, the Cys pairs E543C/T966C and E543C/T1057C were cross-linked by all the MTS reagents tested. The longer space arm reagents produced stronger cross-linking. However, cross-linking was neither augmented nor diminished by PKA treatment, even though CFTR phosphorylation was confirmed by the decreased signals detected using the phosphorylation-sensitive mAb 450 (Fig. 4B). These results indicate that these interfaces remain in tight contact before and after exposure to stimuli that activate CFTR channels.
FIGURE 4.
Phosphorylation of CFTR by PKA does not interfere with NBD1 X-loop binding to CL3 and CL4. Membrane vesicles prepared from HEK cells transiently transfected with Cys pairs introduced at E543C with T966C (CL3) and T1064C (CL4) were pretreated with PKA catalytic subunit in the presence of ATP before incubating with 20 μm MTS reagents. A, CFTR and cross-linked bands were detected with mAb 596. B, CFTR phosphorylation was confirmed with the phosphorylation-sensitive antibody mAb 450. X, C, and B represent the cross-linked, the mature complex glycosylated, and the immature core glycosylated CFTR, respectively.
Unlike the X-loop that is unique in ABC exporters, the contact between the CLs and the Q-loops of NBDs is conserved in importers such as BtuCD, ModBC, HI1470/1, and the maltose transporter as well as exporters such as Sav1866 and CFTR (14–18). From the comparison of the crystal structures of the nucleotide-free and the ATP-bound NBD of the MJ0796 ABC transporter, it has been suggested that the conserved Q-loop connecting the α and β subdomains of the NBDs moves along with the ABC signature upon ATP binding (31). To verify whether the two CFTR Q-loops (Gln-493 and Gln-1291) undergo conformational changes in response to channel gating stimuli, we first tested whether the cross-linking at the CL4/NBD1 and CL2/NBD2 interfaces involving the Q-loops was affected by PKA activating stimuli, and then we tested whether the interface between the two Q-loops themselves was changed by PKA. As shown in Fig. 5, treatment of the cells with PKA stimulation mixture before and during cross-linking had no effect on cross-linking between either M498C/G1061C (Fig. 5A, left panel) or N268C/F1294C (Fig. 5B, left panel), although phosphorylation-sensitive CFTR antibody mAb 450 confirmed the phosphorylation of CFTR (Fig. 5, A and B, right panels). However, when a Cys mutation was introduced in each Q-loop (W496C/K1292C), the PKA stimulation mixture significantly increased the cross-linking of this Cys pair with all MTS reagents tested (Fig. 5C). The very weak cross-linking observed in control cells may represent the basal PKA activity. Enhanced interaction between the two NBDs promoted by PKA phosphorylation was also observed by Mense et al. (32) in split halves of CFTR.
FIGURE 5.
Phosphorylation brings together the Q-loops of NBD1 and NBD2. Transiently transfected HEK cells were pretreated with phosphorylation stimulation mixture (10 μm forskolin, 100 μm DiBu-cAMP, and 1 mm 3-isobutyl-1-methylxanthine) for 10 min. Cross-linking was carried out in the presence of phosphorylation stimuli and detected as described in the legend to Fig. 2. Cys pairs were introduced at: A interface between NBD1 Q-loop and CL4 (M498C/G1061C); B, interface between NBD2 Q-loop and CL2 (N268C/F1294C); or C, both Q-loops (W496C and K1292C). X, C, and B, represent the cross-linked, the mature complex glycosylated, and the immature core glycosylated CFTR, respectively.
