Abstract
The mitochondrial genome is highly susceptible to damage by reactive oxygen species (ROS) generated endogenously as a byproduct of respiration. ROS-induced DNA lesions, including oxidized bases, abasic (AP) sites, and oxidized AP sites, cause DNA strand breaks and are repaired via the base excision repair (BER) pathway in both the nucleus and mitochondria. Repair of damaged bases and AP sites involving 1-nucleotide incorporation, named single nucleotide (SN)-BER, was observed with mitochondrial and nuclear extracts. During SN-BER, the 5′-phosphodeoxyribose (dRP) moiety, generated by AP-endonuclease (APE1), is removed by the lyase activity of DNA polymerase γ (pol γ) and polymerase β in the mitochondria and nucleus, respectively. However, the repair of oxidized deoxyribose fragments at the 5′ terminus after strand break would require 5′-exo/endonuclease activity that is provided by the flap endonuclease (FEN-1) in the nucleus, resulting in multinucleotide repair patch (long patch (LP)-BER). Here we show the presence of a 5′-exo/endonuclease in the mitochondrial extracts of mouse and human cells that is involved in the repair of a lyase-resistant AP site analog via multinucleotide incorporation, upstream and downstream to the lesion site. We conclude that LP-BER also occurs in the mitochondria requiring the 5′-exo/endonuclease and pol γ with 3′-exonuclease activity. Although a FEN-1 antibody cross-reacting species was detected in the mitochondria, it was absent in the LP-BER-proficient APE1 immunocomplex isolated from the mitochondrial extract that contains APE1, pol γ, and DNA ligase 3. The LP-BER activity was marginally affected in FEN-1-depleted mitochondrial extracts, further supporting the involvement of an unidentified 5′-exo/endonuclease in mitochondrial LP-BER.
The mammalian mitochondrion contains 5–15 copies of the circular 16-kb mitochondrial (mt)2 genome, and each mammalian cell thus may contain a thousand or more copies of the mt genome (1). MtDNA, encoding 13 subunits of the electron transport chain and containing genes for ribosomal RNAs and tRNAs (2), is extremely susceptible to oxidative damage, presumably because of the lack of protective histones and proximity to reactive oxygen species (ROS), which are endogenously generated by the electron transport complexes (3, 4). Such damage includes several dozen oxidized bases, abasic (AP) sites, and oxidation products of AP sites leading to DNA strand breaks (5). Endogenous mutations in mtDNA, likely to arise from these lesions, were shown to be considerably higher than in the nuclear genome (6). Approximately 10,000 AP sites were estimated to be generated per nuclear genome per day (7). Analysis of the release of 5-methylene-2-furanone, the product of β-, γ-elimination of 2-deoxyribonolactone, an oxidized AP site, causing DNA strand breakage, suggests that this ribonolactone could account for 70% of the total sugar damage in DNA (8, 9). The oxidized AP sites, whose level is likely to be high especially in the mtDNA, should block replication and transcription and would hence be cytotoxic and mutagenic.
Although the mitochondria lack the enzymes to repair UV-photoproducts in the genome (10), efficient repair of oxidative damage in mtDNA was shown to occur primarily via the DNA base excision repair (BER) pathway. Some of the mitochondrial isoforms of nuclear BER enzymes have been characterized (11), and in vivo evidence for BER in mitochondria was shown (4, 12). Details of the BER pathway, based on studies with the nuclear BER enzymes, have been recently reviewed (13–15). Briefly, repair of damage or abnormal bases is initiated with their excision by a DNA glycosylase. A monofunctional glycosylase, e.g. uracil-DNA glycosylase (UDG), excises U from the DNA to generate an AP site, which is then cleaved by AP-endonuclease (APE1) in the mammalian cell, leaving a 3′-OH group and a nonligatable 5′-deoxyribose phosphate (dRP) residue. In the nucleus, this 5′-blocking group could be removed by DNA polymerase β via its intrinsic dRP lyase activity. In the mitochondria, the DNA polymerase γ (pol γ) with similar dRP lyase activity is also able to remove the dRP moiety (16). In the case of oxidized base repair by DNA glycosylases with associated AP lyase activity, e.g. 8-oxoguanine-DNA glycosylase (OGG1), base excision is coupled to strand cleavage at the AP site with generation of 5′-phosphate and 3′-blocking phospho-α,β unsaturated aldehyde (derived from deoxyribose), which is subsequently removed by the intrinsic 3′-phosphodiesterase activity of APE1, leaving a 3′-OH group as a primer terminus for repair synthesis. The absence of an aldehyde group in the oxidized deoxyribose fragment at the 5′ terminus after DNA strand break precludes removal of these lesions by the dRP lyase activity of pol β in the nucleus. In such a case the 5′-blocking group is removed by flap endonuclease 1 (FEN-1), a 5′-exo/endonuclease (17, 18). Thus, the resulting gap filling by a DNA polymerase and nick-sealing by DNA ligase during BER could proceed via two subpathways: SN-BER where only the damage base is replaced or LP-BER where 2–6 additional nucleotides at the 5′ terminus are removed by FEN-1 followed by resynthesis. In the nucleus, DNA ligase 3 (lig3α) is involved in SN-BER after pol β fills in the single nucleotide gap. FEN-1-mediated gap is likely to be filled in by replicative DNA polymerases δ/ε followed by nick-sealing with DNA ligase 1 (lig1). In contrast to the situation in the nucleus with multiple enzymes, pol γ and lig3α are involved in both replication and BER of mtDNA (19). In this case, 5′-dRP generated after AP site cleavage is removed by the dRP lyase activity of pol γ, similar to that of pol β for the nuclear SN-BER. With mitochondrial extracts of rat liver, the repair of uracil was shown to occur only via the SN-BER process in contrast to the nucleus where LP-BER activity was also documented (4). However, how an oxidized AP site lacking the aldehyde group could be repaired in the mitochondria has not been investigated. Such repair would require the presence of FEN-1 or another 5′-exo/endonuclease in the mitochondria. In this study, we analyzed the repair of a reduced AP site analog and APE1 substrate tetrahydrofuran, which has been extensively used in in vitro BER studies. We provide evidence for LP-BER in human and mouse mitochondria mediated primarily by an unidentified 5′-exo/endonuclease.
