Abstract
Cholesterol is required for normal cellular and physiological function, yet dysregulation of cholesterol metabolism is associated with diseases such as atherosclerosis. Cholesterol biosynthesis is regulated by end product negative feedback inhibition where the levels of sterols and oxysterols regulate the expression of cholesterologenic enzymes. Sterol regulatory element-binding protein-2 is responsive to both sterols and oxysterols and has been shown to mediate the transcriptional response of the cholesterologenic enzymes to these lipids. Here, we show that the nuclear hormone receptor for oxysterols, the liver X receptor α (LXRα), regulates cholesterol biosynthesis by directly silencing the expression of two key cholesterologenic enzymes (lanosterol 14α-demethylase (CYP51A1), and squalene synthase (farnesyl diphosphate farnesyl transferase 1)) via novel negative LXR DNA response elements (nLXREs) located in each of these genes. Examination of the CYP51A1 gene revealed that both the SRE and nLXRE are required for normal oxysterol-dependent repression of this gene. Thus, these data suggest that LXRα plays an important role in the regulation of cholesterol biosynthesis.
Cholesterol is essential for maintenance of cellular membrane fluidity and is a precursor for the production of steroid hormones and bile acids. Although cholesterol is required for normal cellular and physiological function, dysregulation of cholesterol metabolism is associated with atherosclerosis and increased risk of heart disease (1). Thus, cholesterol metabolism is under strict biological regulation, and drugs inhibiting cholesterol biosynthesis such as the statins, inhibitors of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR)2 (see Fig. 1a), have been successfully utilized clinically to reduce the risk of atherosclerosis. The biosynthesis of cholesterol has long been known to be regulated by end product feedback inhibition, and this regulation has been attributed to direct regulation of the expression of several cholesterol biosynthetic genes by the sterol sensing sterol regulatory element-binding protein-2 (SREBP-2) (2). The SREBPs are responsive to both alterations in cholesterol levels as well as oxidized cholesterol metabolites known as oxysterols (3).
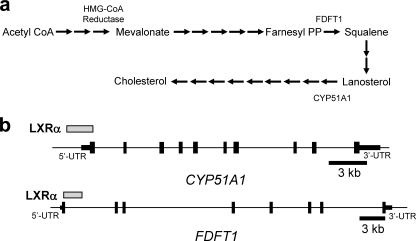
FIGURE 1.
Identification of LXRα occupancy sites in the regulatory regions of cholesterol biosynthetic enzyme genes. a, schematic outlining the cholesterol biosynthetic process identifying the position of HMGCR, FDFT1, and CYP51A1 in the process. b, position of LXRα occupancy within the CYP51A1 and FDFT1 genes as determined by ChIP-on-chip.
The nuclear hormone receptors LXRα (NR1H3) and LXRβ (NR1H2) have been demonstrated to be physiological receptors for oxysterols (4–6) and regulate components of the cholesterol metabolic pathway including reverse cholesterol transport and cholesterol elimination (7). No role for the LXRs in direct regulation of cholesterol biosynthesis has been described. A recent study by Wong et al. (8) suggested that 24,25-epoxycholesterol was important for “fine-tuning” acute control of cellular cholesterol biosynthesis. Because this endogenous oxysterol is known to be one of the more potent natural oxysterol ligands for the LXRs, it is possible that some of its activity may be mediated by these receptors. Here, we show that two key cholesterologenic enzyme genes are LXR target genes and that LXR plays a role in the regulation of cholesterol biosynthesis.
EXPERIMENTAL PROCEDURES
Plasmid Construction—The CYP51A1 promoter (–2217 to +155) was amplified from genomic DNA isolated from HepG2 cells and cloned into pTAL-Luc luciferase report vector (Clontech, CA) by introducing KpnI and BglII into the primers. The proximal portion (–2217 to –1749) containing LXRE and the distal portion (–1749 to +155) containing the SRE of the promoter were amplified from the CYP51A1 promoter vector and cloned into pTAL-Luc vector using the same restriction sites. Three copies of CYP51A1 LXRE, FDFT1 LXRE, and NR1H3 LXRE were cloned into pTAL-Luc through MluI and BglII. LXRα and RXRα were cloned into pDEST14 (Invitrogen) using Gateway® technology (Invitrogen) for EMSA analysis. LXRα and LXRβ were cloned into pcDNA3.1 vector for overexpression analysis. The LXRE mutant CYP51A1 promoter reporter was created using QuikChange® II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Cell Culture and Cotransfections—HepG2 cells were maintained and routinely propagated in minimum essential medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. HEK293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. In experiments where lipids and sterols were depleted, cells were maintained on charcoal treated serum (10% fetal bovine serum) and treated with 7.5 μm lovastatin and 100 μm mevalonic acid. 24 h prior to transfection, HepG2 or HEK293 cells were plated in 96-well plates at a density of 15 × 103 cells/well. Each transfection contained 100 ng of the pTAL-Luc reporter, 50 ng of pGL4.73 reporter, and 50 ng of receptor (as indicated) as described in the figure legend using Lipofectamine™ 2000 (Invitrogen). 16 h post-transfection, the cells were treated with LXR ligands. 24 h post-treatment, the luciferase activity was measured using the Dual-Glo™ luciferase assay system (Promega). The values indicated represent the means ± S.E. from three independently transfected wells. The experiments were repeated at least three times, and representative experiments are shown.
mRNA and Protein Measurement—cDNA synthesized from mRNA was quantified by quantitative PCR analysis using SYBR green technology. The primers for quantitative PCR are: CYP51A1, CAGAACTCCTCAGACTGTGG (forward) and GTCTTTGATTGACAGTGGGA (reverse).; FDFT1, CCGTGACTATCTGGAAGAC (forward) and CATACCTGCTCCAAACCTC (reverse); LXRα, GGAGGTACAACCCTGGGAGT (forward) and AGCAATGAGCAAGGCAAACT (reverse).
