Abstract
Class I and II histone deacetylases (HDACs) play vital roles in regulating cardiac development, morphogenesis, and hypertrophic responses. Although the roles of Hdac1 and Hdac2, class I HDACs, in cardiac hyperplasia, growth, and hypertrophic responsiveness have been reported, the role in the heart of Hdac3, another class I HDAC, has been less well explored. Here we report that myocyte-specific overexpression of Hdac3 in mice results in cardiac abnormalities at birth. Hdac3 overexpression produces thickening of ventricular myocardium, especially the interventricular septum, and reduction of both ventricular cavities in newborn hearts. Our data suggest that increased thickness of myocardium in Hdac3-transgenic (Hdac3-Tg) mice is due to increased cardiomyocyte hyperplasia without hypertrophy. Hdac3 overexpression inhibits several cyclin-dependent kinase inhibitors, including Cdkn1a, Cdkn1b, Cdkn1c, Cdkn2b, and Cdkn2c. Hdac3-Tg mice did not develop cardiac hypertrophy at 3 months of age, unlike previously reported Hdac2-Tg mice. Further, Hdac3 overexpression did not augment isoproterenol-induced cardiac hypertrophy when compared with wild-type littermates. These findings identify Hdac3 as a novel regulator of cardiac myocyte proliferation during cardiac development.
Histone acetyl transferases and histone deacetylases (HDACs)3 are critical regulators of chromatin remodeling (1). Modifications of core histones, around which DNA is packaged, play prominent roles in the regulation of gene expression (2). For instance, core histones are acetylated by histone acetyl transferases, leading to unwinding and relaxation of the chromatin structure and subsequent gene activation. This enzymatic acetylation step is reversed by HDAC-dependent deacetylation, leading to chromatin condensation and gene repression (3). Histone modifications provide marks for interactions with transcriptional coactivator and repressor proteins to regulate gene expression (4). The mammalian HDACs are classified into four subfamilies based on their highly conserved homologous sequences and functional similarities (3). Class I HDACs include HDAC1, -2, -3, and -8; class II HDACs include HDAC4, -5, -6, -7, -9, and -10; class III HDACs are known as sirtuins (5); and the lone class IV HDAC is HDAC11 (6). Conventional HDAC inhibitors, such as trichostatin A, inhibit both class I and class II HDACs (7).
Various class I and II Hdac gene deletion and overexpression analyses in mice have suggested important roles for histone modification and HDAC function in cardiac development and in the regulation of cardiac hypertrophy (8–13). For example, class II HDACs, including Hdac5 and Hdac9, act as negative regulators of cardiac growth (14). Inactivation of either Hdac5 or Hdac9 results in mice that are sensitive to hypertrophic stress, whereas the simultaneous loss of both Hdac5 and Hdac9 produces mice with grossly enlarged hearts (15). Loss of Hdac7 results in abnormal thinning of myocardial walls of the ventricular chambers and dilation of atria (16). Deletion of the class I HDAC, Hdac1, results in embryonic lethality by embryonic day 10.5 due to severe proliferation defects (17). Recently, our group and others have shown that global loss of Hdac2 results in partial or complete perinatal lethality due to severe cardiac developmental defects that include cardiac myocyte hyperplasia (12, 18). Hdac2-null mice are resistant to hypertrophic stimuli due to activation of glycogen synthase kinase 3 β (GSK3β), whereas Hdac2-Tg mice are sensitive to hypertrophic stimuli (12). Interestingly, Hdac1 and Hdac2 display evidence of redundancy with regard to the regulation of cardiac development and morphogenesis; combined loss of Hdac1 and Hdac2 in cardiac myocytes results in complete perinatal lethality due to severe cardiac defects in excess of that seen in either individual knock-out (18).
The overlapping functions of Hdac1 and Hdac2 may be due, at least in part, to their co-existence within the same Sin3A, NuRD, and CoREST co-repressor complexes (19, 20). Hdac3, which is structurally related to Hdac1 and Hdac2, is a component of the NCoR-SMRT co-repressor complex, which is distinct from co-repressor complexes that typically contain Hdac1 and Hdac2 (21). A series of recent reports, including those from our group, have demonstrated striking anti-hypertrophic effects of broad spectrum chemical HDAC inhibitors, but the molecular targets of these agents in the heart remain unclear (8–11). Whether Hdac3 participates in mediating the effects of HDAC inhibitors in the heart or plays an independent role in cardiac development, growth, and hypertrophic response is presently unknown. Therefore to further elucidate the role of Hdac3 in cardiac growth, development, and hypertrophic responses, we generated myocyte-specific Hdac3-Tg mice. Two independent Hdac3-Tg mice lines revealed increased cardiac myocyte proliferation and down-regulation of CKIs. Unlike Hdac2-Tg mice, these mice did not show spontaneous cardiac hypertrophy or increased sensitivity to hypertrophic stimuli. Our data suggest a novel and distinct role of Hdac3 in myocyte proliferation and cardiac growth distinct from Hdac1 and Hdac2.
