Abstract
During endochondral ossification, chondrocytes undergo hypertrophic differentiation and die by apoptosis. The level of inorganic phosphate (Pi) elevates at the site of cartilage mineralization, and when chondrocytes were treated with Pi, they underwent rapid apoptosis. Gene silencing of the proapoptotic Bcl-2 homology 3-only molecule bnip3 significantly suppressed Pi-induced apoptosis. Conversely, overexpression of Bcl-xL suppressed, and its knockdown promoted, the apoptosis of chondrocytes. Bnip3 was associated with Bcl-xL in chondrocytes stimulated with Pi. Bcl-xL was expressed uniformly in the growth plate chondrocytes, whereas Bnip3 expression was exclusively localized in the hypertrophic chondrocytes. Finally, we generated chondrocyte-specific bcl-x knock-out mice using the Cre-loxP recombination system, and we provided evidence that the hypertrophic chondrocyte layer was shortened in those mice because of an increased apoptosis of prehypertrophic and hypertrophic chondrocytes, with the mice afflicted with dwarfism as a result. These results suggest the pivotal role of Bcl-2 family members in the regulation of chondrocyte apoptosis.
Endochondral ossification is an essential process for skeletal development, fracture healing, and pathologic conditions such as osteoarthritis and ectopic ossification (1). In this process, chondrocytes first proliferate and then differentiate into mature hypertrophic chondrocytes, which mineralize the surrounding matrix that is finally replaced by bone (1). There is controversy as to the cell fate of hypertrophic chondrocytes, and several studies have shown that they undergo apoptosis after terminal differentiation (2–4). Apoptosis is a form of programmed cell death that is characterized by specific morphological and biochemical features, and is tightly regulated by extracellular stimuli and intracellular signaling pathways (5). Morphologically, apoptosis is characterized by a series of structural changes in dying cells as follows: blebbing of the plasma membrane, condensation of the cytoplasm and the nucleus, and cellular fragmentation into membrane apoptotic bodies. Biochemically, apoptosis is characterized by the degradation of chromatin, initially into large fragments of 50–300 kb and subsequently into smaller fragments that are monomers and multimers of 200 bases. Not only does apoptosis regulate various aspects of the biological activity, but it also can trigger cancer, autoimmune diseases, and degenerative disorders (6). Several molecules such as Sox5, -6, and -9 and Runx2 have been reported to regulate proliferation and hypertrophic differentiation of chondrocytes (7–9). However, the physiologic and pathologic significance of chondrocyte apoptosis and key molecules that regulate this process remain to be elucidated.
Previous studies have shown the possible involvement of cytokines, such as tumor necrosis factor-α and Fas ligand, and hormones, such as glucocorticoids and parathyroid hormone-related peptide (PTHrP),2 in the localized hypoxia and increased generation of reactive oxygen species in chondrocyte apoptosis. Shapiro and co-workers (10–12) and Poole and co-workers (13) reported that the terminal differentiation of chondrocytes is accompanied by marked accumulation of extracellular phosphate ions, which may lead to chondrocyte apoptosis through a plasma membrane phosphate transporter mechanism. The importance of phosphate ions in chondrocyte apoptosis was also confirmed by Demay and co-workers (14), who reported that hypophosphatemia leads to impaired apoptosis of hypertrophic chondrocytes and subsequent expansion of the late hypertrophic chondrocyte layer in vitamin D receptor-null mice, which was reversed by feeding with a high phosphate diet. Although these reports indicate that Pi entry into the cells induces apoptosis in growth plate chondrocytes, the precise molecular mechanism that results in the apoptosis of chondrocytes is still an enigma.
There are two distinct signaling pathways of apoptosis in mammals. One is initiated by death receptors (death receptor pathways) (15), and the other is regulated by anti- and pro-apoptotic Bcl-2 family members and involves release of cytochrome c from mitochondria into the cytoplasm (mitochondrial pathways) (16, 17). The anti-apoptotic Bcl-2 family members include mammalian Bcl-2, Bcl-xL, and Mcl-1, and more than 20 pro-apoptotic Bcl-2 family proteins have been identified to date in mammals, which are divided into two groups as follows: multidomain members (Bax, Bak, Bok/Mtd, etc.) and BH3 domain-only members (Bid, Bad, Bim, Bik, Puma, Noxa, Bmf, Hrk, Bnip3, Nix, etc.) (18). Although the BH3-only family members display tissue-specific distribution patterns, multidomain pro-apoptotic members are ubiquitously expressed, indicating that BH3-only proteins play a tissue/cell-specific and a stimulus-specific role in apoptosis and that the other members play an essential role further downstream (18).
We report here that the balance between the anti-apoptotic Bcl-2 family member Bcl-xL and the pro-apoptotic family member Bnip3 critically regulates the apoptosis of terminally differentiated chondrocytes both in vitro and in vivo. We first showed that the Bcl-xL/Bnip3 axis plays an essential role in Pi-induced chondrocyte apoptosis in vitro. Subsequently, we generated mutant mice conditionally deficient in the bcl-x gene using the Cre-loxP recombination system, and we found that the hypertrophic chondrocyte layer was shortened in these mice because of increased apoptosis.
EXPERIMENTAL PROCEDURES
Plasmids and Viral Vectors—For the production of retrovirus, full-length cDNA of mouse bcl-xL, bcl-2, mcl-1, and bnip3 was amplified by PCR, subcloned into pCR-TOPO II vectors (Invitrogen), and inserted into pMx vectors (19). The production of the retroviral vectors was performed as reported previously (20). Briefly, Plat-E cells (2 × 106 cells) were plated in 60-mm dishes and transfected with 2 μg of pMx vector using FuGENE (Roche Applied Science) on the following day. After 24 h, the medium was replaced with fresh medium, which was collected and used as the retroviral supernatant 48 h after the transfection. The puromycin-resistant gene and blasticidin-resistant gene were inserted into a pMx vector for the selection of stable cells. For gene silencing, RNAi sequences were designed for each of the mouse Bcl-2 family genes. Targeting sequences used were as follows: GCGTTCAGTGATCTAACATCC for Bcl-xL; GGATGCCTTTGTGGAACTATA for Bcl-2; GGACTGGCTTGTCAAACAAAG for Mcl-1; GGTAGGACAGAAACTAGAT for Bim; GGGTCAGCTATTATCTCAA for Bid; GAGCCAAACCTGACCACTA for Puma; GGTGGAGAGTTCAATTAAG for Bik; GAAGAGAAGTTGAAAGTAT for Bnip3; GGAAGAAAGTAAGTCTGAT for Nix; GGAAGGAAGCAGAATTGTA for Hrk; GCTCAAAGAAAGATGGCTT for Bmf; and GCTACGTCCAGGAGCGCACCA for green fluorescent protein. RNAi expression vectors for these genes were constructed with piGENEmU6 vector (for mouse) (iGENE Therapeutics) as described (21). For retrovirus expressing RNAi, the U6 promoter and inserts in piGENE vectors were cloned into pMx vectors. The adenovirus vector expressing FADDDN, which lacks the N-terminal domain that is responsible for recruiting and activating caspase-8 at the death receptor complex (22), was created as described previously (7). Adenovirus vectors expressing bacterial Cre recombinase was a generous gift from Kojiro Ueki (University of Tokyo). Adenoviruses were amplified in HEK293 cells and purified with the AdenoX virus purification kit (Clontech). Viral titers were determined by the end point dilution assay, and the viruses were used at 50 multiplicities of infection.