CYP51A1, FDFT1 HMG-CoA, and LXRα protein levels were assessed by Western blot using antibodies from Abnova (Taiwan). LXRα and control siRNA was obtained from Ambion (Austin, TX). The transfection was performed using siPORT™ NeoFX SiRNA transfection reagents in accordance with the manufacturer's instructions.
Cholesterol Quantification and Biosynthesis Measurement—Cholesterol quantification was performed using a cholesterol/cholesterol ester quantification kit (BioVision). The cholesterol level was normalized to protein content. Cholesterol de novo biosynthesis was measured by TLC. HepG2 cells were treated with siRNA for 45 h, and the media were replaced with growth media supplemented with 1 μCi/ml of 14C sodium acetate (Amersham Biosciences). After 3 h of incubation at 37 °C under 5% CO2, total lipid was extracted and dissolved in chloroform:methanol (2:1). TLC was performed in heptane:isopropylether:acetic acid (160 ml/40 ml/2 ml) for 1 h. The TLC plate was scanned using Bioscan AR2000 Imaging Scanner (Bioscan, Washington, D. C.). The number of counts/min was normalized to protein content.
ChIP-on-chip—ChIP-on-chip procedures were performed as previously described (9).
Microarray—Microarray identification of genes altered by knock-down of LXR expression in mice was previously described (10).
Electrophoretic Mobility Shift Assays—LXRα and RXRα were expressed using TnT® quick coupled transcription/translation systems (Promega). EMSAs were performed using the [α-33P]dATP-labeled CYP51A1 and FDFT1 LXRE oligonucleotide. Competition assays were performed using various amounts of the unlabeled CYP51A1, FDFT1, ABCA1, and mutant CYP51A1 oligonucleotide as previously described (5×, 25×, and 100× molar excess) (11).
HMG-CoA Reductase Enzyme Assay—HMG-CoA reductase activity was measured by spectrophotometric assay as described by Takahashi et al. (12). The reaction buffer consisted of 25 mm potassium phosphate (pH 7.2), 50 mm KCl, 1 mm EDTA, and 5 mm dithiothreitol. Control and siRNA-treated HepG2 cells were cultivated in 6-well plates. After treatment, the cells were harvested in 100 μl of reaction buffer/well. The assay was performed in reaction buffer with 0.3 mm NADPH, 0.3 mm R,S-HMG-CoA and 2 μl protein in a final volume of 200 μl. The kinetics program in the Benchmark Plus™ microplate spectrophotometer was used, and the reaction was monitored every 20 s for up to 10 min at 37 °C. One unit of HMG-CoA reductase activity is defined as the amount of the enzyme that caused oxidation of 1 μmol of NADPH/min at 37 °C.
RESULTS
Identification of Novel LXRα Target Genes—To identify novel biochemical pathways regulated by LXRα, we performed LXRα chromatin immunoprecipitation followed by whole genome analysis of the enriched DNA fragments by microarray experiment (ChIP-on-chip) (9). Regions of the genome with significant LXRα occupancy (termed promising active regions (PARs)) were screened for putative LXREs using a previously described algorithm (13, 14). PARs were then queried against the human genome data base, and 1304 unique genes were identified with a PAR within 1 kb of a gene. We then compared these data with microarray data obtained from the livers of mice treated with a LXR-directed antisense oligonucleotide that significantly reduced LXRα expression (10). This allowed us to identify genes that were significantly altered by LXRα knock-down that also demonstrated LXRα occupancy and contained a defined LXRE. Fifty-seven genes were identified as having met these criteria and included the well characterized LXR target genes ABCA1 and NR1H3, indicating that the method was successful. Upon closer examination of the putative target genes, it was noted that there were examples of genes whose expression was increased in response to a decrease in LXRα expression, suggesting that LXRα may play a direct repressive role in regulation of these genes. We found this particularly intriguing and novel given that LXR has not been previously characterized as a direct silencer of gene expression.