EXPERIMENTAL PROCEDURES
Hdac3-Transgenic Mice—A cDNA encoding murine FLAG-tagged Hdac3 was cloned into an expression plasmid containing the Myh6 (encoding α-myosin heavy chain) promoter, and transgenic mice were generated by standard techniques. Genotyping was performed by PCR analysis of genomic DNA, and cardiac-specific expression of Hdac3 was revealed by qRT-PCR and Western blot analysis using antibodies to Hdac3 and FLAG (Sigma).
Quantitative Real-time PCR—Total RNA was isolated from dissected mouse hearts using TRIzol (Invitrogen). RNA was reverse-transcribed using random hexamers and the SuperScript first strand synthesis kit (Invitrogen). Gene expression was then evaluated by qRT-PCR (ABI PRISM 7900) using SYBR Green (Applied Biosystems). Signals were normalized to corresponding glyceraldehyde-3-phosphate dehydrogenase controls, and the ratios were expressed as -fold changes when compared with wild-type controls. PCR conditions and primer set sequences are available upon request.
Western Blotting—We used antibodies to Hdac3 (1:1000 dilution, Sigma), α-tubulin (1:5000 dilution, Sigma), and acetylhistone H4 (1:200 dilution, Upstate Biotechnology). Primary antibody binding was visualized by using the Westernbreeze Kit (Invitrogen) as described earlier and according to the manufacturer's instructions (12).
HDAC Activity Assay—HDAC activity was measured as described previously (22). In brief, total extract was prepared from hearts of post-natal day 1 (P1) wild-type and Hdac3-Tg mice (two independent lines). 50 μg of total extract was incubated with HDAC assay substrate and incubated at 30 °C for 60 min. After the addition of diluted activator reagent, samples were incubated at room temperature for 15 min, and readings were taken by fluorescent microplate reader at an excitation of 360 nm and emission of 450 nm. A standard curve was performed according to the manufacturer's protocol.
Histology and Immunohistochemistry—Protocols for hematoxylin and eosin staining and immunohistochemistry have been described (12). P1 wild-type and Hdac3-Tg heart sections were immunostained with anti-phospho-histone H3 antibody (1:2000, Cell Signaling), Ki67 antibody (1:50), and MF-20 (1:25, HybridomaBank). Using ImageJ software, phospho-histone H3-positive myocyte nuclei and the total number of nuclei were counted in eight different sections of three independent heart samples. The average of phospho-histone H3-positive nuclei was divided by the total number of nuclei to determine percentages of phospho-histone H3-positive myocytes. Similarly, Ki67-positive nuclei in MF-20-expressing myocytes were counted. The average of MF-20-positive nuclei was divided by the total number of nuclei in MF-20-expressing cells to determine the percentages of Ki67-positive myocytes.
Assessment of Cardiomyocyte Size—TRITC-labeled wheat germ agglutinin (Sigma) staining (100 μg/ml) was performed on sections of neonatal (P1) and 3-month-old hearts from wild-type and Hdac3-Tg mice to determine the cell boundaries. ImageJ software was used to determine the cross-sectional area of the cardiac myocytes. Approximately 500 cardiac myocytes were counted from P1 and 2-month-old hearts.
Isoproterenol Treatment—Isoproterenol (ISO, Sigma, I5627) was delivered to 12-week-old mice by implanting a micro-osmotic pump (Alzet, Durect; model 1002) subcutaneously under isoflurane anesthesia. ISO (30 mg/kg/d) or vehicle (Dulbecco's phosphate-buffered saline, Invitrogen) was infused subcutaneously for 14 days. Mice were sacrificed after 14 days, and heart-to-body-weight ratios and heart-weight-to-tibial-length ratios were determined.
Statistics—All measurement data are expressed as mean ± S.D. The statistical significance of differences between groups was analyzed by Student's t test. Differences were considered significant at a p value <0.05.