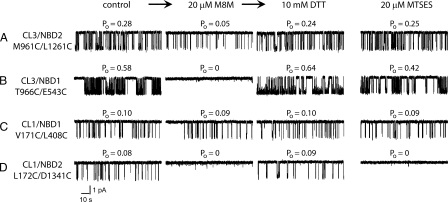
Influence of CL/NBD Cross-linking on Channel Gating—We found previously that cross-linking across either of the domain-swapping interfaces between NBDs and CLs in opposite halves of CFTR rapidly and reversibly arrested channel gating (19). We have now begun to analyze the influence of cross-linking across the newly detected interfaces between NBDs and CLs on the same side of the molecule in addition to the domain-swapping interfaces, and quite different effects were observed. First, cross-linking between residues M961C and L1261C at the CL3/NBD2 interface changed channel gating behavior substantially but did not arrest it completely (Fig. 6A). The channel open probability (Po) gradually decreased from 0.28 to 0.05 because of reduction in open burst duration from 450 to 140 ms. Simple chemical modification of both available sulfhydryl groups by the negatively charged monofunctional MTSES reagent did not affect channel gating in control experiments. However, the brief openings of 140-ms duration at 30 °C observed after the treatment with the M8M cross-linker are typical of ΔNBD2 CFTR channel gating (9). Thus, the cross-linking between CL3 and NBD2 has an effect similar to that of the complete absence of NBD2. In contrast, the cross-linking between residues T996C and E543C at the CL3/NBD1 interface rapidly and reversibly arrested channel gating (Fig. 6B) exactly as we had observed previously with the CL4/NBD1 interface (19). The control treatment of this cysteine pair with the MTSES sulfhydryl reagent did not cause any substantial changes in the channel gating. Turning from the effects of cross-linking CL3 to those of its counterpart, CL1 in the N-terminal MSD, its cross-linking to NBD1 i.e. V171C/L408C had essentially no influence on gating (Fig. 6C). On the other hand, the cross-linking between L172C of CL1 and D1341C of NBD2 rapidly and reversibly inhibited channel gating (Fig. 6D) very similar to what we had observed previously for the CL2/NBD2 interface (19). However, in the case of the completely reversible cessation of gating caused by L172C/D1341C cross-linking, the role of modification of each of these cysteines individually will have to be studied in detail because treatment with monofunctional MTSES also inhibited gating. Nevertheless, the present data together with those reported earlier demonstrate that fixation of the two sides of these interdomain joints with respect to each other has major but varied effects on channel activity. The coordinates of the CFTR model are available from our earlier report (19).
FIGURE 6.
Role of interdomain cross-linking in CFTR channel gating. All single channel measurements began with 20-min control recordings of each Cys pair CFTR variant. Open probabilities (Po) were calculated from the all points histogram for the last 10 min of the recordings. However, only 2 min of control single channel recording are shown in the first column. The functional state after 30 min of exposure to 20 μm M8M from the cis side of the bilayer is shown in the second column. The result of sulfhydryl group restoration 40 min after exposure to 10 mm DTT subsequent to the M8M treatment is shown in the third column. In each panel, the first three columns represent the results of the same 3-step experimental protocol. The effect of sulfhydryl modification by monofunctional MTSES is shown in the fourth column. Both experimental and control protocols were repeated at least three times for all Cys-less variants. A, M961C/L1261C at the CL3/NBD2 interface; B, T966C/E543C at the CL3/NBD1 interface; C, V171C/L408C at the CL1/NBD1 interface; D, L171C/D1341C at the CL1/NBD2 interface.
DISCUSSION
Knowledge of the three-dimensional structure of CFTR will lead to a fuller understanding of how disease-associated mutations compromise its maturation and channel activity and facilitate the development of drugs to rescue maturation and restore function. As of yet, there is no known high resolution structure of eukaryotic ABC transporters; thus, we had constructed a CFTR three-dimensional structure using molecular modeling based on Sav1866, whose structure has been determined to a resolution of 3.0 Å by x-ray crystallography. Several aspects of the model have been experimentally confirmed (19), including the crucial interface between Phe-508 in NBD1 and CL4 of MSD2 and that between NBD2 and CL2. We have established that these interfaces are important in both the stabilization of CFTR structure during biosynthesis and the regulation of channel gating. Using independent techniques such as chemical cross-linking and REFER analysis, we found that the CL2/NBD2 and CL4/NBD1 interfaces may act as joints that coordinate movements of NBDs and MSDs during channel gating. The CL4/NBD1 interface has also been identified by cross-linking in the multidrug resistance P-glycoprotein (33). However, counterparts of none of the other CL/NBD interfaces observed in Sav1866 structure have been confirmed in any eukaryotic ABC proteins. In this study, we have detected the interaction of all four CLs with both NBD1 and NBD2 (Fig. 1) and analyzed the effect of PKA phosphorylation and ATP binding on these interactions. We also observed distinctive effects of cross-linking of various CL/NBD interfaces on channel gating.