DNA Transfection—For the gene transduction or knockdown of Bcl-2 family molecules, growth plate chondrocytes or ATDC5 cells (3 × 105 per well) were seeded onto 60-mm cell culture dishes. After 24 h, the cells were left untreated or treated with the retrovirus vectors together with Polybrene at 8 μg/ml. Thereafter, the medium was replaced with fresh medium containing 1 μg/ml of puromycin and/or 10 μg/ml of blasticidin until they became confluent. Selective overexpression or inhibition of Bcl-2 family member molecules was confirmed by real time RT-PCR or Western blotting.
Cell Culture—Growth plate chondrocytes were isolated from the ribs (excluding the sternum) of C57BL6 or bcl-xfl/fl mice embryos (E18.5). About 1 × 105 cells can be obtained from one mouse. They were cultured in high glucose Dulbecco's modified Eagle's medium (Sigma) containing 10% fetal bovine serum (Sigma) and 1% penicillin/streptomycin (Sigma). They are mainly composed of proliferating chondrocytes and were differentiated into hypertrophic chondrocytes and underwent apoptosis in the presence of Pi. Mouse chondrogenic ATDC5 cells were obtained from the RIKEN Cell Bank (Saitama, Japan). The cells were cultured in Dulbecco's modified Eagle's medium/F-12 (1:1) (Sigma) with 5% fetal bovine serum and 1% penicillin/streptomycin. To induce hypertrophic differentiation, ATDC5 cells were cultured in the presence of ITS supplement (10 μg/ml bovine insulin (I), 5.5 μg/ml human transferrin (T), and 5 ng/ml sodium selenite (S)) (Sigma) for 14 days. To induce apoptosis in ATDC5 cells, the medium was changed to α-minimum Eagle's medium, 5% fetal bovine serum supplemented with ascorbic acid 2-phosphate (0.05 mm) and phosphate (NaH2PO4) at concentrations of 0–20 mm for 24–48 h. These cells were maintained at 37 °C in a humidified 5% CO2, 95% air atmosphere. The medium was replaced every other day.
Cell Viability Assay—Cell viability was determined by Cell Count Kit-8 (Dojindo) to count the living cells. Briefly, the cells were placed (1 ×105 cells per well) in a 96-well plate. After incubation with the indicated concentrations of Pi for 24 h, 10 μl of kit reagent was added and incubated for an additional 3 h. Cell viability was determined by scanning the samples with a microplate reader at 450 nm.
Western Blotting—Western blot analysis was performed with cell extracts from growth plate chondrocytes or ATDC5 cells. Cells were washed twice with ice-cold phosphate-buffered saline, and proteins were extracted with an M-PER, NE-PER (Pierce), or ApoAlert cell fractionation kit (Clontech), according to the manufacturer's instructions. Protein concentrations of the cell lysates were measured with a Protein Assay kit II (Bio-Rad). For Western blot analysis, lysates were fractionated by SDS-PAGE with 7.5–15% Tris-glycine gradient gel or 15% Tris-glycine gel and transferred onto nitrocellulose membranes (Bio-Rad). After blocking with 6% milk/TBS-T, membranes were incubated with primary antibodies to Bcl-xL, Puma, Bik, Bid, cleaved caspase-3 and -7, cleaved lamin A (Cell Signaling Technology), Mcl-1, Hrk (Santa Cruz Biotechnology), Bcl-2, Bim (Pharmingen), Bnip3, Nix, Bmf, FLAG, β-actin (Sigma), cytochrome c, and Cox4 (Clontech), followed by horseradish peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (Promega). Immunoreactive bands were visualized with ECL Plus (Amersham Biosciences), according to the manufacturer's instructions. For co-immunoprecipitation, cells were lysed in 0.2% Nonidet P-40 isotonic buffer (0.2% Nonidet P-40, 142.5 mm KCl, 5 mm MgCl2, 1 mm EGTA, 20 mm HEPES, pH 7.5). Cell lysates were immunoprecipitated with anti-Bcl-xL antibody (Cell Signaling Technology) and separated by SDS-PAGE with a 15% Tris-glycine gradient gel.
Real Time Quantitative RT-PCR—Total RNA was extracted with ISOGEN (Wako Pure Chemicals Industries, Ltd.), and an aliquot (1 μg) was reverse-transcribed using a Takara RNA PCR kit (avian myeloblastosis virus) version 2.1 (Takara Shuzo Co., Ltd.) to make single-stranded cDNA. PCR was performed on an ABI Prism 7000 sequence detection system (Applied Biosystems Inc.) using QuantiTect SYBR Green PCR Master Mix (Qiagen) according to the manufacturer's instructions. After data collection by the ABI Prism 7000 sequence detection system, the mRNA copy number of a specific gene in the total RNA was calculated, and a standard curve generated with serially diluted plasmids containing PCR amplicon sequences, and normalized to rodent total RNA (Applied Biosystems) with mouse β-actin as an internal control. Standard plasmids were synthesized with a TOPO TA cloning kit (Invitrogen), according to the manufacturer's instruction.
Primer Information—Each primer sequence of mouse target genes is described as follows: 5′-TGCAGAGGATGATTGCTGAC-3′ and 5′-GATCAGCTCGGGCACTTTAG-3′ for Bax; 5′-AGGTGACAAGTGACGGTGGT-3′ and 5′-AAGATGCTGTTGGGTTCCAG-3′ for Bak; 5′-ACATGGGGCAAGGTAGTGTC-3′ and 5′-GCTGACCACACACTTGAGGA-3′ for Bok; 5′-TTCGGTGTGATGTACCTGGA-3′ and 5′-CCAGGATGTTGTCACTGTCG-3′ for Bcl-Rambo; 5′-CAGACCCTCAGTCCAGCTTC-3′ and 5′-CGTATGAAGCCGATGGAACT-3′ for Bmf; 5′-ACAACCGCGAGCCAGGTA-3′ and 5′-CAGGGCATAGAACTCGGAAG-3′ for Bcl2l12; 5′-CGGGCAGAGCTACCACCT-3′ and 5′-CGAGCGTTTCTCTCATCACA-3′ for Noxa; 5′-CTCAGCTTGGCAGAACACAT-3′ and 5′-GCAGACACAGGTCCATCTCA-3′ for Bik; 5′-GCCCAGACATTTGGTCAGTT-3′ and 5′-TGCACACACACACAGAGGAA-3′ for Bim; 5′-CGAGCAACAGGTTAGCGAAA-3′ and 5′-TGCACAGGTACGGAATTTTG-3′ for Hrk; 5′-CCACAGCTCTCAGTCAGAAGAA-3′ and 5′-GTGTGCTCAGTCGTTTTCCA-3′ for Nix; 5′-GCCCAGCAGCACTTAGAGTC-3′ and 5′-TGTCGATGCTGCTCTTCTTG-3′ for Puma; 5′-GCTTGGGGATCTACATTGGA-3′ and 5′-TCAGGAACACCGCATTTACA-3′ for Bnip3; 5′-CTCTGCGTTCAGCTTGAGTG-3′ and 5′-CAGAAGCCCACCTACATGGT-3′ for Bid; 5′-AGGGATGGAGGAGGAGCTTA-3′ and 5′-TAGAGTTCCGGGATGTGGAG-3′ for Bad; 5′-GCCAAGACCTGAAACTCTGC-3′; and 5′-GCCATAGCTGAAGTGGAAGC-3′ for col2; 5′-CATAAAGGGCCCACTTGCTA-3′ and 5′-TGGCTGATATTCCTGGTGGT-3′ for col10; 5′-AGATGTGGATCAGCAAGCAG-3′;and 5′-GCGCAAGTTAGGTTTTGTCA-3′ for β-actin.