RESULTS
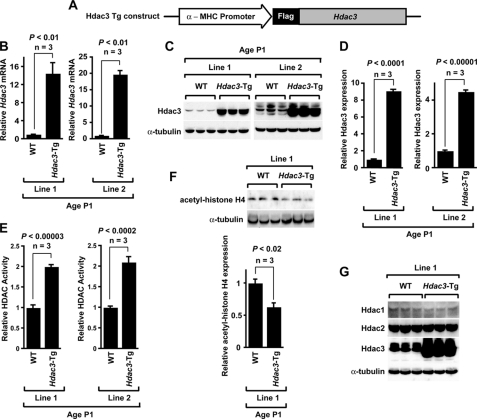
Cardiac Myocyte-specific Hdac3-Tg Mice—To understand the role of Hdac3 in cardiac development and the regulation of hypertrophy, we used the Myh6 (encoding α-myosin heavy chain) promoter to overexpress Hdac3 in cardiac myocytes (Fig. 1A). We confirmed overexpression at the mRNA level in two independent Hdac3-Tg lines by real-time PCR. Our data show 15–20-fold induction of Hdac3 mRNA in P1 hearts of Hdac3-Tg mice when compared with wild-type littermates (Fig. 1B). We also confirmed overexpression of Hdac3 in P1 hearts by Western blotting in both independent Hdac3-Tg lines (Fig. 1C). In both lines, densitometry analysis by ImageJ software determined that Hdac3 protein was overexpressed ∼4–8-fold (Fig. 1D). To determine the functionality of Hdac3 overexpression, we performed HDAC activity assays on P1 heart lysates. As shown in Fig. 1E, both lines of Hdac3-Tg mice demonstrate 2-fold increases in HDAC activity. Furthermore, levels of acetyl-histone H4 in P1 Hdac3-Tg hearts were reduced by almost 40% (Fig. 1F), consistent with a moderate increase in HDAC activity. Further, we determined whether Hdac3 overexpression alters class I HDACs, Hdac1 and Hdac2, expression in neonatal myocardium. Our data show that Hdac3 overexpression has no effect on Hdac1 and Hdac2 protein levels (Fig. 1G).
FIGURE 1.
Generation and characterization of cardiac myocyte-specific Hdac3-Tg mice. A, schematic of transgenic construct used to generate Hdac3-Tg mice. α-MHC, α-myosin heavy chain. B, Hdac3 mRNA expression analysis in two different Hdac3-Tg mice lines. Transcripts for Hdac3 were detected by qRT-PCR in P1 hearts from wild-type (WT) and Hdac3-Tg mice. Three mice in each group were tested, and values are expressed as the -fold change in transcript abundance (± S.D.) when compared with wild-type mice. C, endogenous and transgenic Hdac3 protein were detected with an Hdac3 antibody. All Western blots were performed at least three times with similar results. D, Hdac3 expression in myocardium of P1 wild-type and Hdac3-Tg mice was quantified by using ImageJ software. E, increased HDAC activity in Hdac3-Tg mice. Lysates from P1 hearts were assayed for HDAC activity. Data are the average results (± S.D.) from three separate experiments. F, decreased acetylation of histone H4 in Hdac3-Tg mice. Western blot analysis of acetylated-histone H4 in myocardium from P1 wild-type and Hdac3-Tg mice using anti-acetyl-histone H4 antibody was performed. ImageJ software was used to quantify -fold change in acetylation. G, Hdac1 and Hdac2 levels are not changed in Hdac3-Tg mice. Western blot analysis of myocardium lysates from three P1 wild-type and three Hdac3-Tg mice using anti-Hdac1 and anti-Hdac2 antibody was performed. α-Tubulin is used as a loading control.