Unlike solute importers such as BtuCD, ModBC, HI1470/1, and the maltose transporter, all of which show MSDs that are in contact with only one NBD, the architecture of the exporter Sav1866 shows each MSD in contact with both NBDs. Like P-glycoprotein, Sav1866 is a member of the multidrug resistance class of ABC transporters. Sequence alignment of P-glycoprotein with Sav1866 and biochemical evidence confirm that CL4 of MSD2 is in close proximity with NBD1, as Arg-905 and Ser-909 of CL4 can be chemically cross-linked to Ser-474 and Leu-443 of NBD1, respectively, when these residues are replaced with cysteines in a Cys-less P-glycoprotein (33). As a member of the exporter subclass of ABC proteins, CFTR may share many structural features with Sav1866 and P-glycoprotein. In this study, we show that the CL and NBD interfaces involve aromatic clusters that may stabilize the interface, and the Q-loops and the X-loops that are close to ATP binding sites (Fig. 2). Each NBD is in close contact with three CLs. These stable interactions between NBDs and CLs facilitate their ability to couple the signals of ATP binding and hydrolysis at NBDs to the channel gating at MSDs. The presence of the unique X-loop in exporters but not importers suggests a distinct coupling mechanism between the different subclasses of ABC transporters and that CFTR apparently employs that of the exporters.
The contact interfaces between NBD1 with CL3 and CL4 involve an aromatic cluster, the Q-loop, and the X-loop (Fig. 2). These close interactions between NBD1 with CL3 and CL4, both in MSD2, may explain the observation that MSD2 is required for CFTR maturation (9). Stepwise truncation of the C-terminal region shows that CFTR is able to mature and acquire channel activity when truncated back to residue Glu-1172, just C-terminal of MSD2 (9). Younger et al. (34) also demonstrated the requirement of MSD2 for wild-type CFTR to avoid ER quality control and degradation. They show that a folding defect in ΔF508 detected by RMA1 involves the inability of MSD2 to interact with N-terminal domains. Analogous to the NBD1 interaction with three CLs (CL1, CL3, and CL4), NBD2 also interacts with three CLs (CL1, CL2, and CL3). Moreover, similar to the interactions formed by NBD1, NBD2 contacts with the CLs also involve an aromatic cluster, the Q-loop and the X-loop (Fig. 2). However, our observations that ΔNBD2-CFTR can still mature, reach the plasma membrane, and retain channel function, although not as active as full-length CFTR (9), suggest that the interfaces involving NBD2 are not as important in promoting CFTR conformational maturation as those formed by NBD1 with the CLs. This finding also rationalizes the existence of far fewer maturation-compromising, disease-associated mutations in NBD2 than in NBD1. However, NBD2 becomes more sensitive to trypsin digestion when mutations are introduced at CL4/NBD1 interfaces (9), indicating that the proper folding of NBD2 itself requires a correct assembly of the preceding domains.
While the NBD2/CL interfaces appear to be not as important in the biogenesis of CFTR as those at NBD1/CL, both sets are still integral to the transmission of channel gating signals from the nucleotide-binding sites to the transmembrane pore. This is partially supported by the observation that co-expression of an NBD2 fragment with ΔNBD2-CFTR increases the channel activity of ΔNBD2-CFTR.4 This difference in function of the various contact interfaces may be due to the fact that the CFTR protein comprises pairs of similar but not identical membrane and cytoplasmic domains in a single polypeptide; thus corresponding interfaces from the two halves of the protein are not exactly identical.
Signals of ATP binding and hydrolysis at NBDs need to be transmitted to MSDs to regulate channel gating. Q-loops and X-loops are in close proximity to CLs connecting MSDs and also to the ATP-binding sites. The close contacts of the Q-loops with both the CLs and the ATP binding sites suggest that these structural elements are appropriately located to transmit the impact of ATP binding to the MSDs. In the multidrug ABC transporter BmrA, cross-linking experiments showed that the Q-loop disengages from CL1 during its catalytic cycle (29). In yet another transporter, MJ0796, the comparison of the crystal structures of the nucleotide-free and the ATP-bound NBD suggest that the Q-loop moves along with the LSGGQ motif such that the amide side chain of Gln-90 at the N terminus of the γ-phosphate linker moves ∼5 Å to contact the Na+ cofactor and putative hydrolytic water in the active site (31). In P-glycoprotein, the trapping of AMP-PNP or ADP plus vanadate at NBD reduces the cross-linking of L443C and S909C, suggesting that conformational changes occur at the NBD1/MSD2 interface during the ATP catalytic cycle (33). In our study, we find that the Q-loops of CFTR NBD1 and NBD2 form contacts with CL4 and CL2, respectively. These contacts are formed before and maintained during and after channel activation. While the two NBDs come closer together during the stimulation that activates CFTR channels, the interfaces between CLs and NBDs remain in proximity and may coordinate larger conformational changes on both sides of the contacts. These results, consistent with our previous findings, suggest that these contact interfaces move in unison in response to channel gating signals, and act as connecting joints between the NBD/MSD interfaces. We cannot however rule out the possibility that cross-linking is unable to resolve the small conformational changes at these interfaces during channel gating.