EXPERIMENTAL PROCEDURES
Materials—4-Month-old BALB/c mice were purchased from the National Institute on Aging. All animal experiments were performed according to the NIH Guidelines for Care and Use of Laboratory Animals and approved by the UTMB Animal Care and Use Committee (no. 00-01-007A). Reversed-phase HPLC-purified oligonucleotides were purchased from the Midland Certified Reagent Company, and the sequence of each oligonucleotide is presented in Table 1. FEN-1-specific and control siRNA were purchased from Ambion and 32P-labeled nucleotides from GE Healthcare.
TABLE 1.
Oligonucleotides used in this study
Preparation of Cell and Mitochondrial Extracts—Total cell extracts were prepared by homogenizing liver, kidney, or human colorectal cancer line HCT116 in 20 mm Tris-HCl, pH 8.8, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 12 mm sodium deoxycholate. Mitochondria from mouse liver and kidney and HCT116 cells were isolated using the Mitochondrial Isolation Kit for Tissue or Cultured Cells (Pierce), respectively, according to the manufacturer's recommendation or as described before (20). To remove externally adhering contaminants, intact mitochondria (1 mg/ml) were treated with trypsin (10 μg/ml) for 20 min at room temperature and followed by the addition of an equivalent amount of bovine trypsin inhibitor (Invitrogen) to stop trypsin activity (21, 22). The mitochondrial suspensions were then lysed in 20 mm HEPES-KOH (pH 7.4), 1 mm EDTA, 1 mm dithiothreitol, 300 mm KCl, 5% glycerol, and 0.5% Triton X-100. Protein concentration was determined with the Bio-Rad reagent using bovine serum albumin as the standard.
Western Blot Analysis—Western analysis was performed as described earlier (23). After electroblotting of proteins, the membranes were sequentially probed with antibodies against calregulin (Santa Cruz Biotechnology), β-actin (Sigma), FEN-1 (Bethyl Laboratories or GeneTex, Inc.), lactate dehydrogenase (Chemicon International), subunit β of complex V (Molecular Probes), APE1, EndoG (Novus Biological), pol γ (Abcam or Ref. 16), Lig3 (QED Bioscience Inc.), and PCNA (Bethyl Laboratories). The bound primary antibodies were quantitated by chemiluminescence using horseradish peroxidase-conjugated secondary antibody (anti-mouse or anti-rabbit IgG-HRP; Amersham Biosciences). The membranes were washed, incubated in ECL reagent (Amersham Biosciences), visualized, and analyzed using Gel Logic 2200 (Kodak) and Kodak Molecular Imaging Software (Kodak), respectively.
DNA Repair Synthesis Assay—DNA repair assay was carried out as described earlier, with slight modifications (24). Briefly, the assay mixture (20 μl) contained 40 mm HEPES (pH 7.6), 0.1 mm EDTA, 5 mm MgCl2, 0.2 mg/ml bovine serum albumin, 50 mm KCl, 1 mm dithiothreitol, 2 mm ATP, 3% glycerol, 20 μm of each unlabeled dNTPs, 4 μCi of one of the [α-32P]dNTPs, mitochondrial protein extract (5–10 μg) and various duplex oligo substrates (Table 1). After incubation at 37 °C for various times, the substrates and products were separated by electrophoresis in 20% acrylamide/7 m urea gel. The radioactivity in these bands was quantitated in a PhosphorImager (Molecular Dynamics) using ImageQuant software. Preliminary enzyme assays were carried out to ensure the linearity of reaction with respect to both time and amount of extract.