Mice—Mice carrying the bcl-x gene with two loxP sequences at the promoter region and the second intron (bcl-xfl/fl mice) were generated as reported previously (23). The mice are on a 129SvEv and C57BL6 mixed background. The presence of the floxed bcl-x gene was determined by PCR around the 5′ loxP site using the primers 5′-CGG TTGCCT AGC AAC GGG GC-3′ and 5′-CTC CCA CAG TGG AGA CCTCG-3′, giving a wild-type band of 200 bp and a floxed gene product of 300 bp. These bcl-xfl/fl mice have been used successfully to examine the role of Bcl-xL in a variety of cell types, including those in the liver, ovary, mammary gland, and substantia nigra, as well as in erythroid cells and dendritic cells (23–28). To generate chondrocyte-specific bcl-x knock-out mice, bcl-xfl/fl mice were crossed with type II collagen (Col2a1)-Cre transgenic mice, which express the Cre recombinase gene under the control of the col2a1 gene promoter (29). Col2a1-Cre+/- bcl-xfl/fl (cKO), and Col2a1-Cre-/- bcl-xfl/fl (normal littermates) mice were generated by mating Col2a1-Cre+/-bcl-xfl/+ male mice with Col2a1-Cre-/- bcl-xfl/fl female mice. All animals were housed under specific pathogen-free conditions and treated with humane care under approval from the Animal Care and Use Committee of the University of Tokyo.
Histological Analysis—Tissues were fixed in 4% paraformaldehyde/phosphate-buffered saline, decalcified in 10% EDTA, embedded in paraffin, and cut into sections of 5-μm thickness. Hematoxylin and eosin staining as well as von Kossa staining were performed according to the standard procedure. For immunohistochemistry, sections were incubated overnight at 4 °C with primary antibodies against Bcl-xL (1:200; Cell Signaling Technology), Bnip3 (1:200; Sigma), COL10 (1:500; LSL) and cleaved caspase-7 (1:100; Cell Signaling Technology). The localization of the antigens was visualized by incubation with horseradish peroxidase-conjugated secondary antibodies (Promega) followed by incubation with 3,3′-diaminobenzidine according to the manufacturer's protocol. For fluorescent visualization, a secondary antibody conjugated with Alexa 488 (Molecular Probes) was used. A bone radiograph of whole bodies, femurs, and tibiae was taken with a soft x-ray apparatus (SOFTEX, CMB-2, Tokyo, Japan).
Statistical Analysis—Statistical analyses were performed using two-tailed unpaired Student's t test for the real time PCR and cell viability assay. A p value of <0.05 was considered to be statistically significant. The results are presented as means ± S.D.
RESULTS
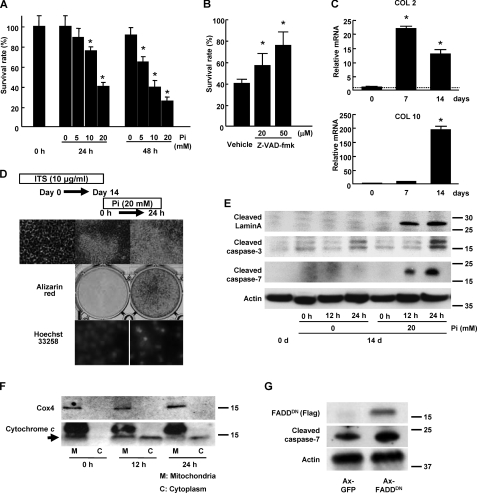
Pi Induces Apoptosis of Chondrocytes through Mitochondrial Pathways—We first evaluated the effect of Pi on the viability of primary chondrocytes in vitro. When mouse primary chondrocytes were stimulated with Pi, a dose- and time-dependent increase in cell death was observed (Fig. 1A). Only about 40% of the chondrocytes survived 24 h after stimulation with 20 mm Pi. Phosphonoformic acid, which inhibits Pi entry into the cells, almost completely restored Pi-induced cell death, as reported previously (data not shown), indicating that the intracellular Pi transport stimulated chondrocyte cell death. In addition, administration of the caspase inhibitor Z-VAD-fmk dose-dependently blocked cell death (Fig. 1B), indicating that Pi-induced cell death is a caspase-dependent process, i.e. apoptosis.
FIGURE 1.
Pi-induced chondrocyte apoptosis. A, Pi-induced cell death of primary chondrocytes in a dose- and time-dependent manner. The results are expressed as the means ± S.D. of six cultures. Experiments were repeated at least three times, and the representative data are presented. *, significantly different from untreated group, p < 0.05. B, pan-caspase inhibitor Z-VAD-fmk dose-dependently inhibited Pi-induced cell death of primary chondrocytes. The cells were treated with either vehicle or Z-VAD-fmk for 30 min before Pi stimulation (20 mm). The results are expressed as the means ± S.D. of six cultures. Experiments were repeated at least three times, and the representative data are presented. *, significantly different from vehicle-treated group, p < 0.01. C, expression of type II (Col 2) and type X collagen (Col 10) expression in ITS-treated ATDC5 cells as determined by real time PCR. Type II collagen expression was increased by day 7 of ITS treatment, which was followed by an increase in a hypertrophic chondrocyte marker, type X collagen. The results are expressed as the means ± S.D. of three samples. Experiments were repeated at least three times, and the representative data are presented. *, significantly different from day 0, p < 0.01. D, Pi stimulation induced the mineralization and apoptosis of differentiated ATDC5 cells. ATDC5 cells exhibited nodule formation when treated with ITS for 14 days, whereas neither mineralization (middle panel) nor chromatin condensation (bottom panel) was observed in the absence of Pi treatment. Pi stimulation (20 mm) of the cells rapidly induced mineralization of the surrounding matrix and the nuclear condensation characteristic of apoptotic cells. E, Western blot analysis of ATDC5 cells. Pi stimulation at 20 mm after ITS treatment for 14 days induced caspase 3 and 7 activation and Lamin proteolysis in ATDC5 cells in the 12–24-h period. F, subcellular fractionation of ATDC5 cells. Cytochrome c, which was exclusively localized in mitochondrial fraction (M), was released into the cytoplasmic fraction (C) in response to Pi stimulation (arrow). Cox4 is a marker of the mitochondrial fraction. Upper bands in mitochondrial fraction are nonspecific bands. G, overexpression of FADDDN did not affect Pi-induced caspase 7 activation of ATDC5 cells, which were pretreated with ITS for 14 days. Adenovirus of green fluorescent protein GFP or FADDDN was introduced 48 h before Pi stimulation (20 mm).