In our previous study, we found that cross-linking of the domain-swapping interfaces at either CL2/NBD2 or CL4/NBD1 reversibly arrested channel gating (19). Similar results are found in this study on cross-linking of domain-swapping interfaces at CL3/NBD1 and CL1/NBD2 (Fig. 6, B and D). However, cross-linking of Cys pairs between CLs and NBDs in the same half of the molecule (CL3/NBD2 and CL1/NBD1) has different effects on channel gating (Fig. 6, A and B). In the case of CL1/NBD1, cross-linking has no substantial effect at all, while for CL3/NBD2, channel gating is not completely arrested by cross-linking but is substantially diminished due to a reduction in open burst duration from 450 to 140 ms. Brief openings of 140-ms duration are typical for ΔNBD2 CFTR channel gating (9). We speculate that CL3/NBD2 cross-linking precludes the influence of NBD2 on the ion channel gating. A specific feature of ΔNBD2 CFTR channel gating is its independence of nucleotide type. The ion channel gating ligand specificity (24) is a property of NBD24 and could be used as an independent test of the type of channel gating after CL3 and NBD2 cross-linking. These data confirm our previous conclusion about corresponding domain-swapping interactions between NBDs and CLs (19) and establish an important potential role of CL3 in allosteric regulation between nucleotide binding and channel gating.
The three-dimensional CFTR model we constructed is based on the crystal structures of Sav1866 (14) and human CFTR NBD1 (10) and a homology model of NBD2 (35). The model accommodates the experimental data on the orientation and packing of transmembrane helices (36, 37), the inter-NBD cross-linking (32), and our cross-linking experiments between CLs and NBDs (19). A particular feature, the NBD1 RI loop, adopts different conformations in several crystal structures of CFTR NBD1 (26, 38), which suggests that the loop is highly dynamic. In our CFTR model that we constructed following the Sav1866 nucleotide bound conformation, the RI loop is “flipped” away from the NBD1-NBD2 interface to avoid serious clashes with NBD2 (19). Attempts to verify this conformation by cross-linking CL1 and RI did not yield positive results (data not shown). On the other hand, another viable conformation for the RI region is suggested by the mouse CFTR NBD1, which shows its RI “flipped” toward the NBD1/NBD2 interface and the CL1 (26). We have verified the latter conformation by introducing Cys pairs in CL1 and different residues in RI (V171C/E407C, V171C/L408C) and showing that these pairs can be cross-linked by M8M (Fig. 3B). It is noteworthy that the RI loop, when positioned in the NBD1-NBD2 interface, does not clash with NBD2 when the CFTR NBDs associate according to the closed nucleotide-free conformation of MsbA (25). Although there is one report that CL1 binds to NBD1 in a PKA-dependent manner using a synthetic CL1 peptide and purified NBD1 (30), in our experiments with functional membrane-bound CFTR, the binding of CL1 with RI is not affected by PKA phosphorylation (supplemental Fig. 2B).
We have identified the interfaces between CFTR nucleotide binding domains and the cytoplasmic loops of the membrane-spanning domains. These interfaces are significantly involved in the stabilization of interdomain contacts and regulation of the channel gating. Our results shed new light on the structure and mechanism of action of CFTR, the only known ABC transporter shown to function as an ion channel. Identification of these interdomain interfaces and understanding of how they are perturbed by disease-associated mutations may also aid efforts to develop new therapeutic strategies to treat cystic fibrosis.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK051619 (to J. R. R.). This work was also supported by Cystic Fibrosis Foundation Grant DOKHOL07I0 (to N. V. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: CFTR, cystic fibrosis transmembrane conductance regulator; ABC, ATP-binding cassette; CF, cystic fibrosis; CL, cytoplasmic loop; DTT, dithiothreitol; MSD, membrane-spanning domain; MTS, methanethiosulfonate; MTSES, sodium (2-sulfonatoethyl) methanethiosulfonate; M1M, 1,1-methanediyl bismethanethiosulfonate; M3M, 1,3-propanediyl bismethanethiosulfonate; M8M, 1,5-pentanediyl bismethanethiosulfonate; M17M, 3,6,9,12,15-pentaoxaheptadecane-1,17-diylbis-methanethiosulfonate; NBD, nucleotide binding domain; PKA, cAMP-dependent kinase; REFER, rate equilibrium-free energy relationship; DTT, dithiothreitol; mAb, monoclonal antibody; AMP-PNP, adenosine 5′-(β,γ-imino)triphosphate.