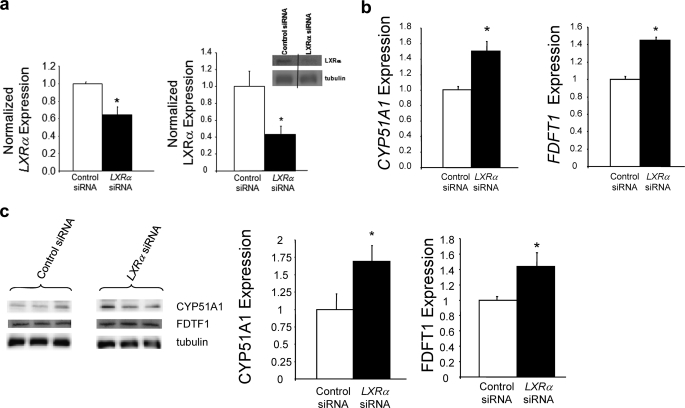
Interestingly, two of the negatively regulated genes encode key enzymes within the cholesterol biosynthesis pathway. The two genes identified were squalene synthase (FDFT1) and lanosterol 14α-demethylase (CYP51A1), and their positions within the cholesterologenic pathway are illustrated in Fig. 1a. Both have regions of significant LXRα binding (spanning 2–3 kb) as determined by ChIP-on-chip either in the 5′ region or in the intronic region of the genes (Fig. 1b). To investigate the putative role of LXRα in regulation of cholesterologenesis, we decreased LXRα expression in the human hepatoma cell line, HepG2, by 37% (LXRα protein decreased 56%) using specific siRNA (Fig. 2a) (9). As illustrated in Fig. 2b, decreasing LXRα expression led to a significant increase in expression of both identified genes. CYP51A1 and FDFT1 expression increased by ∼50 and ∼42%, respectively. Protein levels of CYP51A1 and FDFT1 were also examined following LXRα knock-down and as illustrated in Fig. 2c, CYP51A1 expression increased ∼70% and FDFT1 increased ∼42%. These data suggest that the dominant influence of LXRα on the expression of these genes is negative because “removal” of a significant amount of LXRα results in increased expression of these genes.
FIGURE 2.
Regulation of expression of CYP51A1 and FDFT1 expression by LXRα. a, specific LXRα siRNA treatment depletes both LXRα mRNA and LXRα protein expression in HepG2 cells. b, LXRα depletion results in increased expression of CYP51A1 and FDFT1 mRNA in HepG2 cells. C, LXRα depletion results in increased expression of CYP51A1 and FDFT1 protein. A Western blot showing LXRα expression levels from an example experiment is illustrated. The lanes are from the same gel, and we treated and analyzed it in an identical manner. *, p < 0.05 versus control as determined by Student's t test.
Regulation of Cholesterologenic Enzyme Expression by LXRα—Both FDFT1 and CYP51A1 encode enzymes that are essential for cholesterologenesis. FDFT1 encodes the enzyme squalene synthase that catalyzes the production of squalene from farnesyl phyrophosphate and is the first committed step in sterol synthesis (Fig. 1a). Inhibitors of this enzyme have been shown to be effective inhibitors of cholesterol synthesis (15, 16). CYP51A1 encodes lanosterol 14α-demethylase, which catalyzes removal of the 14α-methyl group from the cholesterol intermediate lanosterol (17) (Fig. 1a). Inhibition of CYP51A1 activity by ketoconazole in humans results in a 27% decrease in total serum cholesterol and a 41% decrease in serum low density lipoprotein levels, illustrating the importance of this enzyme in the cholesterol biosynthesis pathway (18).
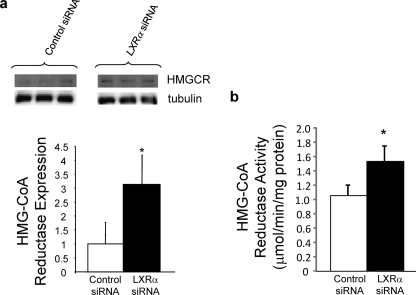
Inhibition of CYP51A1 activity results in accumulation of lanosterol in humans (18), and lanosterol has been suggested to regulate HMGCR proteasomal degradation (19). A more recent study indicated that dihydrolanosterol and not lanosterol is responsible for regulation of HMGCR proteosomal degradation (20). Because CYP51A1 utilizes both dihydrolanosterol and lanosterol as a substrate (17), regulation of CYP51A1 activity has similar effects on the levels of both of these sterols (18), and in fact, it appears that inhibition of CYP51A1 activity has a much larger effect on the level of dihydrolanosterol relative to lanosterol (18). Thus, we hypothesized that LXRα may regulate HMGCR expression via modulation of dihydrolanosterol levels by its direct ability to repress CYP51A1 expression. Indeed, we found that depletion of LXRα expression resulted in a 3-fold increase in HMGCR expression (Fig. 3a) and a 46% increase in HMGCR activity (Fig. 3b). Thus, LXRα regulates the expression of three key enzymes in the cholesterol biosynthesis pathway.
FIGURE 3.
Regulation of HMGCR expression and activity by LXRα. a, LXRα depletion causes an increase in HMGCR protein expression in HepG2 cells. b, LXRα depletion results in an increase in HMGCR activity in HepG2 cells. *, p < 0.05 versus control as determined by Student's t test.
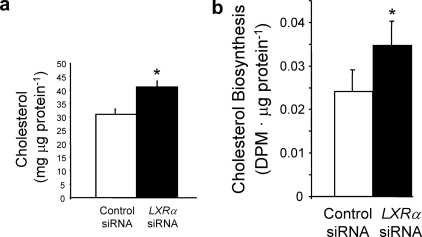
LXRα Is Required for Normal Levels of Cholesterologenesis—Based on these data, we predicted that LXRα knock-down would result in increased cholesterol content in HepG2 cells. Fig. 4a shows that this is indeed the case. When LXRα expression is decreased by 56%, intracellular cholesterol levels increase ∼30%. We also examined de novo cholesterologenesis in this model and noted a 44% increase in cholesterol biosynthesis in LXRα-depleted cells (Fig. 4b). These data are consistent with a model where LXRα regulates the expression of CYP51A1 and FDFT1 and the stability of HMGCR, resulting in modulation of cholesterol biosynthesis.