Next, we investigated the molecular mechanisms that lead to hypertrophic differentiation, mineralization, and apoptosis of chondrocytes, using a chondrogenic cell line, ATDC5. ATDC5 cells express markers of hypertrophic chondrocytes and mineralize the surrounding matrix under ITS treatment and subsequent Pi stimulation (30, 31). During ITS treatment, the expression of a chondrocyte marker type II collagen increased earlier than day 7 and remained high levels on day 14. In contrast, the expression of a hypertrophic chondrocyte marker type X collagen was low on day 7, and gradually increased thereafter, and was highly expressed on day 14 (Fig. 1C). Neither mineralization nor caspase activation was observed (Fig. 1D). After Pi stimulation, the cells underwent mineralization and apoptosis within 24 h (Fig. 1E). To examine the localization of cytochrome c in the course of the apoptosis of ATDC5 cells, we performed cell fractionation experiments. Cytochrome c was mainly detected in the mitochondrial fractions, which is positive for Cox4, before Pi stimulation, and was released into the cytoplasm in response to Pi stimulation (Fig. 1F). The differentiation and apoptosis of chondrocytes are independently regulated since overexpression of ranx2 or dominant negative ranx2 did not affect Pi-induced apoptosis. Adenovirus vector-mediated introduction of dominant negative FADD (FADDDN), which blocks death receptor-initiated apoptosis, almost completely abolished tumor necrosis factor-α-induced cell death (data not shown) but did not affect Pi-induced apoptosis in ATDC5 cells (Fig. 1G). These results suggest that the mitochondrial pathway, but not the death receptor pathway, is mainly involved in the Pi-induced apoptosis of ATDC5 cells.
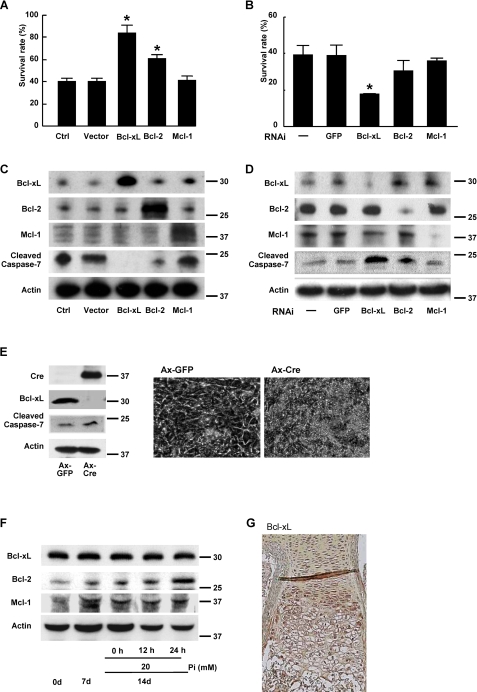
Bcl-xL Regulates Chondrocyte Apoptosis—Because mitochondrial pathways are primarily regulated by Bcl-2 family member proteins, we next examined the expression levels and functions of Bcl-2 family molecules during the differentiation and apoptosis of chondrocytes. We first examined the effect of gain or loss of function of anti-apoptotic Bcl-2 family members, Bcl-2, Bcl-xL, and Mcl-1, on the cell viability of ATDC5 cells. Among these three molecules, Bcl-xL most efficiently suppressed Pi-induced chondrocyte apoptosis when overexpressed by retroviral vectors (Fig. 2A). Conversely, the knockdown of bcl-xL gene through RNAi efficiently reduced the cell viability compared with the knockdown of the bcl-2 or mcl-1 gene (Fig. 2B). Western blot analysis revealed that cleaved caspase-7 expression was suppressed by Bcl-xL overexpression and increased by its knockdown in differentiated ATDC5 cells treated with Pi (Fig. 2, C and D). The important role of Bcl-xL was further confirmed by the experiments using primary chondrocytes obtained from bcl-xfl/fl mice. Adenovirus vector-mediated overexpression of Cre recombinase efficiently down-regulated bcl-x gene in primary chondrocytes obtained from bcl-xfl/fl mice and induced caspase-7 activation (Fig. 2E, left) in the cells, and they became apoptotic morphologically (Fig. 2E, right). These results suggest that Bcl-xL primarily maintains the viability of chondrocytes. However, the expression levels of Bcl-xL did not appear to change in the course of chondrocytic differentiation or Pi-induced apoptosis in ATDC5 cells (Fig. 2F). In addition, immunohistological examination revealed that Bcl-xL is uniformly expressed in growth plate chondrocytes (Fig. 2G). It should be noted that the expression of another antiapoptotic member Bcl-2 increased in response to Pi stimulation, suggesting an important role of the molecule in regulating chondrocyte apoptosis (Fig. 2F). However, the increased level of Bcl-2 was not enough to suppress Pi-induced apoptosis of ATDC5 cell.
FIGURE 2.
Role of anti-apoptotic Bcl-2 family member proteins in Pi-induced apoptosis (20 mm). A and B, effects of anti-apoptotic Bcl-2 family member proteins on Pi-induced apoptosis of primary chondrocytes. Overexpression of Bcl-xL and Bcl-2 significantly suppressed the Pi-induced cell death of primary chondrocytes (A), whereas gene silencing of Bcl-xL alone promoted it (B). The results are expressed as the means ± S.D. of six cultures. Ctrl, control; GFP, green fluorescent protein. C and D, effects of anti-apoptotic Bcl-2 family proteins on caspase activation in ATDC5 cells after Pi stimulation as determined by Western blot analysis. Pi-induced caspase-7 activation as determined by cleave caspase-7 blotting was most efficiently suppressed by Bcl-xL transduction. Bcl-2 transduction had a milder activity, and Mcl-1 transduction had no effect (C). Gene silencing of Bcl-xL markedly increased Pi-induced caspase-7 activation, whereas that of Bcl-2 had milder effect, and Mcl-2 had no effect (D). Mcl-2 was detected as diffuse bands. E, adenovirus vector-mediated overexpression of Cre recombinase efficiently down-regulated bcl-x gene in primary chondrocytes obtained from bcl-x fl/fl mice and induced caspase-7 activation (left) in the cells, and they underwent morphological apoptosis (right). F, expression levels of anti-apoptotic Bcl-2 family member proteins in ATDC5 cells during ITS treatment and Pi stimulation. No significant change was observed. G, immunohistological examination of a murine metatarsal bone (E18.5) with anti-Bcl-xL antibody. Bcl-xL was uniformly expressed in the growth plate chondrocytes.