5′and 3′-Terminal Labeling of Oligonucleotides—Oligo5 was labeled at the 5′-end with [γ-32P]ATP and T4 polynucleotide kinase (New England BioLabs) as described earlier (25). 3′-End labeling was performed for an oligonucleotide with the same sequence as oligo5 but one nucleotide shorter at the 3′ terminus. After annealing with the complementary oligo, the 1-nt 5′-overhang was used as the template for filling with [α-32P]dTMP at the 3′-end using 3′-exo-deficient DNA polymerase (Klenow Fragment, New England BioLabs).
Immunoprecipitation—Immunoprecipitation was performed using the standard procedure after lysing mitochondria or nuclei in nondenaturing lysis buffer (NLB) containing 1% Triton X-100, 50 mm Tris-HCl pH 7.4, 300 mm NaCl, 5 mm EDTA, 0.02% sodium azide, and Complete protease inhibitor mixture. FLAG-specific immunoprecipitation was performed using 70 μl of anti-FLAG M2-agarose from mouse, washed twice with TBS buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5), and then incubated with ∼1 mg of mitochondrial extract isolated from HCT116 cells that were transfected with human APE1 cDNA cloned into a C-terminal pFLAG-CMV-5.1 vector (Sigma), for 4 h at 4 °C in a tube rotator. To remove nonspecific proteins, four washes with TBS buffer were performed, and the final pellet was resuspended in SDS sample buffer for Western analysis or in PBS for repair activity assay. Immunoprecipitation with FEN-1 antibody was performed by first immobilizing the antibody on protein A-agarose (Invitrogen) in PBS buffer for 2 h at room temperature in a tube rotator. Unbound antibody was removed by three washes with PBS and once with NLB. Nuclear or mitochondrial extract was then added to immobilized FEN-1 antibody and further incubated overnight at 4 °C in a tube rotator. After removing nonspecific proteins by four washes with PBS, the final pellet was resuspended in SDS sample buffer.
FEN-1 Down-regulation with siRNA—HCT116 cells were transfected with 25 nm FEN-1-specific or control siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendation. To optimize the siRNA concentration, preliminary transfections were carried out with 1–100 nm FEN-1-specific siRNA.
RESULTS
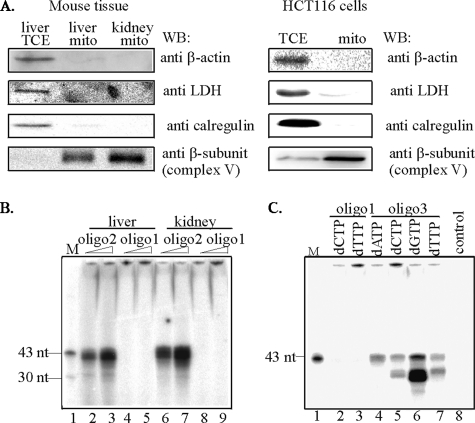
Purity of Mitochondrial Extracts—It is now evident that DNA repair proteins are not exclusively localized in the nucleus or mitochondria, and that a variable but significant fraction could also be found in the cytoplasm. It is thus critical to isolate mitochondrial fraction free of adherent cytosolic proteins when examining mitochondria-specific repair. Several methods have been used to ensure the purity of mitochondria. In our laboratory, to obtain pure mitochondrial extracts, we routinely treat purified mitochondria with trypsin. In previous reports, we showed the importance of this treatment to remove cellular contamination, mainly from the endoplasmic reticulum (26, 27). Western analysis of mitochondrial fractions of mouse liver and kidney and human HCT116 cells confirmed the absence of cytoplasmic (lactate dehydrogenase, LDH), endoplasmic reticulum (calregulin), and cytoskeletal (β-actin) contamination in our mitochondrial preparations (Fig. 1A). Only mitochondrial extracts verified by Western analysis were used in further studies.
FIGURE 1.
Functional properties of mitochondrial extracts. A, purity of mitochondrial extracts analyzed by Western blot of total cell (TCE) or mitochondrial (mito) extracts from young mouse liver and kidney (left panel), and human HCT116 cells (right panel) probed with antibody against: lactate dehydrogenase, LDH (cytoplasmic), calregulin (ER), β-actin (cytoskeletal), β-subunit of complex V (mitochondrial). B, SN-BER of uracil-containing oligo2 duplex using two different amounts of mitochondrial extracts of liver or kidney of young mice. C, LP-BER of THF-containing oligo3 duplex using mitochondrial extracts from HCT116 cells. M, marker (43 and 30 nt). control, no extract.