FIGURE 4.
Regulation of cholesterol biosynthesis by LXRα. a, LXRα depletion increases cholesterol content in HepG2 cells. b, LXRα depletion results in an increase in cholesterol biosynthesis in HepG2 cells. *, p < 0.05 versus control as determined by Student's t test.
Identification of Negative LXR DNA Response Elements—Because CYP51A1 and FDFT1 were identified based on LXRα occupancy in ChIP-on-chip analysis in addition to the requirement for a putative LXRE within the occupancy site, these data indicate that these may be the first observations of “negative” LXREs in the genome. Analysis of the PARs from CYP51A1 and FDFT1 (Fig. 1b) by the LXRα ChIP-on-chip study revealed putative LXREs. We performed EMSAs on all putative LXREs and identified two LXREs (one in each gene) that bound to the LXRα/RXRα heterodimer effectively (Fig. 5). The novel LXREs exhibited similarity to other well characterized “positive” LXREs identified in the ABCA1 and NR1H3 genes (Fig. 2a). One is a DR4 element, whereas other is a very unique DR2 element. No LXREs with a DR2 organization have been previously identified. RXRα heterodimerization was required for LXRα binding to both novel LXREs (Fig. 5b). To assess the functionality of these novel LXREs, we cloned three copies of each upstream of a minimal promoter-luciferase reporter construct and examined their activities in the absence and presence of either synthetic (GW3965 or T0901317 (T1317)) or natural (oxysterol; OHC) LXR ligands in HepG2 cells. The positive LXRE and CYP51A1 and FDFT1 LXREs both affected basal transcription but with opposing effects (Fig. 6a). The NR1H3 positive LXRE increased transcription by 200%, whereas the two other LXREs decreased basal transcription by 42–56% (Fig. 6a). The addition of ligands had no effect on basal transcription from a reporter lacking a LXRE (data not shown). In cells transfected with the positive LXRE, a further increase in transcription was noted with treatment with either of the two synthetic ligands (5–6-fold induction) (Fig. 6b). Both LXREs derived from the CYP51A1 and FDFT1 showed minimal responsiveness to the synthetic LXR ligands (Fig. 6, c and d), and quite novel was our observation that the nLXRE reporters from each of the both cholesterologenic enzyme genes conferred oxysterol-dependent repression of transcription to the reporter construct (Fig. 6, c and d). We termed these LXREs “negative” LXREs (nLXREs) based on their ability to confer negative responsiveness to LXR. Both of these nLXREs were responsive to oxysterols that are known agonists of LXR such as 24,25-epoyxycholesterol and 25-hydroxycholesterol but were unresponsive to well characterized agonist such as 22(R)-hydroxycholesterol. This unusual ligand selectivity, taken together with their relative lack of responsiveness to synthetic LXR ligands, indicates that these novel nLXREs may confer differential ligand sensitivity to the receptor.
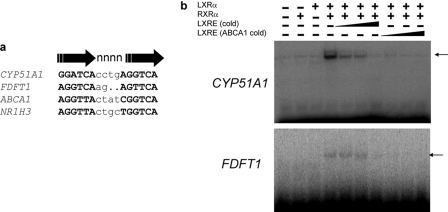
FIGURE 5.
Identification of LXR-binding sites (LXREs) within the CYP51A1 and FDFT1 genes. a, location and sequence of the putative LXRE identified in the CYP51A1 promoter and the first intron of FDFT1 compared with the LXREs of the NR1H3 and ABCA1 genes. b, electrophoretic mobility shift assay illustrating the ability of LXRα/RXRα to bind to the CYP51A1 and FDFT1 LXREs. A competition experiment was performed with the either the CYP51A1 or FDFT1 LXRE as the labeled probe, and the binding was competed with either cold CYP51A1 or FDFT1 oligonucleotide (5×, 25×, and 100× molar excess) or ABCA1 LXRE oligonucleotide (5×, 25×, and 100× molar excess).
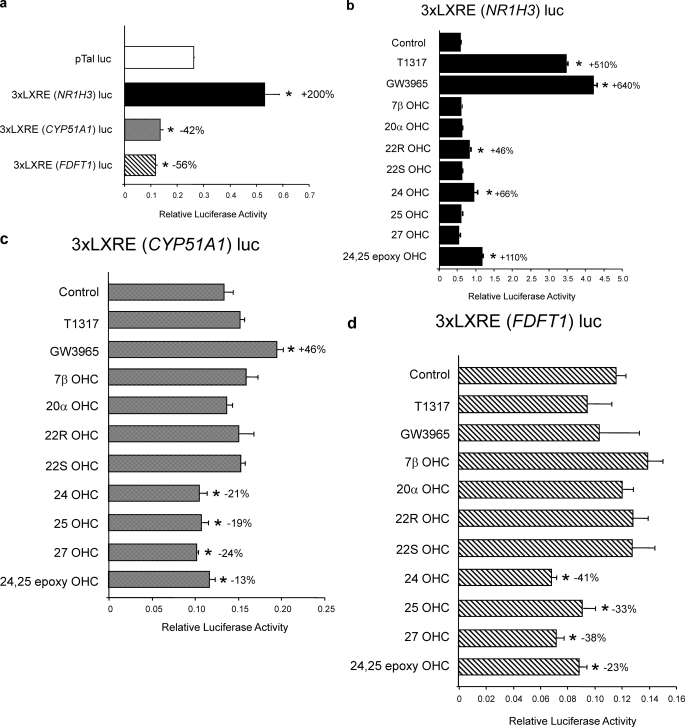
FIGURE 6.