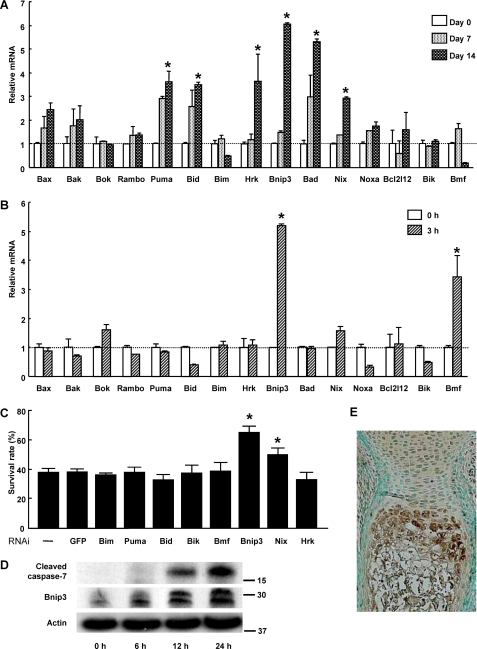
Bnip3, a Pro-apoptotic Bcl-2 Family Member Protein, Promotes the Pi-induced Apoptosis of Chondrocytes—The fact that the expression level of Bcl-xL was not affected by Pi stimulation and that Bcl-xL is uniformly expressed in the chondrocyte layers prompted us to investigate the role of pro-apoptotic molecule(s) that may inhibit the anti-apoptotic function of Bcl-xL. Among 15 pro-apoptotic Bcl-2 family members, six molecules (Puma, Bid, Hrk, Bnip3, Bad, and Nix) increased in expression level during the course of hypertrophic differentiation (Fig. 3A), and two of them (Bnip3 and Bmf) in response to Pi stimulation (Fig. 3B). As shown in Fig. 3C, gene silencing of the proapoptotic BH3-only molecule bnip3 significantly suppressed Pi-induced apoptosis. Western blot analysis revealed that the protein levels of Bnip3 increased in response to Pi stimulation, followed by caspase-7 activation (Fig. 3D). Immunohistological examination of murine growth plates revealed that Bnip3 expression was exclusively localized in the prehypertrophic and hypertrophic layers of chondrocytes (Fig. 3E). Although overexpression of Bnip3 increased Pi-induced chondrocyte apoptosis, its overexpression alone failed to induce chondrocyte apoptosis in the absence of Pi (data not shown).
FIGURE 3.
Role of pro-apoptotic Bcl-2 family member proteins in Pi-induced cell death. A, expression patterns of pro-apoptotic Bcl-2 family member proteins in ATDC5 cells during ITS treatment as determined by time and RT-PCR. The results are expressed as the means ± S.D. of three cultures. Experiments were repeated at least three times, and the representative data are presented. *, significantly different from day 0, p < 0.05. B, expression patterns of pro-apoptotic Bcl-2 family member proteins in ATDC5 cells after Pi stimulation as determined by real time PCR. ATDC5 cells were pretreated with ITS for 14 days (0 h) before Pi stimulation (20 mm). The results are expressed as the means ± S.D. of three cultures. Experiments were repeated at least three times, and the representative data are presented. *, significantly different from time 0, p < 0.05. C, effect of gene silencing of pro-apoptotic Bcl-2 family member proteins on cell viability of primary chondrocytes 24 h after Pi stimulation. Gene silencing of Bnip3 and Nix significantly promoted the cell viability, p < 0.05. The results are expressed as the means ± S.D. of six cultures. Experiments were repeated at least three times, and the representative data are presented. GFP, green fluorescent protein. D, Western blot analysis after Pi stimulation. Protein levels of Bnip3 increased in ATDC5 cells after 6 h of Pi stimulation, which was followed by caspase-7 activation. Bnip3 was detected as two bands. E, immunohistological examination of a murine metatarsal bone (E18.5) with anti-Bnip3 antibody. Bnip3 expression was primarily localized in the hypertrophic layers of chondrocytes.
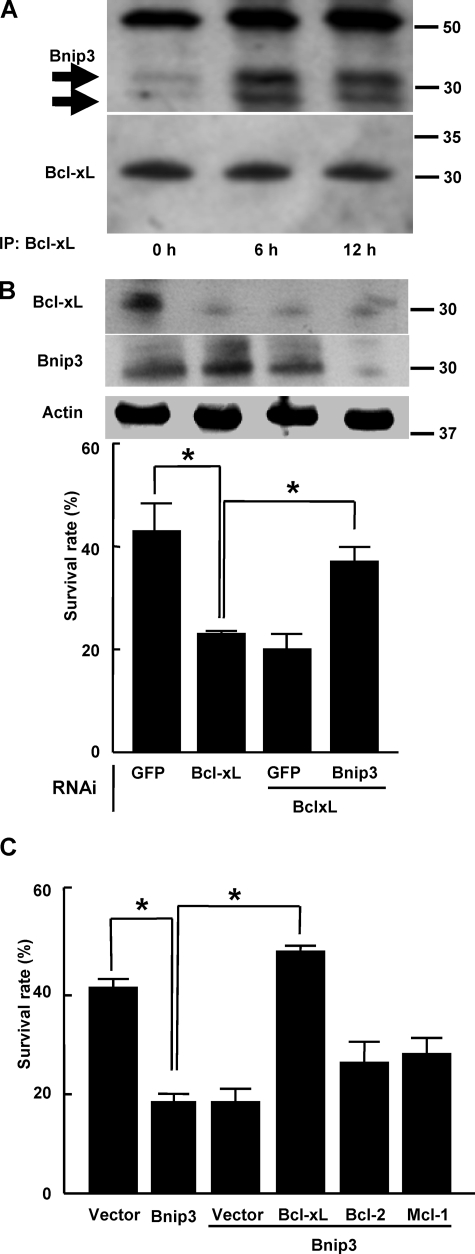
Bnip3 Associates with Bcl-xL and Attenuates the Anti-apoptotic Effect of Bcl-xL on Chondrocytes—We then investigated the molecular interaction between Bcl-xL and Bnip3 in chondrocytes. Earlier studies have shown that Bnip3 heterodimerizes with Bcl-2 and Bcl-xL and facilitates cell death via mitochondrial pathways (32). As shown in Fig. 4A, Bnip3 was co-immunoprecipitated with Bcl-xL in Pi-treated chondrocytes. The susceptibility to apoptosis by bcl-xL knockdown was partially restored by simultaneous silencing of bnip3 (Fig. 4B). Conversely, overexpression of Bnip3 promoted Pi-induced cell death, which was almost completely rescued by the simultaneous transduction of Bcl-xL but not by that of Bcl-2 or Mcl-1 (Fig. 4C). Taken together, Bnip3 is up-regulated and associates with Bcl-xL in chondrocytes in response to Pi stimulation, impairs the anti-apoptotic function of Bcl-xL, and consequently causes apoptosis in these cells.
FIGURE 4.
Molecular interaction between Bcl-xL and Bnip3 in chondrocytes. A, Bcl-xL was immunoprecipitated (IP) from ATDC5 cells left untreated or treated with 20 mm Pi. The immunoprecipitated materials were Western blotted with anti-Bnip3 antibody. Bnip3 was associated with Bcl-xL after 6 h of Pi treatment. Bnip3 was detected as two bands (arrows). The top band is the immunoglobulin heavy chain. B, susceptibility to apoptosis of primary chondrocytes by Bcl-xL knockdown was partially restored by the simultaneous silencing of Bnip3. Western blot analysis shows efficient gene knockdown of Bcl-xL and Bnip3 through RNAi. The results are expressed as the means ± S.D. of six cultures. Experiments were repeated at least three times, and the representative data are presented. *, significantly different, p < 0.05. GFP, green fluorescent protein. C, overexpression of Bnip3 promoted the Pi-induced cell death of primary chondrocytes, which was rescued by simultaneous transduction of Bcl-xL, whereas overexpression of Bcl-2 or Mcl-1 had little effect. The results are expressed as the means ± S.D. of six cultures. Experiments were repeated at least three times and the representative data are presented. *, significantly different, p < 0.05.