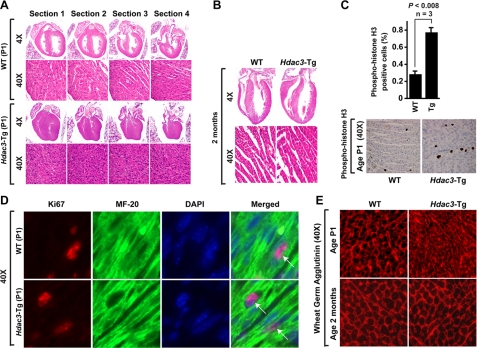
Hdac3 Overexpression Causes Myocyte Hyperplasia during Development—To determine the role of Hdac3 overexpression in cardiac development, we examined wild-type and Hdac3-Tg hearts at birth (P1). Histological analysis demonstrated near or complete loss of right and left ventricular lumens in Hdac3-Tg mice when compared with wild-type littermates (Fig. 2A). We also observed thickened ventricular walls and interventricular septa in serial sections of the P1 Hdac3-Tg hearts (Fig. 2A). Hematoxylin and eosin staining revealed increased nucleation of cardiac myocytes (Fig. 2A). We performed phospho-histone H3 staining to assess proliferation and to help to distinguish between cardiac myocyte hyperplasia and hypertrophy as the cause of increased wall thickness. Hdac3-Tg hearts showed a 2–3-fold increase in phospho-histone H3 staining throughout the myocardium when compared with wild-type hearts at P1 (p < 0.008, Fig. 2C). Similarly, Ki67 staining was also increased 2-fold in Hdac3-Tg hearts when compared with wild type (wild type 30.9% ± 2.9%, n = 3, Hdac3-Tg 54.8% ± 3.6%, n = 7, Fig. 2D). Increased proliferation was not observed in other tissue examined, including lung and skeletal muscle (data not shown). We performed Ki67 and MF-20 co-immunostaining on wild-type and Hdac3-Tg P1 heart sections to determine whether proliferating cells are myocytes. Our data suggest that ∼90% of the proliferating cells are cardiac myocytes (Fig. 2D). Wheat germ agglutinin staining to assess cardiac myocyte size revealed no significant difference in cardiac myocytes in Hdac3-Tg mice at P1 age when compared with wild-type littermates (wild type, 11.9 ± 1.2 μm2, Hdac3-Tg P1 myocytes, 12.0 ± 1.7 μm2, Fig. 2D). Interestingly, by 2 months of age, cardiac myocytes of Hdac3-Tg mice appeared relatively normal in size, and no increase in proliferative index was evident (Fig. 2, B and D). Histological analysis with trichrome stains revealed no evidence of excessive cardiac fibrosis in Hdac3-Tg mice at birth or at 2 months of age (data not shown).
FIGURE 2.
Cardiac defects in Hdac3-Tg neonates. A, hematoxylin and eosin-stained sections at successive levels of P1 hearts from wild-type (WT) and Hdac3-Tg mice demonstrate a thicker septum, thicker ventricular walls, and diminished ventricular lumens. B, hematoxylin and eosin-stained sections of adult hearts from wild-type and Hdac3-Tg mice demonstrate normal ventricular septum, walls, and lumens. C, increased proliferation of cardiomyocytes in P1 Hdac3-Tg mice. Immunohistochemistry for phospho-histone H3 was performed on heart sections from P1 mice. Quantification of phospho-histone H3-positive cells was performed on eight sections from three individual hearts and averaged. D, co-immunofluorescent staining for Ki67 (red), MyHC (green, MF-20 staining), and 4′,6-diamidino-2-phenylindole (DAPI) (blue) was performed on heart sections from P1 wild-type and Hdac3-Tg mice. E, wheat germ agglutinin staining shows decreased myocyte size in Hdac3-Tg when compared with wild-type P1 hearts.
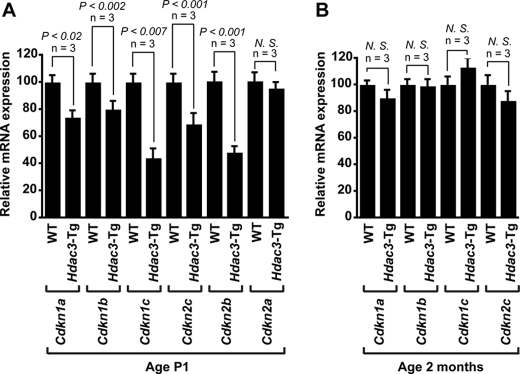
Hdac3 Overexpression Inhibits Cyclin-dependent Kinase Inhibitor Expression—Loss of Hdac3 has been shown to inhibit cell proliferation in human colon cancer cells via up-regulation of Cdkn1a (p21WAF/CIP1) (23). Conversely, Hdac3 overexpression has been shown to repress Cdkn1a and Cdkn2b transcription in human embryonic kidney cells (24). To determine the mechanism by which Hdac3 overexpression induces cardiac myocyte proliferation, we examined expression of CKIs in Hdac3-Tg P1 hearts. RT-PCR analysis indicates that Hdac3 suppresses expression of Cdkn1a, Cdkn1b (p27Kip1), Cdkn1c (p57Kip2), Cdkn2c (p18INC4c), and Cdkn2b (p15INC4b) at the transcriptional level (Fig. 3A). Hdac3 overexpression had no effect on expression of Cdkn2a (p16INC4a) in P1 hearts (Fig. 3A). These data suggest specificity of Hdac3 in the regulation of CKIs. At 2 months of age, no changes in CKI expression levels were detected (Fig. 3B).