The nLXREs from the CYP51A1 and FDFT1 genes confer oxysterol-dependent repression to a reporter gene in HepG2 cells. a, the nLXREs from both the CYP51A1 and FDFT1 genes mediate repression of basal transcription when cloned upstream of a minimal promoter luciferase reporter, whereas the positive LXRE mediates moderate stimulation of transcription in transfected HepG2 cells. b, effects of various LXR ligands on a luciferase reporter harboring three copies of the NR1H3 LXRE upstream of a minimal promoter when transfected in to HepG2 cells. c, effects of various LXR ligands on a luciferase reporter harboring three copies of the CYP51A1 LXRE upstream of a minimal promoter when transfected in to HepG2 cells. d, effects of various LXR ligands on a luciferase reporter harboring three copies of the FDFT1 LXRE upstream of a minimal promoter when transfected in to HepG2 cells. All of the experiments within this figure utilized lipid depleted media as described under “Experimental Procedures.” Synthetic LXR ligands T1317 and GW3965 were tested at 1 μm, and the oxysterols were tested at 10 μm. Me2SO concentrations in all wells were held constant at 0.1%. *, p < 0.05 versus control as determined by Student's t test.
We also examined the activity of these three LXRE containing reporter constructs in HEK293 cells. Like HepG2 cells, HEK293 cells also express LXR, and the LXR responsiveness of reporter genes can be enhanced by overexpression of LXR (21). We transfected the reporters into HEK293 cells in the absence or presence of a vector directing the overexpression of either LXRα or LXRβ. As illustrated in Fig. 7a, the reporter containing the positive LXREs from the NR1H3 gene displayed enhanced transcription (53%), and overexpression of either LXR resulted in further enhancement of transcription (Fig. 7a). Consistent with our data from the HepG2 cells, transfection of the cells with the reporters containing the nLXREs derived from the CYP51A1 or FDFT1 genes led to decreased basal transcription (44–50%) that was further suppressed upon overexpression of either LXRα or LXRβ. We next examined the ability of LXR ligands to alter the transcriptional activity of LXRα transfected into HEK293 cells along with the nLXRE reporters derived from either the CYP51A1 or FDFT1 genes. The results were very similar to those obtained in HepG2 cells in that the identical oxysterols that repressed transcription in HepG2 cells also repressed in HEK293 cells (Fig. 7, b and c). These data are consistent with our observations that the nLXREs mediate LXR-dependent repression of transcription.
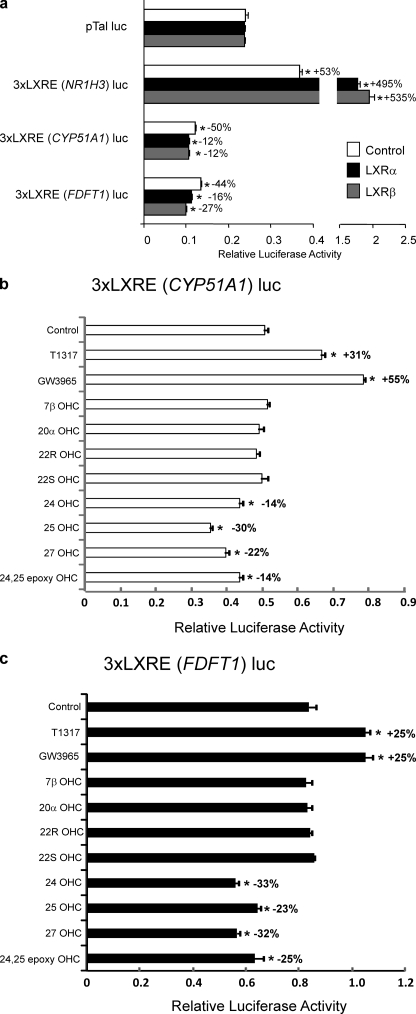
FIGURE 7.
The nLXREs from the CYP51A1 and FDFT1 genes confer oxysterol-dependent repression to a reporter gene in HEK293 cells. a, HEK 293 cells were transfected with either LXRα or LXRβ and reporter genes containing either a positive LXRE or negative LXREs. The nLXREs from both the CYP51A1 and FDFT1 genes mediate repression of basal transcription when cloned upstream of a minimal promoter luciferase reporter, whereas the positive LXRE mediates stimulation of transcription. Control cells received empty expression vector instead of a vector directing the expression of LXR. b, effects of various LXR ligands on a luciferase reporter harboring three copies of the CYP51A1 LXRE upstream of a minimal promoter when transfected in HEK293 cells along with LXRα. c, effects of various LXR ligands on a luciferase reporter harboring three copies of the FDFT1 LXRE upstream of a minimal promoter when transfected in to HEK293 cells along with LXRα. All of the experiments within this figure utilized lipid depleted medium as described under “Experimental Procedures.” Synthetic LXR ligands T1317 and GW3965 were tested at 1 μm, and the oxysterols were tested at 10 μm. Me2SO concentrations in all wells were held constant at 0.1%. *, p < 0.05 versus control as determined by Student's t test.