SN-BER and LP-BER Activities in Mitochondrial Extracts—To confirm repair competence of mitochondrial extracts, oligo2 was annealed to the complementary oligo containing G opposite to U (Table 1), and the duplex was incubated only with [α-32P]dCTP and mitochondrial extracts (Fig. 1B). Mitochondrial extracts of mouse liver and kidney (lanes 2, 3 and 6, 7, respectively) catalyzed incorporation of [α-32P]dCMP in the absence of other dNTPs, indicating that the purified extract carried out SN-BER of uracil. The absence of [α-32P]dCMP incorporation into control oligo1 without U (lanes 4, 5 and 8, 9) indicated specificity of DNA repair. To examine the ability of mitochondria to carry out LP-BER, the THF-containing oligo3 duplex (Table 1) was used as the substrate. Fig. 1C shows DNA repair assay with mitochondrial extracts from HCT116 cells. Incorporation of all four deoxynucleotides in the substrate indicated multibase repair patch (Fig. 1C, lanes 4–7). Again, control oligo1 duplex with normal bases did not incorporate [α-32P]dCMP or [α-32P]dTMP, confirming specific repair synthesis (Fig. 1C, lanes 2 and 3). As expected, no radioactivity was incorporated into the substrate in the absence of mitochondrial extract (Fig. 1C, lane 8). Furthermore, the presence of radioactivity in the unligated product was observed primarily when [α-32P]dGTP was used (Fig. 1C, lane 6).
Authentic LP-BER and Not Strand Displacement Synthesis with Mitochondrial Extracts—A potential complication of analyzing LP-BER using mitochondrial extracts is the robust strand displacement activity of pol γ as a result of which the downstream fragment in the oligo duplex after a strand break could be removed and followed by resynthesis of the entire strand. This could be mistaken for repair and is of particular concern with short oligo substrates, which does not require the 5′-end processing activity of a 5′-exo/endonuclease. We tested for such a possibility and the presence of authentic LP-BER activity in the mitochondrial extracts several ways. First, we used an oligo duplex with 10 additional bases on the 3′-side of the THF lesion. Strand displacement/resynthesis would be reflected in comparable incorporation of deoxynucleotides at distal versus proximal sites relative to the lesion. Secondly, we radiolabeled the THF-containing strand at the 3′ terminus. Once the strand is cleaved by APE1, the label would be permanently lost if strand displacement/synthesis occurs. On the other hand, in the case of true repair, the radiolabeled 21-nt fragment generated by APE1 would be converted into the full-length oligo after resynthesis of a short (2–8 nt) gap by pol γ.
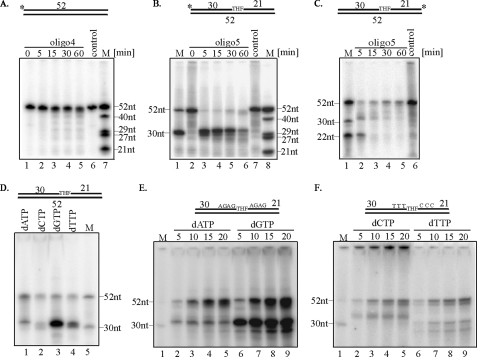
To characterize LP-BER activity of mitochondrial extracts, the repair of THF-containing oligo5 labeled at the 5′ or 3′ terminus with 32P was carried out (Table 1). First, we analyzed the size of 5′-end-labeled oligo4, which does not contain any damage, after its incubation with mitochondrial extract of young liver, and we could not detect any qualitative changes (Fig. 2A). We then analyzed the size of 5′-end-labeled oligo5 after incubation with mouse liver mitochondrial extracts from young mice. The appearance of a 30-nt band indicated cleavage of the substrate by mitochondrial APE1 at the THF site. Slow restoration of the 52-nt band indicated repair (Fig. 2B, lanes 2–6). We detected up to 2–3-fold increase in complete repair product formation as a function of incubation time. The 30-nt band was not observed when control oligo4 was used, confirming the synthesis to be damage-specific (Fig. 2A, lanes 1–5). In a parallel experiment, 3′-end-labeled oligo5 duplex was incubated with liver mitochondrial extracts isolated from young mice. Again the appearance of a 21-nt band after 5 min of incubation indicated strand cleavage at the THF site by APE1 (Fig. 2C, lane 2), and time-dependent restoration of the 52-nt band indicated complete repair (Fig. 2C, lanes 2–5). Some intermediate sized bands were also visible. As expected, the smaller bands were absent in the control containing no mitochondrial extract (Fig. 2, B and C, lanes 7 and 6, respectively). In all experiments, some loss of the 52-nt band occurred, which was likely due to nonspecific degradation. This should perhaps be expected because of the presence of multiple nucleases in crude mitochondrial extracts. This is also evident from of undamaged control oligo (Fig. 2A). In particular, the 3′- and 5′-terminally radiolabeled oligos could be readily removed by the exonuclease. Nevertheless, the restoration of the 52-nt full-length oligo after incubation with both 5′- and 3′-terminally radiolabeled substrates (Fig. 2, B and C) was consistent with initial strand break at the THF site followed by repair synthesis and ligation. Fig. 2C also strongly argues against strand displacement synthesis in which the 3′-terminal label would be irreversibly lost. Similar experiments were carried out with unlabeled oligo5 substrate in the presence of each radiolabeled dNTPs and mitochondrial extracts from liver (Fig. 2D). Incorporation of radioactivity confirmed that the mitochondrial extracts carried out LP-BER (Fig. 2D, lanes 1–4). It needs to be emphasized that similar experiments were performed using mitochondrial extracts isolated from young mouse kidney and HCT116 cells with similar results (data not shown). Taken these data together, we conclude that mitochondrial extracts from various mammalian tissues and human cells could perform LP-BER.