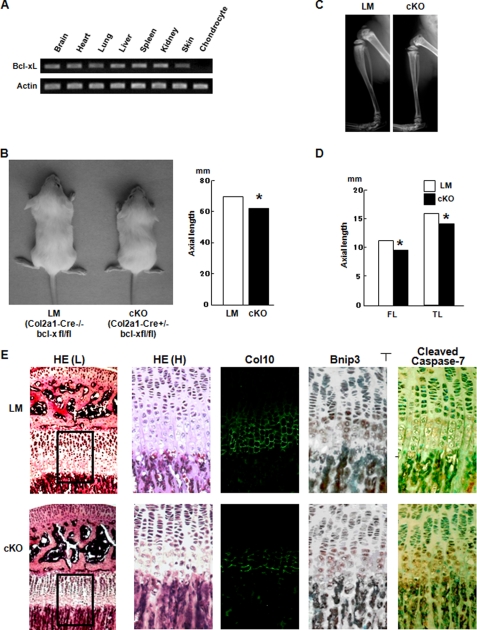
Chondrocyte-specific Bcl-x Knock-out Mice Exhibit a Reduction in the Hypertrophic Layer of Growth Plate Chondrocytes—To further confirm the essential role of the Bcl-xL/Bnip3 axis in chondrocytes, we generated chondrocyte-specific conditional knock-out (cKO) mice of the bcl-x gene by mating bcl-xfl/fl mice with Col2a1-Cre transgenic mice, in which Cre recombinase is specifically expressed in chondrocytes under the control of the col2a1 promoter. The cKO mice (Col2a1-Cre+/- bcl-xfl/fl mice) were born at approximately a Mendelian frequency. Bcl-xL expression was markedly reduced in chondrocytes of the cKO mice, whereas its expression in other cells or tissues was comparable with that in their normal littermates (LM) that did not carry the col2a1-cre gene (Col2a1-Cre-/- bcl-xfl/fl mice) (Fig. 5A). bcl-x cKO mice exhibited dwarfism, with shortened limbs and trunk compared with their normal littermates (bcl-xfl/fl mice) (Fig. 5B). The trunk of the cKO mice was about 10% shorter than that of bcl-xfl/fl mice at 5 weeks of age. The longitudinal length of the femur and tibia, which are formed through endochondral ossification, were also significantly shorter in cKO mice (Fig. 5, C and D). Histological analysis of the growth plates of the proximal tibia showed that the hypertrophic chondrocyte layer was markedly shortened in cKO mice, probably because of an increased apoptosis of prehypertrophic and hypertrophic chondrocytes, as determined by immunohistochemical analysis of cleaved caspase-7 (Fig. 5E). No abnormality in vascular invasion was observed (data not shown). Bnip3 was predominantly expressed in the prehypertrophic and hypertrophic layers of chondrocytes, and its expression level was not different between the cKO mice and their normal littermates. Taking these histological findings together, Bcl-xL is essential for the survival of prehypertrophic and hypertrophic chondrocytes by antagonizing the pro-apoptotic function of Bnip3.
FIGURE 5.
Chondrocyte-specific bcl-x knock-out mice. A, RT-PCR of Bcl-xL expression. Bcl-xL expression was specifically reduced in chondrocytes. B, gross appearance of a chondrocyte-specific bcl-x-deficient mouse (cKO) and a normal littermate (LM) at 5 weeks of age (left panel). Total axial length (from the nose to the tail end) of cKO mice was significantly reduced in cKO mice, p < 0.05. The results are expressed as the means ± S.D. of three samples. C, representative x-ray images of hind limbs of LM and cKO mice at 5 weeks of age. The total bone length of the tibia as well as the growth plate length of cKO mice was shorter than that of their normal littermates. D, bone lengths of LM and cKO mice at 5 weeks of age. FL, femoral length; TL, tibial length. The results are expressed as the means ± S.D. of three samples. *, significantly different, p < 0.05. E, histological analysis of the growth plates of the proximal tibia at 5 weeks of age. In cKO mice, the hypertrophic layers, as evidenced by type X collagen staining, were shorter than those in LM mice. Although Bnip3 expression levels showed no difference between LM and cKO mice, the number of cleaved caspase-7-positive cells in the prehypertrophic and hypertrophic area was increased in cKO mice. HE, hematoxylin and eosin staining. HE(H) is the higher magnification of the rectangular areas in HE(L).
DISCUSSION
Although the molecular mechanism and pathophysiologic role of chondrocyte apoptosis have not been fully elucidated yet, the possible involvement of Pi in this process has been suggested. It was reported that both intracellular and extracellular calcium and phosphate levels increase in the hypertrophic area where terminally differentiated chondrocytes mineralize the surrounding matrix and subsequently die (11, 30, 33). In vitro studies have also made clear that Pi induces the mineralization and cell death of chondrocytes (11, 12). Magne et al. (30) thoroughly investigated the involvement of Pi and Ca2+ in chondrocyte maturation using ATDC5 cells, and showed that Pi is a key regulator of chondrocyte maturation, mineralization, and apoptosis. In addition, disorders in Pi homeostasis result in abnormal endochondral ossification, and hypophosphatemia caused by inefficient phosphate reabsorption by the kidney is associated with defective mineralization of the skeleton, which manifests as rickets or osteomalacia (34, 35). Sabbagh et al. (14) reported that hypophosphatemia, but not hypocalcemia or hyperparathyroidism, is responsible for the reduced chondrocyte apoptosis and the subsequent rachitic changes in vitamin D receptor-null mice and Hyp mice. All of these observations indicate a crucial role of Pi in controlling the cell fate of hypertrophic chondrocytes.
This study shows that hypertrophic chondrocytes are susceptible to Pi-induced apoptosis, which is regulated by the balance maintained between pro-apoptotic and anti-apoptotic Bcl-2 family members. Among the anti-apoptotic members of the Bcl-2 family, overexpression of an anti-apoptotic member Bcl-xL suppressed and, conversely, its down-regulation by RNAi stimulated Pi-induced chondrocyte apoptosis most efficiently. Although overexpression of Bcl-2 also suppressed chondrocyte apoptosis, its suppression by RNAi was not as efficient as Bcl-xL knockdown was in inducing chondrocyte apoptosis. In addition, although Bcl-2-deficient mice exhibit abnormal skeletal development because of the accelerated maturation of chondrocytes (36), no apparent increase in chondrocyte apoptosis was observed in these mice (data not shown). Overexpression or down-regulation of Mcl-1 did not affect Pi-induced chondrocyte apoptosis. These results suggest that Bcl-xL is mainly implicated in the survival of chondrocytes, although it is also likely that other anti-apoptotic Bcl-2 family members have additive or synergistic roles with Bcl-xL. Because the skeletal phenotypes of bcl-x gene knock-out mice have not been investigated because of their embryonic lethality, we generated chondrocyte-specific bcl-x gene knock-out mice using the Cre-loxP recombination system. The mice exhibited dwarfism and immunohistological examination of the growth plate revealed a shortening of the hypertrophic layer because of the massive apoptosis of hypertrophic chondrocytes, confirming the pivotal role of Bcl-xL in hypertrophic chondrocyte survival. Amizuka et al. (37) previously reported that the apoptosis of chondrocytes was increased in the hypertrophic zone of PTHrP-deficient mice. Park et al. (38) recently reported that the transcriptional repressor Nkx3.2/Bapx1, which acts downstream of parathyroid hormone/PTHrP receptor signaling, enhances chondrocyte survival by constitutively activating RelA in a ligand-independent manner. However, because the morphological features of growth plates of bcl-x cKO mice were quite different from those of PTHrP-deficient mice, in which the number of proliferating chondrocytes was markedly reduced because of apoptosis and accelerated endochondral ossification was observed, we speculated that the apoptosis of hypertrophic chondrocytes in bcl-x cKO mice is not because of a defect in the PTHrP receptor/Nkx3.2 axis. Colnot et al. (39) demonstrated that anti-apoptotic protein Galectin 3 regulates the survival of chondrocytes, and its deficiency in mice results in an accumulation of empty lacuna at the junction between avascular cartilage and vascular bone, without affecting endochondral ossification. From these results, they concluded that the apoptosis of hypertrophic chondrocytes does not affect the longitudinal bone growth. In contrast, bcl-x cKO mice displayed a dwarf phenotype, which was considered to be caused by the increased apoptosis of hypertrophic chondrocytes. The reason for the discrepancy between their results and ours remains elusive, and further investigation will be required to clarify the relationship between Bcl-xL and Galectin 3.