Both the nLXRE and the SRE Are Required for Oxysterol Regulation of CYP51A1 Expression—Because the CYP51A1 promoter contains a SRE that is also responsive to oxysterols via SREBP, we assessed the LXRα-derived component of repression by depleting LXRα expression with siRNA and examining the 25-OHC responsiveness in HepG2 cells cultured in lipid and oxysterol-depleted conditions. Depletion of LXRα resulting in a significant loss of the repressive effects of 25-OHC, suggesting that a significant component of the repressive effects is mediated by this receptor (p < 0.05) (Fig. 8a). Based on previous publications illustrating that SREBP-1a/2 plays an important role in CYP51A1 expression (22) as well as our data indicating that LXRα also is important, we hypothesized that both the nLXRE and the SRE play a role in oxysterol responsiveness of the gene. To investigate this, we constructed a CYP51A1 promoter reporter by cloning ∼2kbofthe CYP51A1 promoter upstream of a luciferase reporter and as shown in Fig. 3b. When this construct was transfected into HepG2 cells, transcription is repressed ∼20% relative to the control (Fig. 8b). Treatment with 25-OHC results in further repression (∼75% relative to control; Fig. 8b). We manufactured CYP51A1 promoter-reporter constructs in which we deleted the nLXRE or the SRE as illustrated in Fig. 8b. Interestingly, removal of the nLXRE resulted in an increase in transcription (Fig. 8b), suggesting a basal repressive function of this element consistent with what we have seen in heterologous systems such as our previous cotransfection studies (Figs. 6 and 7). 25-OHC treatment represses transcription driven by this CYP51A1 promoter fragment containing only the SRE, but not to the same absolute level as that observed when both the nLXRE and the SRE elements were present (Fig. 8b). Deletion of the SRE results in basal expression similar to control and 25-OHC treatment represses transcription, but the magnitude of repression is clearly less efficacious than when the nLXRE is present. We examined the role of the nLXRE more closely by creating a mutant LXRE CYP51A1 promoter (Fig. 8c) where the LXRE is unable to bind to LXR. We confirmed that the mutant LXRE is unable to bind to LXRα in an EMSA (Fig. 8d). Both the wild type promoter and the LXRE mutant promoter were transfected into HepG2 cells, and the ability of 25-OHC to repress transcription was examined. As illustrated in Fig. 8e, mutation of the LXRE within the CYP51A1 promoter results in significant loss (but not entire loss) of the ability of the oxysterol to inhibit transcription. Thus, these data clearly indicate that both the nLXRE and SRE play a role in mediating the oxysterol responsiveness of transcription of this gene and additionally indicate that the nLXRE also has a basal repressor effect on transcription of this gene.
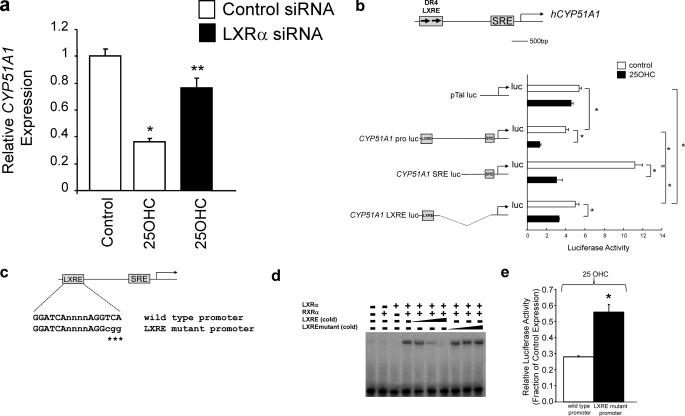
FIGURE 8.
Examination of the role of the nLXRE in the CYP51A1 promoter. a, LXRα depletion by siRNA treatment results in significant loss of the ability of 25-OHC to repress CYP51A1 expression in HepG2 cells. b, deletion analysis suggests that both the nLXRE and the SRE mediate a component of the ability of 25-OHC to repress transcription of a CYP51A1 promoter driven luciferase reporter in HepG2 cells. c, schematic illustrating the LXRE mutant that was created to eliminate LXR binding to the CYP51A1 promoter reporter construct. d, EMSA demonstrating that the mutant LXRE does not bind to LXRα. A competition experiment was performed with the wild type LXRE as the labeled probe, and the binding was competed with either cold wild type oligonucleotide (5×, 25×, and 100× molar excess) or mutant oligonucleotide (5×, 25×, and 100× molar excess). e, elimination of LXR binding results in reduced ability of 25-OHC to repress transcription of the CYP51A1 reporter. HepG2 cells were treated with 10 μm 25-OHC resulting in ∼72% repression of transcription of the wild type promoter but only ∼45% inhibition of transcription from the LXRE mutant CYP51A1 promoter. All of the experiments within this figure utilized lipid depleted medium as described under “Experimental Procedures.” Me2SO concentrations in all wells were held constant. *, p < 0.05 versus control as determined by Student's t test.