J. R. Riordan, unpublished results.
References
- 1.Quinton, P. M. (2007) Physiology (Bethesda) 22 212–225 [DOI] [PubMed] [Google Scholar]
- 2.Matsui, H., Randell, S. H., Peretti, S. W., Davis, C. W., and Boucher, R. C. (1998) J. Clin. Investig. 102 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarran, R., Grubb, B. R., Parsons, D., Picher, M., Hirsh, A. J., Davis, C. W., and Boucher, R. C. (2001) Mol. Cell 8 149–158 [DOI] [PubMed] [Google Scholar]
- 4.Joo, N. S., Lee, D. J., Winges, K. M., Rustagi, A., and Wine, J. J. (2004) J. Biol. Chem. 279 38854–38860 [DOI] [PubMed] [Google Scholar]
- 5.Cheng, S. H., Gregory, R. J., Marshall, J., Paul, S., Souza, D. W., White, G. A., O'Riordan, C. R., and Smith, A. E. (1990) Cell 63 827–834 [DOI] [PubMed] [Google Scholar]
- 6.Ward, C. L., Omura, S., and Kopito, R. R. (1995) Cell 83 121–127 [DOI] [PubMed] [Google Scholar]
- 7.Serohijos, A. W., Hegedus, T., Riordan, J. R., and Dokholyan, N. V. (2008) PLoS Comput. Biol. 4 e1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du, K., Sharma, M., and Lukacs, G. L. (2005) Nat. Struct. Mol. Biol. 12 17–25 [DOI] [PubMed] [Google Scholar]
- 9.Cui, L., Aleksandrov, L., Chang, X. B., Hou, Y. X., He, L., Hegedus, T., Gentzsch, M., Aleksandrov, A., Balch, W. E., and Riordan, J. R. (2007) J. Mol. Biol. 365 981–994 [DOI] [PubMed] [Google Scholar]
- 10.Lewis, H. A., Zhao, X., Wang, C., Sauder, J. M., Rooney, I., Noland, B. W., Lorimer, D., Kearins, M. C., Conners, K., Condon, B., Maloney, P. C., Guggino, W. B., Hunt, J. F., and Emtage, S. (2005) J. Biol. Chem. 280 1346–1353 [DOI] [PubMed] [Google Scholar]
- 11.Aleksandrov, L., Aleksandrov, A. A., Chang, X. B., and Riordan, J. R. (2002) J. Biol. Chem. 277 15419–15425 [DOI] [PubMed] [Google Scholar]
- 12.Aleksandrov, A. A., Aleksandrov, L. A., and Riordan, J. R. (2007) Pflugers. Arch. 453 693–702 [DOI] [PubMed] [Google Scholar]
- 13.Winter, M. C., and Welsh, M. J. (1997) Nature 389 294–296 [DOI] [PubMed] [Google Scholar]
- 14.Dawson, R. J., and Locher, K. P. (2006) Nature 443 180–185 [DOI] [PubMed] [Google Scholar]
- 15.Hollenstein, K., Frei, D. C., and Locher, K. P. (2007) Nature 446 213–216 [DOI] [PubMed] [Google Scholar]
- 16.Locher, K. P., Lee, A. T., and Rees, D. C. (2002) Science 296 1091–1098 [DOI] [PubMed] [Google Scholar]
- 17.Pinkett, H. W., Lee, A. T., Lum, P., Locher, K. P., and Rees, D. C. (2007) Science 315 373–377 [DOI] [PubMed] [Google Scholar]
- 18.Oldham, M. L., Khare, D., Quiocho, F. A., Davidson, A. L., and Chen, J. (2007) Nature 450 515–521 [DOI] [PubMed] [Google Scholar]
- 19.Serohijos, A. W., Hegedus, T., Aleksandrov, A. A., He, L., Cui, L., Dokholyan, N. V., and Riordan, J. R. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 3256–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seibert, F. S., Linsdell, P., Loo, T. W., Hanrahan, J. W., Clarke, D. M., and Riordan, J. R. (1996) J. Biol. Chem. 271 15139–15145 [DOI] [PubMed] [Google Scholar]