Because the expression levels of Bcl-xL and other anti-apoptotic members did not appear to change in the course of the hypertrophic differentiation and the apoptosis of chondrocytes, we speculated that the expression of pro-apoptotic molecule(s) is up-regulated in the hypertrophic chondrocytes and impairs the anti-apoptotic effect of Bcl-xL. By screening the pro-apoptotic Bcl-2 family members, we identified Bnip3 as a putative candidate pro-apoptotic gene in chondrocytes. Bnip3 belongs to the BH3-only subgroup members and facilitates both caspase-dependent and -independent apoptosis by antagonizing Bcl-2 or Bcl-xL activity (32, 40). Bnip3 is ubiquitously expressed, and its expression levels change in a number of tumors (35, 41). Interestingly, it was reported that the interaction between Bnip3 and Bcl-xL, and indeed the ability of BNIP3 to induce apoptosis, does not depend on the presence of a BH3 domain in BNIP3. Bnip3 is also involved in ischemia-induced cardiac myocyte cell death by acting downstream of the transcriptional hypoxia-inducible factor 1a (HIF-1a) (42). We found that the expression of Bnip3 increased in accordance with hypertrophic differentiation and during Pi-induced apoptosis in ATDC5 cells, which corresponded to the expression patterns of Bnip3 in the growth plate, as demonstrated by immunohistological examinations. Bnip3 increases its expression levels in response to Pi stimulation and binds to Bcl-xL in Pi-treated chondrocytes. In addition, knockdown of Bnip3 blocked Pi-induced apoptosis of chondrocytes, and therefore we concluded that Bnip3 determines the cell fate of hypertrophic chondrocytes by impairing the anti-apoptotic function of Bcl-xL, although the involvement of other BH3-only proteins cannot be completely excluded. Overexpression of Bnip3 increased Pi-induced chondrocyte apoptosis but failed to induce apoptosis in the absence of Pi (data not shown), indicating that other molecules induced by Pi stimulation is also required for chondrocyte apoptosis. Previous reports have shown an increase in HIF-1a levels in hypertrophic chondrocytes (43), which may stimulate Bnip3 expression.
Because Pi accelerates the terminal differentiation of chondrocytes, as evidenced by increased expression levels of type X collagen, it is possible that Pi induces chondrocyte apoptosis by simply accelerating terminal differentiation of the cells. However, overexpression of Bnip3 induced the apoptosis of chondrocytes without affecting their hypertrophic differentiation, indicating that the differentiation and the apoptosis of chondrocytes are distinctly regulated. While we were preparing the manuscript, Diwan et al. (44) recently reported the phenotype of Bnip3 knock-out mice with no increase in mortality or apparent physical abnormalities. Detailed analysis of the growth plates is necessary to clarify whether there is any abnormality in chondrocyte phenotypes in the KO mice, and whether Bnip3 functions as a bona fide BH3-only protein in chondrocytes remains to be determined.
Recently, Bnip3 has been reported to be involved in autophagy in hypoxic cardiac cells (42). Autophagy is primarily considered a pro-survival rather than pro-death mechanism. However, recent studies have revealed that it is also linked to programmed cell death, which is inhibited by Bcl-xL (45). Moreover, terminally differentiated chondrocytes are reported to exhibit autophagic characteristics (46). Because growth plate chondrocytes are exposed to hypoxic stress (43), autophagy may emerge to help survive such conditions. Although the precise mechanism of autophagic cell death remains unclear, the apoptotic cell death and the autophagic cell death might be intricately intertwined with each other in growth plate chondrocytes and regulated by Bcl-xL and Bnip3.
In summary, the apoptosis of hypertrophic chondrocytes is at least partly regulated by the balance between anti-apoptotic Bcl-xL and pro-apoptotic Bnip3, in which Pi appears to play a critical role. Further studies will be required to fully detail the molecular events regulating chondrocyte apoptosis, which have begun to be uncovered here.
Supplementary Material
Acknowledgments
We thank R. Yamaguchi (Dept. of Orthopaedic Surgery, University of Tokyo) who provided expert technical assistance. Pacific Edit reviewed the manuscript prior to submission.
This work was authored, in whole or in part, by National Institutes of Health staff. This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and health science research grants from the Ministry of Health, Labor, and Welfare of Japan (to S. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: PTHrP, parathyroid hormone-related peptide; RT, reverse transcription; cKO, conditional knock-out; RNAi, RNA interference; BH3, Bcr homology domain 3; Z, benzyloxycarbonyl; fmk, fluoromethyl ketone; LM, littermate; BH3, Bcl-2 homology 3.