FIGURE 3.
Hdac3 overexpression inhibits cyclin-dependent kinase (CDK) inhibitors Cdkn1a (p21WAF/CIP1), Cdkn1b (p27Kip1), Cdkn1c (p57Kip2), Cdkn2c (p18INC4c), Cdkn2b (p15INC4b), and Cdkn2a (p16INC4a). A and B, transcript for Cdkn1a, Cdkn1b, Cdkn1c, Cdkn2c, Cdkn2b, and Cdkn2a were detected by qRT-PCR in hearts from P1 (A) and 2-month-old (B) wild-type and Hdac3-Tg mice. Three mice in each group were tested, and values are expressed as the -fold change in transcript abundance (±S.D.) when compared with wild-type mice. N.S., not significant.
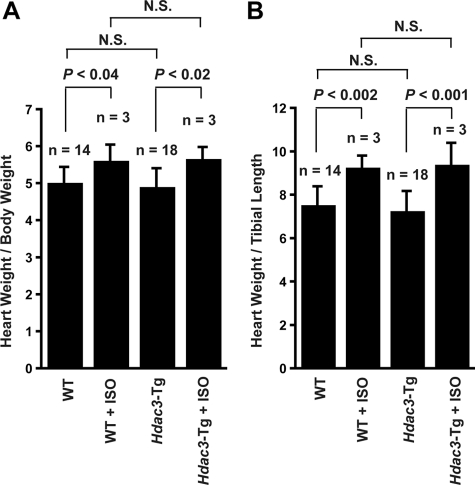
Hypertrophic Response to Adrenergic Stimulus in Hdac3-Tg Mice—Previously, our laboratory has demonstrated that myocyte-specific overexpression of another class I HDAC, Hdac2, induces cardiac hypertrophy and cardiac fibrosis by 2 months of age (12). To determine whether Hdac3 overexpression is sufficient to regulate cardiac hypertrophy, we determined heart-to-body-weight ratios and heart-weight-to-tibia-length ratios in 3-month-old Hdac3-Tg mice (n = 18) and when compared with wild-type (n = 14) littermates. No significant differences in heart-weight-to-body or heart-to-tibia-length measurements were present (Fig. 4, A and B). Further, we examined whether Hdac3 overexpression augments hypertrophic responsiveness to adrenergic stimulation. Wild-type and Hdac3-Tg littermates at 3 months of age were treated with a constant infusion of saline or ISO for 2 weeks. As predicted, wild-type mice exhibited marked cardiac hypertrophy, as revealed by an increase in the heart-to body-weight (and heart-to-tibia-length) ratios (Fig. 4, A and B). Hypertrophy of Hdac3-Tg mice in response to ISO was similar to wild type, and there was no apparent augmentation of hypertrophic response when compared with wild type (Fig. 4, A and B).
FIGURE 4.
A and B, heart-to-body-weight ratios (A) and heart-weight-to-tibial-length ratios (B) of wild-type and Hdac3-Tg mice at the age of 12 weeks with or without ISO treatment.
DISCUSSION
Hdac3 Regulates Cardiac Hyperplasia—Cardiac myocytes rapidly proliferate during embryonic and fetal life but generally lose their ability to proliferate shortly after birth (25). The withdrawal of postnatal cardiomyocytes from the cell cycle is linked to significant down-regulation of cyclins, CDKs, and E2F transcription factors and to up-regulation of the negative regulators of cell cycle progression including Cdkn1a, Cdkn1b, Cdkn1c, and Cdkn2c (26). Loss or down-regulation of these CKIs causes increased proliferation of neonatal myocytes. For example, mice lacking Cdkn1b show a 2–3-fold increase in myocyte proliferation and prolonged cell cycle (27). FOXO1-dependent activation of Cdkn1a, Cdkn1b, and Cdkn1c expression results in decreased myocyte proliferation (28). Although the roles of CKIs in the regulation of cardiomyocyte proliferation are well documented, their transcriptional regulation during cardiac development is not well defined. Our data suggest a novel regulator of CKI transcription in neonatal cardiac myocytes. We show that Hdac3 overexpression in cardiac myocytes suppresses Cdkn1a, Cdkn1b, Cdkn1c, Cdkn2b, and Cdkn2c mRNA expression neonatal P1 hearts. It will be interesting in future studies to determine whether Hdac3 directly or indirectly regulates the transcription of these genes.