- 21.Seibert, F. S., Jia, Y., Mathews, C. J., Hanrahan, J. W., Riordan, J. R., Loo, T. W., and Clarke, D. M. (1997) Biochemistry 36 11966–11974 [DOI] [PubMed] [Google Scholar]
- 22.Seibert, F. S., Linsdell, P., Loo, T. W., Hanrahan, J. W., Riordan, J. R., and Clarke, D. M. (1996) J. Biol. Chem. 271 27493–27499 [DOI] [PubMed] [Google Scholar]
- 23.Chang, X.-B., Tabcharani, J. A., Hou, Y.-X., Jensen, T. J., Kartner, N., Alon, N., Hanrahan, J. W., and Riordan, J. R. (1993) J. Biol. Chem. 268 11304–11311 [PubMed] [Google Scholar]
- 24.Cui, L., Aleksandrov, L., Hou, Y. X., Gentzsch, M., Cen, J. H., Riordan, J. R., and Aleksandrov, A. A. (2006) J. Physiol. 572 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward, A., Reyes, C. L., Yu, J., Roth, C. B., and Chang, G. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis, H. A., Buchanan, S. G., Burley, S. K., Conners, K., Dickey, M., Dorwart, M., Fowler, R., Gao, X., Guggino, W. B., Hendrickson, W. A., Hunt, J. F., Kearins, M. C., Lorimer, D., Maloney, P. C., Post, K. W., Rajashankar, K. R., Rutter, M. E., Sauder, J. M., Shriver, S., Thibodeau, P. H., Thomas, P. J., Zhang, M., Zhao, X., and Emtage, S. (2004) EMBO J. 23 282–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaitseva, J., Jenewein, S., Wiedenmann, A., Benabdelhak, H., Holland, I. B., and Schmitt, L. (2005) Biochemistry 44 9680–9690 [DOI] [PubMed] [Google Scholar]
- 28.Hung, L. W., Wang, I. X., Nikaido, K., Liu, P. Q., Ames, G. F., and Kim, S. H. (1998) Nature 396 703–707 [DOI] [PubMed] [Google Scholar]
- 29.Dalmas, O., Orelle, C., Foucher, A. E., Geourjon, C., Crouzy, S., Di Pietro, A., and Jault, J. M. (2005) J. Biol. Chem. 280 36857–36864 [DOI] [PubMed] [Google Scholar]
- 30.Baker, J., Kanelis, V., Hudson, R. P., Thibodeau, P. H., Choy, W.-Y., Sprangers, R., Dorwart, M. R., Thomas, P. J., and Forman-Kay, J. D. (2006) Peds. Pulmonol. Suppl. 29 97–99 [Google Scholar]
- 31.Smith, P. C., Karpowich, N., Millen, L., Moody, J. E., Rosen, J., Thomas, P. J., and Hunt, J. F. (2002) Mol. Cell 10 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mense, M., Vergani, P., White, D. M., Altberg, G., Nairn, A. C., and Gadsby, D. C. (2006) EMBO J. 25 4728–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zolnerciks, J. K., Wooding, C., and Linton, K. J. (2007) FASEB J. 21 3937–3948 [DOI] [PubMed] [Google Scholar]
- 34.Younger, J. M., Chen, L., Ren, H. Y., Rosser, M. F., Turnbull, E. L., Fan, C. Y., Patterson, C., and Cyr, D. M. (2006) Cell 126 571–582 [DOI] [PubMed] [Google Scholar]
- 35.Callebaut, I., Eudes, R., Mornon, J. P., and Lehn, P. (2004) Cell Mol. Life Sci. 61 230–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, E. Y., Bartlett, M. C., Loo, T. W., and Clarke, D. M. (2004) J. Biol. Chem. 279 39620–39627 [DOI] [PubMed] [Google Scholar]
- 37.Wang, Y., Loo, T. W., Bartlett, M. C., and Clarke, D. M. (2007) J. Biol. Chem. 282 33247–33251 [DOI] [PubMed] [Google Scholar]
- 38.Thibodeau, P. H., Brautigam, C. A., Machius, M., Tomchik, D. R., and Thomas, P. J. (2004) Ped. Pulmonol. Suppl. 27 189 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.