References
- 1.Kronenberg, H. M. (2003) Nature 423 332-336 [DOI] [PubMed] [Google Scholar]
- 2.Farnum, C. E., and Wilsman, N. J. (1987) Anat. Rec. 219 221-232 [DOI] [PubMed] [Google Scholar]
- 3.Gibson, G. J., Kohler, W. J., and Schaffler, M. B. (1995) Dev. Dyn. 203 468-476 [DOI] [PubMed] [Google Scholar]
- 4.Hatori, M., Klatte, K. J., Teixeira, C. C., and Shapiro, I. M. (1995) J. Bone Miner. Res. 10 1960-1968 [DOI] [PubMed] [Google Scholar]
- 5.Kerr, J. F., Wyllie, A. H., and Currie, A. R. (1972) Br. J. Cancer 26 239-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson, C. B. (1995) Science 267 1456-1462 [DOI] [PubMed] [Google Scholar]
- 7.Ikeda, T., Kamekura, S., Mabuchi, A., Kou, I., Seki, S., Takato, T., Nakamura, K., Kawaguchi, H., Ikegawa, S., and Chung, U. I. (2004) Arthritis Rheum. 50 3561-3573 [DOI] [PubMed] [Google Scholar]
- 8.Inada, M., Yasui, T., Nomura, S., Miyake, S., Deguchi, K., Himeno, M., Sato, M., Yamagiwa, H., Kimura, T., Yasui, N., Ochi, T., Endo, N., Kitamura, Y., Kishimoto, T., and Komori, T. (1999) Dev. Dyn. 214 279-290 [DOI] [PubMed] [Google Scholar]
- 9.Yoshida, C. A., Yamamoto, H., Fujita, T., Furuichi, T., Ito, K., Inoue, K., Yamana, K., Zanma, A., Takada, K., Ito, Y., and Komori, T. (2004) Genes Dev. 18 952-963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyde, A., and Shapiro, I. M. (1980) Histochemistry 69 85-94 [DOI] [PubMed] [Google Scholar]
- 11.Mansfield, K., Rajpurohit, R., and Shapiro, I. M. (1999) J. Cell. Physiol. 179 276-286 [DOI] [PubMed] [Google Scholar]
- 12.Mansfield, K., Teixeira, C. C., Adams, C. S., and Shapiro, I. M. (2001) Bone (Elmsford) 28 1-8 [DOI] [PubMed] [Google Scholar]
- 13.Mwale, F., Tchetina, E., Wu, C. W., and Poole, A. R. (2002) J. Bone Miner. Res. 17 275-283 [DOI] [PubMed] [Google Scholar]
- 14.Sabbagh, Y., Carpenter, T. O., and Demay, M. B. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 9637-9642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagata, S. (1997) Cell 88 355-365 [DOI] [PubMed] [Google Scholar]
- 16.Gross, A., McDonnell, J. M., and Korsmeyer, S. J. (1999) Genes Dev. 13 1899-1911 [DOI] [PubMed] [Google Scholar]
- 17.Strasser, A., O'Connor, L., and Dixit, V. M. (2000) Annu. Rev. Biochem. 69 217-245 [DOI] [PubMed] [Google Scholar]
- 18.Huang, D. C., and Strasser, A. (2000) Cell 103 839-842 [DOI] [PubMed] [Google Scholar]
- 19.Kitamura, T. (1998) Int. J. Hematol. 67 351-359 [DOI] [PubMed] [Google Scholar]
- 20.Morita, S., Kojima, T., and Kitamura, T. (2000) Gene Ther. 7 1063-1066 [DOI] [PubMed] [Google Scholar]
- 21.Miyagishi, M., and Taira, K. (2004) Methods Mol. Biol. 252 483-491 [DOI] [PubMed] [Google Scholar]
- 22.Newton, K., Harris, A. W., Bath, M. L., Smith, K. G., and Strasser, A. (1998) EMBO J. 17 706-718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rucker, E. B., III, Dierisseau, P., Wagner, K. U., Garrett, L., Wynshaw-Boris, A., Flaws, J. A., and Hennighausen, L. (2000) Mol. Endocrinol. 14 1038-1052 [DOI] [PubMed] [Google Scholar]
- 24.Hon, H., Rucker, E. B., III, Hennighausen, L., and Jacob, J. (2004) J. Immunol. 173 4425-4432 [DOI] [PubMed] [Google Scholar]
- 25.Riedlinger, G., Okagaki, R., Wagner, K. U., Rucker, E. B., III, Oka, T., Miyoshi, K., Flaws, J. A., and Hennighausen, L. (2002) Biol. Reprod. 66 438-444 [DOI] [PubMed] [Google Scholar]
- 26.Savitt, J. M., Jang, S. S., Mu, W., Dawson, V. L., and Dawson, T. M. (2005) J. Neurosci. 25 6721-6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner, K. U., Claudio, E., Rucker, E. B., III, Riedlinger, G., Broussard, C., Schwartzberg, P. L., Siebenlist, U., and Hennighausen, L. (2000) Development (Camb.) 127 4949-4958 [DOI] [PubMed] [Google Scholar]
- 28.Walton, K. D., Wagner, K. U., Rucker, E. B., III, Shillingford, J. M., Miyoshi, K., and Hennighausen, L. (2001) Mech. Dev. 109 281-293 [DOI] [PubMed] [Google Scholar]
- 29.Ovchinnikov, D. A., Deng, J. M., Ogunrinu, G., and Behringer, R. R. (2000) Genesis 26 145-146 [PubMed] [Google Scholar]
- 30.Magne, D., Bluteau, G., Faucheux, C., Palmer, G., Vignes-Colombeix, C., Pilet, P., Rouillon, T., Caverzasio, J., Weiss, P., Daculsi, G., and Guicheux, J. (2003) J. Bone Miner. Res. 18 1430-1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shukunami, C., Shigeno, C., Atsumi, T., Ishizeki, K., Suzuki, F., and Hiraki, Y. (1996) J. Cell Biol. 133 457-468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray, R., Chen, G., Vande Velde, C., Cizeau, J., Park, J. H., Reed, J. C., Gietz, R. D., and Greenberg, A. H. (2000) J. Biol. Chem. 275 1439-1448 [DOI] [PubMed] [Google Scholar]
- 33.Wu, L. N., Ishikawa, Y., Sauer, G. R., Genge, B. R., Mwale, F., Mishima, H., and Wuthier, R. E. (1995) J. Cell. Biochem. 57 218-237 [DOI] [PubMed] [Google Scholar]
- 34.Anderson, H. C., Cecil, R., and Sajdera, S. W. (1975) Am. J. Pathol. 79 237-254 [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, H., and Paik, S. G. (2006) Mol. Cells 21 1-6 [PubMed] [Google Scholar]
- 36.Amling, M., Neff, L., Tanaka, S., Inoue, D., Kuida, K., Weir, E., Philbrick, W. M., Broadus, A. E., and Baron, R. (1997) J. Cell Biol. 136 205-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amizuka, N., Henderson, J. E., Hoshi, K., Warshawsky, H., Ozawa, H., Goltzman, D., and Karaplis, A. C. (1996) Endocrinology 137 5055-5067 [DOI] [PubMed] [Google Scholar]
- 38.Park, M., Yong, Y., Choi, S. W., Kim, J. H., Lee, J. E., and Kim, D. W. (2007) Nat. Cell Biol. 9 287-298 [DOI] [PubMed] [Google Scholar]
- 39.Colnot, C., Sidhu, S. S., Balmain, N., and Poirier, F. (2001) Dev. Biol. 229 203-214 [DOI] [PubMed] [Google Scholar]
- 40.Kubli, D. A., Ycaza, J. E., and Gustafsson, A. B. (2007) Biochem. J., 405 407-415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okami, J., Simeone, D. M., and Logsdon, C. D. (2004) Cancer Res. 64 5338-5346 [DOI] [PubMed] [Google Scholar]
- 42.Hamacher-Brady, A., Brady, N. R., Logue, S. E., Sayen, M. R., Jinno, M., Kirshenbaum, L. A., Gottlieb, R. A., and Gustafsson, A. B. (2007) Cell Death Differ. 14 146-157 [DOI] [PubMed] [Google Scholar]
- 43.Schipani, E. (2006) Ann. N. Y. Acad. Sci. 1068 66-73 [DOI] [PubMed] [Google Scholar]
- 44.Diwan, A., Krenz, M., Syed, F. M., Wansapura, J., Ren, X., Koesters, A. G., Li, H., Kirshenbaum, L. A., Hahn, H. S., Robbins, J., Jones, W. K., and Dorn, G. W. (2007) J. Clin. Investig. 117 2825-2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamacher-Brady, A., Brady, N. R., Gottlieb, R. A., and Gustafsson, A. B. (2006) Autophagy 2 307-309 [DOI] [PubMed] [Google Scholar]
- 46.Srinivas, V., and Shapiro, I. M. (2006) Autophagy 2 215-216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.