FIGURE 2.
Analysis of mitochondrial LP-BER. Time course of repair of nondamaged oligo4 duplex (A) and THF-containing oligo5 duplex (B)5′-end-labeled or (C)3′-end-labeled oligo5 duplex, by mitochondrial extract of young mouse liver. The asterisk indicates 32P-terminal label. DNA repair assay for unlabeled THF-containing oligo5 duplex (D) or oligo6 duplex (E and F) using mitochondrial extracts of young mouse liver or HCT116 cells respectively and individual [α-32P]dNTPs used to monitor repair are indicated. M, marker oligonucleotides of indicated length.
Characterization of Long Patch Repair Product—pol γ, the sole mitochondrial DNA polymerase, is responsible for mtDNA replication and also participates in BER (19). To our knowledge, it is the only mammalian DNA polymerase possessing both 3′-5′ exonuclease and dRP lyase activities (16). Incorporation of multiple nucleotides in the THF oligo substrate could thus be due to the filling of gap generated by the 3′-exonuclease activity of pol γ or a 5′-exo/endonuclease, such as FEN-1, or both. To establish the directionality of repair synthesis, we designed the sequence of oligo6 (Table 1) such that incorporation of [α-32P]dTMP and [α-32P]dCMP would indicate resynthesis of DNA 3′-5′ and 5′-3′ to the THF site, respectively. Fig. 2E shows incorporation of [α-32P]dAMP and [α-32P]dGMP into oligo6, indicating repair of the intact template by HCT116 mitochondrial extracts. In addition, appearance of the radiolabeled 31-nt band, especially when [α-32P]dGTP was used, indicated incorporation of [α-32P]dGMP at the THF site after strand cleavage. Persistence of the cleaved and radiolabeled fragment strongly suggests that its ligation or 3′-exonucleolytic processing of the downstream fragment was rate-limiting. However, the radiolabeled 31-nt band, observed after [α-32P]dAMP incorporation suggests resynthesis of the template 5′ to the THF site, which is removed by the 3′-exonuclease activity of pol γ (Table 1). Radiolabeling of the 52-nt band by [α-32P]dCMP and [α-32P]dTMP indicated repair due to replacement of the nucleotides on both sides of the THF (Fig. 2F). The 31-nt band with [α-32P]dTMP incorporation likely represented the unligated repair intermediate. Fragments that are shorter than 30 nt and radiolabeled with [α-32P]dTMP were likely to represent incomplete synthesis after exonucleolytic degradation by pol γ, and similar fragments were observed after [α-32P]dGMP incorporation (Fig. 2E, lanes 6–9). We could not detect labeling of the 31-nt band with [α-32P]dCMP, which could be only incorporated at the 3′-site of the damage (Table 1). The band longer than 30 nt represented the flap generated after repair synthesis. Similar experiments with mitochondrial extracts from mouse liver and kidney yielded comparable results (data not shown). Summarizing the results, we conclude that mitochondrial repair of an AP site analog involved the replacement of deoxynucleotides on both sides of the damage, which may be unique to mitochondrial LP-BER.
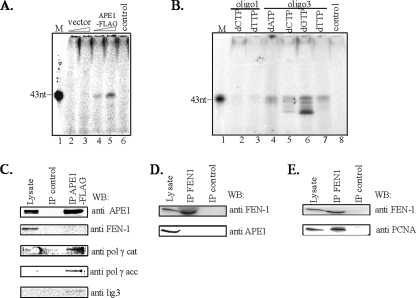
Mitochondrial-specific Proteins Involved in LP-BER—We recently observed that the immunocomplex for a DNA glycosylase could be isolated from nuclear extracts, and that contains all proteins necessary for carrying out complete BER (24). We therefore examined whether similar immunocomplexes could be isolated from mitochondrial extracts. Because of the availability of highly specific FLAG antibody for isolating immunocomplexes, we ectopically expressed C-terminal FLAG-tagged APE1 in HCT116 cells at a level comparable to endogenous APE1. The APE1-FLAG immunocomplex (IP), isolated from the mitochondrial extracts of these cells, was used for repair of THF in oligo3 (Table 1). We assessed LP-BER by monitoring incorporation of [α-32P]dTMP as before. Fig. 3A shows that the IP of APE1-FLAG (lanes 4 and 5) but not of the vector control (lanes 2 and 3) incorporated [α-32P]dTMP, suggesting that the immunocomplex contains all proteins required to carry out LP-BER. To further characterize the repair synthesis, we performed incorporation assays using each of the four [α-32P]dNTPs. All 4 dNMPs were incorporated into the oligo3 duplex (Fig. 3B, lanes 4–7), and intermediates of the repair reaction were visible. DNA synthesis due to repair was confirmed using control oligo1 (Fig. 3B, lanes 2 and 3). The absence of radiolabeled fragments in the control oligo further supports our conclusions that the DNA synthesis observed for the THF-oligo was due to the true repair and not resynthesis of the undamaged DNA after exonucleolytic degradation. These results provide the first evidence that mitochondrial BER is also carried out by a repair complex as we had observed earlier for repair of 5-hydroxyuracil with NEIL2 IP isolated from the nuclear extract (24).