Hdac3 overexpression produced increased cardiac myocyte proliferation at birth, but this effect was no longer evident by 2 months of age, although Hdac3-Tg protein was still detectable. Thus, mechanisms must exist within the myocyte to overcome the pro-proliferative effects of Hdac3 and to produce cell cycle exit. Similar phenomena have been described previously. For example, loss of Hopx or Hdac2 leads to increase in fetal cardiac myocyte proliferation, but these abnormalities abate in adulthood (12, 29). Similarly, Cdkn1b knock-out mice show increased fetal cardiac myocyte proliferation but eventually withdraw from the cell cycle (27). Normalization of cardiac size and myocyte cell number in adult transgenic mice despite early postnatal hyperplasia must be accomplished by an increase in cell loss via apoptosis or other mechanisms.
Previous studies have demonstrated that HDAC inhibitors, which target class I and II HDACs, induce growth arrest, maturation, and apoptosis of several tumors and cancer cell lines, implicating the specific roles of individual HDACs in the maintenance of cell proliferation and survival (30). Among class I and II HDACs, previous studies have demonstrated a role for class I HDACs, Hdac1 and Hdac2 in the regulation of cell proliferation. For example, global loss of Hdac1 results in embryonic lethality before embryonic day 10.5 due to severe proliferation defects and retardation in development (17). Interestingly, loss of Hdac1 up-regulates Cdkn1a and Cdkn1b, which may explain the proliferation defects (17). Loss of Hdac2 in mice results in severe cardiac defects at birth including increased cardiac myocyte proliferation and thickening of the myocardium, although the exact mechanism is not known (12, 18). Recently, it has been shown that global loss of Hdac3 leads to embryonic lethality before embryonic day 9.5 due to decrease in S phase cells and proliferation defects (31). Consistent with these findings, our data support a role for Hdac3 as a regulator of cardiac myocyte proliferation.
Previous studies have suggested a role of Hdac3 in cancer cell proliferation and mitosis. Hdac3 overexpression may inhibit Cdkn1a expression at the transcriptional level by interacting with Sp1, at least in some tumor cell lines (24). Additionally, liver-specific loss of Hdac3 results in 12-fold up-regulation of Cdkn1a at the transcriptional level (32). In accordance with these findings, our data suggest that overexpression of Hdac3 inhibits Cdkn1a transcription in cardiac myocytes. Further, we show that Hdac3 overexpression inhibits other CKIs including Cdkn1b, Cdkn1c, Cdkn2b, and Cdkn2c.
Roles of Class I and II HDACs in Hypertrophic Responsiveness—Most adult cardiac myocytes are unable to proliferate but can undergo hypertrophy in response to appropriate stimulation (26). The role of the HDACs in the regulation of cardiac hypertrophy is complex. Previous studies have demonstrated that class I and II HDACs have opposing roles in the regulation of cardiac hypertrophy (14). For example, inactivation of class II HDACs produces mice with grossly enlarged hearts, suggesting that class II HDACs repress cardiac hypertrophy (15). Recently, we have shown that global loss of the class I HDAC, Hdac2, results in mice that are resistant to hypertrophic stress (12). Furthermore, cardiac myocyte-specific overexpression of Hdac2 induces significant cardiac hypertrophy in 2-month-old mice (12). Together these data suggest that class I HDACs might normally function in the heart to repress anti-hypertrophic pathways.
Although class I HDACs are structurally related, they contribute to different transcriptional repression complexes (33). For example, mSin3A, NuRD, and CoREST complexes contain Hdac1 and Hdac2, whereas Hdac3 is a part of the NCoR-SMRT complex (19–21). Interestingly, class II HDACs can recruit NCoR-SMRT complexes that include Hdac3 (34). This has suggested that Hdac3 might be functionally related to class II HDAC effects with regard to the regulation of cardiac development and hypertrophy. However, our results suggest that overexpression of Hdac3 in the heart does not significantly affect hypertrophic responsiveness, at least under the conditions tested.