DISCUSSION
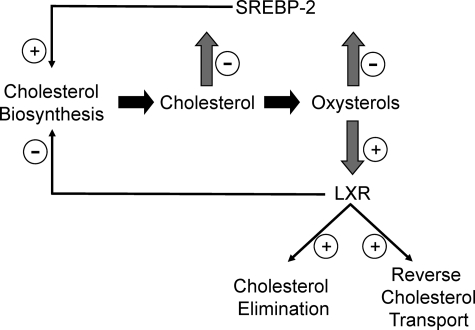
These findings indicate that, beyond the role of LXRα in the regulation of cholesterol transport and elimination, this nuclear receptor also plays an important role in end product inhibition of cholesterol biosynthesis (Fig. 9). It is interesting to note that the two enzymes whose expression is directly repressed by LXRα are in steps of cholesterologenesis where the pathway has committed to sterol synthesis. This is in contrast to SREBP-2, which regulates the expression of enzymes throughout the pathway. Our data suggest that LXRα works in concert with SREBP to regulate the expression of CYP51A1 in response to 25-OHC. 24,25-Epoxycholesterol has been recently shown to be an important for fine-tuning acute control of cellular cholesterol homeostatis (8). Because this endogenous oxysterol is one of the more potent natural ligands of LXR, it is possible that at least some of the activity of 24,25-epoxycholesterol described by these investigators may be mediated via the LXR-dependent mechanism we have described.
FIGURE 9.
Model illustrating the roles of LXR and SREBP in regulation of cholesterol biosynthesis.
All of the genes directly regulated by LXR described to date are under positive regulation by LXR (7) similar to the NR1H3 gene, indicating that the mode of regulation of CYP51A1 and FDFT1 is very unique. Promoter-specific transcriptional repression by unliganded LXR has been reported previously (23), but in this case the addition of an LXR ligand results in activation of transcription not further repression as we have noted for the CYP51A1 LXRE. A recent study indicates that LXRα may repress Ucp1 expression via a mechanism that involves competition with other transcription factors within the promoter(24). LXR has also been shown to repress the C-reactive protein gene, but it is not clear whether this is mediated by an LXRE (25). Additionally, LXR has been demonstrated to repress a number of genes involved in the inflammatory response utilizing a mechanism that has been attributed to antagonism of the NF-κB pathway (26). Thus, the CYP51A1 and FDFT1 LXREs represent a novel class of LXRE (nLXRE) that confers negative responsiveness to LXR and oxysterols. Additionally, the nLXRE is differentially responsive to distinct classes of LXR ligands (e.g. synthetic versus oxysterols). The mechanism responsible for the differential responsiveness is currently unclear, but it has been suggested that synthetic versus oxysterol binding to LXR results in different conformations of the receptor (27). Although our data suggest that these oxysterol-specific repressive effects are mediated via direct binding of the ligands to LXR because we observe dependence on LXR binding to the promoter, it is possible that other effects of these specific oxysterols may also be playing a role (e.g. oxysterol-dependent post-translational modification of the receptor).
It remains to be determined whether there are additional genes that contain a functional nLXRE. Because LXR is a putative target for the development of anti-atherogenic agents (28), the activity of synthetic LXR ligands on nLXRE containing genes such as CYP51A1 and FDFT1 must be considered. These results also suggest putative therapeutic potential of LXR agonists as inhibitors of cholesterol biosynthesis. Our data support a model for oxysterol regulation of cholesterol biosynthesis in which SREBP-2 increases cholesterologenic enzyme expression in response to decreasing oxysterol levels, whereas LXRα responds to increasing oxysterol levels to repress the expression of key cholesterologenic enzymes (Fig. 8). This mode of dual regulation of cholesterologenesis may provide for exceptionally sensitive and stringent control of cholesterol production where its levels must by delicately balanced to support cellular function but also avoid toxicity.
This work was supported, in whole or in part, by National Institutes of Health Grant DK080201 (to T. P. B.). This work was also supported by an award from the American Heart Association (to T. P. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HMGCR, 3-hydroxy-3-methylglutaryl-coenzyme A reductase; SRE, sterol regulatory element; SREBP, SRE-binding protein; FDFT, farnesyl diphosphate farnesyl transferase; EMSA, electrophoretic mobility shift assay; siRNA, small interfering RNA; ChIP, chromatin immunoprecipitation; PAR, promising active region; LXRE, LXR DNA response element; nLXRE, negative LXRE; OHC, oxysterol.