FIGURE 3.
Characterization of HCT116 mitochondrial repair complex for LP-BER. A, repair of unlabeled THF-containing oligo3 duplex with [α-32P]dTTP and two different amounts of mitochondrial APE1-FLAG or control FLAG immunocomplex. B, monitoring of LP-BER with APE1-FLAG immunocomplex with [α-32P]dNTPs. M, marker (43 nt). control, no protein. Western analysis of mitochondrial APE1-FLAG immunocomplex (C). The absence of APE1 (D) and presence of PCNA (E) in FEN-1 immunocomplex isolated from mitochondrial and nuclear extracts, respectively. Lysate, 50 μg of mitochondrial or nuclear extracts.
We then tested for the presence of other BER proteins in the IP. In addition to APE1, both catalytic and accessory subunits of pol γ as well as lig3 could be detected by Western analysis (Fig. 3C). Because removal of the THF residue at the 5′ terminus of APE1-mediated cleaved strand could not be carried out by pol γ alone, a 5′-exo/endonuclease activity must be present in the IP. We tested for the presence of FEN-1, the logical candidate, but could not detect it by Western analysis even though a FEN-1 antibody cross-reacting band was observed after Western analysis of the crude mitochondrial lysate (Fig. 3C). These results strongly suggest that LP-BER in mitochondria, unlike in the nucleus, may not involve FEN-1. To further confirm the absence of FEN-1 in mitochondrial immunocomplexes proficient in LP-BER, several independent immunoprecipitates of mitochondrial and nuclear extracts were isolated using FEN-1-specific antibody (Fig. 3, D and E). Lack of APE1 in the FEN-1 IP from mitochondrial extracts confirmed lack of stable interaction of FEN-1 with this protein (Fig. 3D). On the other hand, as a positive control, the presence of PCNA in FEN-1 IP from the nuclear extract was detected (Fig. 3E), in support of earlier studies (19, 28).
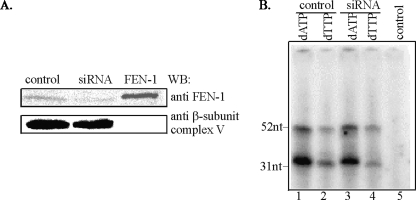
To independently verify the lack of FEN-1 involvement in mitochondrial LP-BER, we down-regulated FEN-1 with specific siRNA. Western analysis showed reduction of the FEN-1-specific band by 80% in the mitochondrial extract of HCT116 cells (Fig. 4A). Repair of oligo5 with this mitochondrial extract was monitored by incorporation of [32P]dAMP and [32P]dTMP (Fig. 4B). It is evident that LP-BER repair activity of the mitochondrial extract was barely reduced after FEN-1 down-regulation. These results strongly suggest that mitochondrial LP-BER is, mostly if not completely, FEN-1-independent, and imply the presence of an unidentified 5′-exo/endonuclease in the mitochondria.
FIGURE 4.
Effect of FEN-1 down-regulation on repair by HCT116 mitochondrial extract. A, Western analysis of FEN-1 in mitochondrial extracts of HCT116 cells transfected with control or FEN-1-specific siRNA. Human recombinant FEN-1 was used as a control. The membrane was reprobed for β-subunit of ATP synthase to assure equal loading. B, repair of THF-containing oligo5 duplex with mitochondrial extracts from control and FEN-1-down-regulated cells. control, no extract.