HDAC inhibitors are in phase I, II, and III clinical trials for hematological and oncological conditions (30, 35). Recently, suberoylanilide hydroxamic acid (also referred to as vorinostat/Zolinza) was approved by the Food and Drug Administration for the treatment of cutaneous T cell lymphoma (36). Elucidation of the specific functions of individual HDACs will be important to determine the molecular targets of these inhibitors and to provide information relevant to understanding and preventing potential cardiac side effects. Furthermore, mechanisms to regulate cardiac myocyte proliferation will be applicable to the exploding field of cardiac progenitor cell expansion and therapy.
Acknowledgments
We thank Yang Luo for technical assistance in the initial phase of the project.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 HL071546 (to J. A. E.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HDAC, histone deacetylase; Tg, transgenic; CKI, cyclin-dependent kinase inhibitors; SMRT, silencing mediator for retinoid and thyroid hormone receptors; NCor, nuclear receptor corepressor; RT-PCR, reverse transcription-PCR; qRT-PCR, quantitative RT-PCR; TRITC, tetramethylrhodamine isothiocyanate; ISO, isoproterenol; CDK, cyclin-dependent kinase; P1, postnatal day 1.
References
- 1.Wolffe, A. P. (1996) Science 272 371-372 [DOI] [PubMed] [Google Scholar]
- 2.Thiagalingam, S., Cheng, K. H., Lee, H. J., Mineva, N., Thiagalingam, A., and Ponte, J. F. (2003) Ann. N. Y. Acad. Sci. 983 84-100 [DOI] [PubMed] [Google Scholar]
- 3.Narlikar, G. J., Fan, H. Y., and Kingston, R. E. (2002) Cell 108 475-487 [DOI] [PubMed] [Google Scholar]
- 4.Ekwall, K. (2005) Trends Genet. 21 608-615 [DOI] [PubMed] [Google Scholar]
- 5.de Ruijter, A. J., van Gennip, A. H., Caron, H. N., Kemp, S., and van Kuilenburg, A. B. (2003) Biochem. J. 370 737-749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregoretti, I. V., Lee, Y. M., and Goodson, H. V. (2004) J. Mol. Biol. 338 17-31 [DOI] [PubMed] [Google Scholar]
- 7.Bolden, J. E., Peart, M. J., and Johnstone, R. W. (2006) Nat. Rev. Drug Discov. 5 769-784 [DOI] [PubMed] [Google Scholar]
- 8.Antos, C. L., McKinsey, T. A., Dreitz, M., Hollingsworth, L. M., Zhang, C. L., Schreiber, K., Rindt, H., Gorczynski, R. J., and Olson, E. N. (2003) J. Biol. Chem. 278 28930-28937 [DOI] [PubMed] [Google Scholar]
- 9.Kee, H. J., Sohn, I. S., Nam, K. I., Park, J. E., Qian, Y. R., Yin, Z., Ahn, Y., Jeong, M. H., Bang, Y. J., Kim, N., Kim, J. K., Kim, K. K., Epstein, J. A., and Kook, H. (2006) Circulation 113 51-59 [DOI] [PubMed] [Google Scholar]
- 10.Kong, Y., Tannous, P., Lu, G., Berenji, K., Rothermel, B. A., Olson, E. N., and Hill, J. A. (2006) Circulation 113 2579-2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kook, H., Lepore, J. J., Gitler, A. D., Lu, M. M., Wing-Man Yung, W., Mackay, J., Zhou, R., Ferrari, V., Gruber, P., and Epstein, J. A. (2003) J. Clin. Investig. 112 863-871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trivedi, C. M., Luo, Y., Yin, Z., Zhang, M., Zhu, W., Wang, T., Floss, T., Goettlicher, M., Noppinger, P. R., Wurst, W., Ferrari, V. A., Abrams, C. S., Gruber, P. J., and Epstein, J. A. (2007) Nat. Med. 13 324-331 [DOI] [PubMed] [Google Scholar]
- 13.Zhang, C. L., McKinsey, T. A., Chang, S., Antos, C. L., Hill, J. A., and Olson, E. N. (2002) Cell 110 479-488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Backs, J., and Olson, E. N. (2006) Circ. Res. 98 15-24 [DOI] [PubMed] [Google Scholar]