References
- 1.Maxfield, F. R., and Tabas, I. (2005) Nature 438 612–621 [DOI] [PubMed] [Google Scholar]
- 2.Goldstein, J. L., DeBose-Boyd, R. A., and Brown, M. S. (2006) Cell 124 35–46 [DOI] [PubMed] [Google Scholar]
- 3.Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002) J. Clin. Investig. 109 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janowski, B. A., Willy, P. J., Devi, T. R., Falck, J. R., and Mangelsdorf, D. J. (1996) Nature 383 728–731 [DOI] [PubMed] [Google Scholar]
- 5.Lehmann, J. M., Kliewer, S. A., Moore, L. B., SmithOliver, T. A., Oliver, B. B., Su, J. L., Sundseth, S. S., Winegar, D. A., Blanchard, D. E., Spencer, T. A., and Willson, T. M. (1997) J. Biol. Chem. 272 3137–3140 [DOI] [PubMed] [Google Scholar]
- 6.Chen, W. L., Chen, G. X., Head, D. L., Mangelsdorf, D. J., and Russell, D. W. (2007) Cell Metab. 5 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zelcer, N., and Tontonoz, P. (2006) J. Clin. Investig. 116 607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong, J., Quinn, C. M., Gelissen, I. C., and Brown, A. J. (2008) J. Biol. Chem. 283 700–707 [DOI] [PubMed] [Google Scholar]
- 9.Stayrook, K. R., Rogers, P. M., Savkur, R. S., Wang, Y., Su, C., Varga, G., Bu, X., Wei, T., Nagpal, S., Liu, X. S., and Burris, T. P. (2008) Mol. Pharmacol. 73 607–612 [DOI] [PubMed] [Google Scholar]
- 10.Hu, T. H., Foxworthy, P., Siesky, A., Ficorilli, J. V., Gao, H., Li, S. Y., Christe, M., Ryan, T., Cao, G. Q., Eacho, P., Michael, M. D., and Michael, L. F. (2005) Endocrinology 146 5380–5387 [DOI] [PubMed] [Google Scholar]
- 11.Burris, T. P., Guo, W. W., Le, T., and McCabe, E. R. B. (1995) Biochem. Biophys. Res. Commun. 214 576–581 [DOI] [PubMed] [Google Scholar]
- 12.Takahashi, S., Kuzuyama, T., and Seto, H. (1999) J. Bacteriol. 181 1256–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varga, G., and Su, C. (2007) BioDrugs 21 117–124 [DOI] [PubMed] [Google Scholar]
- 14.Sandelin, A., and Wasserman, W. W. (2005) Mol. Endocrinol. 19 595–606 [DOI] [PubMed] [Google Scholar]
- 15.Amin, D., Rutledge, R. Z., Needle, S. N., Galczenski, H. F., Neuenschwander, K., Scotese, A. C., Maguire, M. P., Bush, R. C., Hele, D. J., Bilder, G. E., and Perrone, M. H. (1997) J. Pharmacol. Exp. Ther. 281 746–752 [PubMed] [Google Scholar]
- 16.Ugawa, T., Kakuta, H., Moritani, H., Matsuda, K., Ishihara, T., Yamaguchi, M., Naganuma, S., Iizumi, Y., and Shikama, H. (2000) Br. J. Pharmacol. 131 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lepesheva, G. I., and Waterman, M. R. (2004) Mol. Cell. Endocrinol. 215 165–170 [DOI] [PubMed] [Google Scholar]
- 18.Miettinen, T. A. (1988) J. Lipid Res. 29 43–51 [PubMed] [Google Scholar]
- 19.Song, B. L., Javitt, N. B., and DeBose-Boyd, R. A. (2005) Cell Metab. 1 179–189 [DOI] [PubMed] [Google Scholar]
- 20.Lange, Y., Ory, D. S., Ye, J., Lanier, M. H., Hsu, F. F., and Steck, T. L. (2008) J. Biol. Chem. 283 1445–1455 [DOI] [PubMed] [Google Scholar]
- 21.Yoshikawa, T., Ide, T., Shimano, H., Yahagi, N., Amemiya-Kudo, M., Matsuzaka, T., Yatoh, S., Kitamine, T., Okazaki, H., Tamura, Y., Sekiya, M., Takahashi, A., Hasty, A. H., Sato, R., Sone, H., Osuga, J., Ishibashi, S., and Yamada, N. (2003) Mol. Endocrinol. 17 1240–1254 [DOI] [PubMed] [Google Scholar]
- 22.Halder, S. K., Fink, M., Waterman, M. R., and Rozman, D. (2002) Mol. Endocrinol. 16 1853–1863 [DOI] [PubMed] [Google Scholar]
- 23.Wagner, B. L., Valledor, A. F., Shao, G., Daige, C. L., Bischoff, E. D., Petrowski, M., Jepsen, K., Baek, S. H., Heyman, R. A., Rosenfeld, M. G., Schulman, I. G., and Glass, C. K. (2003) Mol. Cell Biol. 23 5780–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, H., Zhang, Y., Yehuda-Shnaidman, E., Medvedev, A. V., Kumar, N., Daniel, K. W., Robidoux, J., Czech, M. P., Mangelsdorf, D. J., and Collins, S. (2008) Mol. Cell Biol. 28 2187–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blaschke, F., Takata, Y., Caglayan, E., Collins, A., Tontonoz, P., Hsueh, W. A., and Tangirala, R. K. (2006) Circ. Res. 99 e88–e99 [DOI] [PubMed] [Google Scholar]
- 26.Joseph, S. B., Castrillo, A., Laffitte, B. A., Mangelsdorf, D. J., and Tontonoz, P. (2003) Nat. Med. 9 213–219 [DOI] [PubMed] [Google Scholar]
- 27.Albers, M., Blume, B., Schlueter, T., Wright, M. B., Kober, I., Kremoser, C., Deuschle, U., and Koegl, M. (2006) J. Biol. Chem. 281 4920–4930 [DOI] [PubMed] [Google Scholar]
- 28.Michael, L. F., Schkeryantz, J. M., and Burris, T. P. (2005) Mini-Rev. Med. Chem. 5 729–740 [DOI] [PubMed] [Google Scholar]