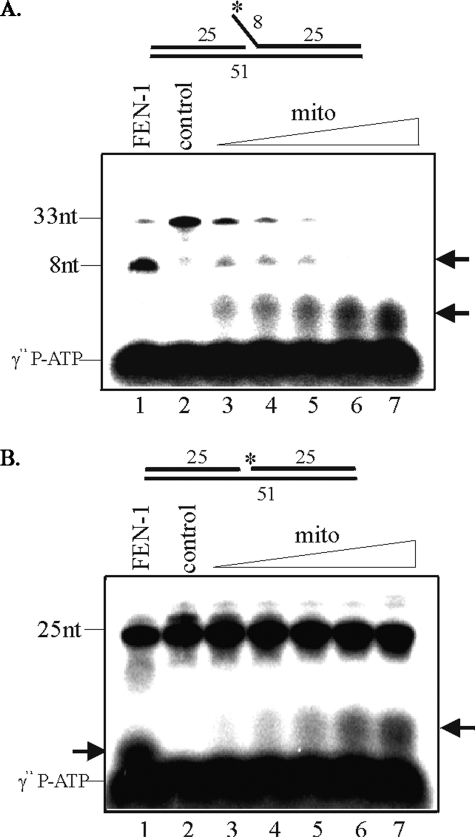
Presence of a Distinct 5′-Endo/exonuclease in the Mitochondria—We tested for the presence of a 5′-exo/endonuclease activity in the mitochondrial extract with oligo8 a FEN-1-specific flap substrate (Table 1). When 5′-32P-labeled oligo8 was incubated with HCT116 mitochondrial extract, the 8-nt fragment, the predicted cleavage product of FEN-1, was not observed. On the other hand, in the positive control experiment with recombinant human FEN-1, the radiolabeled 8-nt fragment was indeed generated (Fig. 5A, lane 1). However, we did observe radiolabeled fragments ∼3–4-nt long after incubation with mitochondrial extract whose level increased with the amount of extract (Fig. 5A, lanes 3–7). Because we could observe a very weak flap-specific activity, it is possible that the flap product was further degraded by the robust activity of the unidentified 5′-exo/endonuclease (Fig. 5A, lanes 3–7). We also used 5′-labeled DNA gap substrate (oligo7, Table 1). A similar 3–4-nt band with a dose-dependent increase was observed with the same extract (Fig. 5B, lanes 3–7). These results strongly indicate that mammalian mitochondria possess a distinct 5′-exo/endonuclease. To rule out the possibility that the detected activity of mitochondrial 5′-exo/endonuclease is actually mitochondrial endonuclease G (EndoG), we fractionated mitochondrial proteins on SP FF cation exchange column using NaCl gradient and tested the fractions for 5′-exo/endonuclease activity with oligo8 and the presence of EndoG by Western analysis. Mitochondrial 5′-exo/endonuclease activity was recovered in the flow-through fraction and did not bind to the cation exchange column unlike EndoG which eluted at 0.4–0.5 m NaCl (data not shown).
FIGURE 5.
Distinct mitochondrial 5′-exo/endonuclease activity in HCT116 cells. Substrates were labeled at the terminus of flap of oligo8 (A) or gap of oligo7 (B) as indicated by an asterisk, and increasing concentration of mitochondrial extracts was used in assays (lanes 3–7). Human recombinant FEN-1 was used as a positive control (lane 1). control, no extract.
In conclusion, we have shown the presence of the LP-BER pathway in mammalian mitochondria that would be required for the repair of oxidized AP sites. Unlike the nuclear repair, this pathway appears to be mostly FEN-1-independent, and the repair patch spans both sides of the damage which is unique to the mitochondria.
DISCUSSION
BER is the only well characterized repair pathway in the mitochondria for repair of oxidized and alkylated bases, uracil and AP sites, and also single strand breaks in DNA. We have shown in this study that an AP site analog can be repaired by the mammalian mitochondrial extract, via the LP-BER pathway. We have shown for the first time at this report the existence of this sub-pathway of BER in mammalian mitochondria in which the SN-BER was characterized earlier (19, 29).
Single nucleotide versus long patch repair were first identified as BER subpathways for the nuclear genome, which utilize distinct set of proteins for DNA synthesis and strand joining. Nuclear SN-BER requires XRCC1, pol β, and lig3α while LP-BER utilizes replication-associated proteins, namely PCNA, FEN-1, pol δ (ε), and lig1 (15). The repair patch size of 2–6 nucleotides for LP-BER is the consequence of replacement of nucleotides 3′ to the damage site in addition to the 1-nucleotide gap generated by removal of the damage itself. It is generally accepted that replicating DNA polymerases displace the 5′-strand during repair synthesis during LP-BER and generate a 2–6-nucleotide flap in a situation analogous to discontinuous strand synthesis during DNA replication where the 5′-RNA primer-containing segments of Okazaki fragments are displaced (14). FEN-1 acts as a 5′-endonuclease to remove the flap, and the resulting nick is sealed by lig1. However, replacement synthesis observed in LP-BER may not necessarily require prior generation of a flap structure by strand displacement. Nuclear SN-BER requires the AP lyase activity of pol β, which removes the 5′-dRP generated by APE1 cleavage of the AP site (18, 30). The lyase reaction requires the presence of the sugar aldehyde; therefore, oxidized deoxyribose or its fragments at the 5′ terminus lacking the aldehyde group could not be removed by pol β while FEN-1 could carry out such 5′-end processing. The mitochondria contain only one DNA polymerase, pol γ, and one DNA ligase, lig3α, both of which are required for mitochondrial genome replication as well as repair. Unlike the nuclear replicative polymerases, pol δ/ε, the mitochondrial pol γ possesses dRP lyase activity, which is necessary for removal of the 5-dRP moiety after AP endonuclease activity in mitochondrial SN-BER (16). However, this dRP lyase activity is 17-fold lower than of pol β (16), and given the oxidative environment, a second pathway for 5′-end processing in the mitochondria seems likely. The second pathway would be essential when the 5′-dRP group is oxidized and could not be removed by lyase reaction. Although not identified