- 15.Chang, S., McKinsey, T. A., Zhang, C. L., Richardson, J. A., Hill, J. A., and Olson, E. N. (2004) Mol. Cell. Biol. 24 8467-8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang, S., Young, B. D., Li, S., Qi, X., Richardson, J. A., and Olson, E. N. (2006) Cell 126 321-334 [DOI] [PubMed] [Google Scholar]
- 17.Lagger, G., O'Carroll, D., Rembold, M., Khier, H., Tischler, J., Weitzer, G., Schuettengruber, B., Hauser, C., Brunmeir, R., Jenuwein, T., and Seiser, C. (2002) EMBO J. 21 2672-2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montgomery, R. L., Davis, C. A., Potthoff, M. J., Haberland, M., Fielitz, J., Qi, X., Hill, J. A., Richardson, J. A., and Olson, E. N. (2007) Genes Dev. 21 1790-1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassig, C. A., Fleischer, T. C., Billin, A. N., Schreiber, S. L., and Ayer, D. E. (1997) Cell 89 341-347 [DOI] [PubMed] [Google Scholar]
- 20.You, A., Tong, J. K., Grozinger, C. M., and Schreiber, S. L. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 1454-1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guenther, M. G., Lane, W. S., Fischle, W., Verdin, E., Lazar, M. A., and Shiekhattar, R. (2000) Genes Dev. 14 1048-1057 [PMC free article] [PubMed] [Google Scholar]
- 22.Trivedi, C. M., Patel, R. C., and Patel, C. V. (2007) Atherosclerosis 195 e50-60 [DOI] [PubMed] [Google Scholar]
- 23.Wilson, A. J., Byun, D. S., Popova, N., Murray, L. B., L'Italien, K., Sowa, Y., Arango, D., Velcich, A., Augenlicht, L. H., and Mariadason, J. M. (2006) J. Biol. Chem. 281 13548-13558 [DOI] [PubMed] [Google Scholar]
- 24.Huang, W., Tan, D., Wang, X., Han, S., Tan, J., Zhao, Y., Lu, J., and Huang, B. (2006) Biochem. Biophys. Res. Commun. 339 165-171 [DOI] [PubMed] [Google Scholar]
- 25.Bicknell, K. A., Coxon, C. H., and Brooks, G. (2007) J. Mol. Cell. Cardiol. 42 706-721 [DOI] [PubMed] [Google Scholar]
- 26.Ahuja, P., Sdek, P., and MacLellan, W. R. (2007) Physiol. Rev. 87 521-544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poolman, R. A., Li, J. M., Durand, B., and Brooks, G. (1999) Circ. Res. 85 117-127 [DOI] [PubMed] [Google Scholar]
- 28.Evans-Anderson, H. J., Alfieri, C. M., and Yutzey, K. E. (2008) Circ. Res. 102 686-694 [DOI] [PubMed] [Google Scholar]
- 29.Chen, F., Kook, H., Milewski, R., Gitler, A. D., Lu, M. M., Li, J., Nazarian, R., Schnepp, R., Jen, K., Biben, C., Runke, G., Mackay, J. P., Novotny, J., Schwartz, R. J., Harvey, R. P., Mullins, M. C., and Epstein, J. A. (2002) Cell 110 713-723 [DOI] [PubMed] [Google Scholar]
- 30.Xu, W. S., Parmigiani, R. B., and Marks, P. A. (2007) Oncogene 26 5541-5552 [DOI] [PubMed] [Google Scholar]
- 31.Bhaskara, S., Chyla, B. J., Amann, J. M., Knutson, S. K., Cortez, D., Sun, Z. W., and Hiebert, S. W. (2008) Mol. Cell 30 61-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knutson, S. K., Chyla, B. J., Amann, J. M., Bhaskara, S., Huppert, S. S., and Hiebert, S. W. (2008) EMBO J. 27 1017-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahringer, J. (2000) Trends Genet. 16 351-356 [DOI] [PubMed] [Google Scholar]
- 34.Fischle, W., Dequiedt, F., Hendzel, M. J., Guenther, M. G., Lazar, M. A., Voelter, W., and Verdin, E. (2002) Mol. Cell 9 45-57 [DOI] [PubMed] [Google Scholar]
- 35.Marks, P., Rifkind, R. A., Richon, V. M., Breslow, R., Miller, T., and Kelly, W. K. (2001) Nat. Rev. Cancer 1 194-202 [DOI] [PubMed] [Google Scholar]
- 36.Garber, K. (2007) Nat. Biotechnol. 25 17-19 [DOI] [PubMed] [Google Scholar]