earlier, we suspected the presence of FEN-1 in the mitochondria. Indeed, our results indicate, for the first time, FEN-1-like activity along with the presence of a FEN-1 antibody cross-reacting protein in the mitochondrial extract. We should note that unequivocally establishing the presence of a protein in the mitochondria is difficult. It is nearly impossible to remove materials adventitiously associated with the mitochondrial outer membrane even after repeated banding by equilibrium centrifugation. The mitochondria are often closely associated with the endoplasmic reticulum, and also perinuclear clustering of mitochondria has been observed. Electron microscopy of mitochondrial sections with gold particles has been used to unequivocally establish the presence of a protein in the mitochondrial matrix. However, such a criteria may not be conclusive for low abundance proteins. To reduce contamination, we routinely treat purified intact mitochondria with trypsin followed by its inactivation with the trypsin inhibitor. Unlike with other proteases such as proteinase K, extensive degradation of the membrane and loss of membrane potential can be prevented by limited trypsin treatment. Because of this stringent purification procedure, we believe that our observation on the presence of FEN-1 in the mitochondrial extract is not an artifact. The presence of FEN-1 or similar 5′-exo/endonuclease in the mitochondria could be also predicted because of its necessity during replication of the mitochondrial genome. At the same time, our results clearly show that LP-BER preferably utilizes an unidentified 5′-exo/endonuclease in the mitochondria.
Isolation of a mitochondrial complex capable of carrying out complete repair similar to what we had observed in the nuclear extract suggests the existence of mitochondrial “repairosome.” So far, we could observe stable interaction among APE1, both subunits of polγ and lig3 in the complex. These proteins are necessary but not sufficient for repair of reduced/oxidized AP residues because THF could not be removed by the dRP lyase activity of polγ. We have detected a 5′-exo/endonuclease activity distinct from FEN-1, and further characterization of this activity will be required. It is worth noting that the mitochondria of Podospora anserine express a 5′-exonuclease with sequence homology to yeast RAD2 (an ortholog of FEN-1), which is active on various substrates including DNA with a flap (31, 32). Finally, while mitochondrial FEN-1 could be dispensable for BER, it may be still required for 5′ terminus processing of Okazaki fragments during mitochondrial replication (33, 34).
BER is distinct from other excision repair pathways in that the processing of the damaged base or AP site generates a strand break that contains a 3′- or 5′-blocking group, which needs to be removed prior to the next step of repair synthesis and strand ligation. The 3′-blocking group, commonly 3′-phospho-α,β-unsaturated aldehyde generated by AP lyase activity of oxidized base-specific DNA glycosylase or 3′-phosphoglycerate generated by oxidation of deoxyribose, is removed by the 3′-phosphodiesterase activity of APE1. Similarly, the 5′-blocking group, an oxidized sugar fragment, needs to be removed by a 5′-exo/endonuclease. Removal of additional nucleotides beyond the sugar fragments would require resynthesis of a longer patch. Thus LP-BER could indicate resynthesis of segments 3′ or 5′ to the damage site or both. The design of our substrate oligo allowed us to establish the directionality of long patch repair synthesis with mitochondrial extracts. It should be noted that, unlike the prototype Xth of Escherichia coli, mammalian APE1 has very weak 3′-exonuclease activity and excises only 1–3 nucleotides at a strand break (35). At the same time, because pol β lacks the 3′-exonuclease activity, the LP-BER in the nucleus occurs primarily through replacement synthesis of the segment 3′ to the damage site. In contrast, pol γ has a robust 3′-exonuclease activity (34), and hence, the long patch repair in the mitochondria could also result from resynthesis of the segment 5′ to the damage site. Our results support the model of mt-specific LP-BER where poly γ incorporates nucleotides both upstream and downstream to the damage. Thus, mitochondrial LP-BER is different from that in the nucleus where only replacement synthesis of the 5′-segment was reported. It is not clear how many nucleotides are replaced on both sides of the damage. Our preliminary studies suggest that, based on specific activity of individual radiolabeled nucleotides, up to 6–9 nt from the site of the damage could be replaced. Further studies are warranted to establish repair patch size in mitochondrial LP-BER.
During the preparation of our manuscript, Akbari et al. published a report providing evidence for LP-BER activity in mammalian mitochondria, but could not detect FEN-1 in these (36). More recently, during revision of our manuscript, Liu et al. (37) showed, in a publication in advance, both the presence of FEN-1 in human mitochondria and its involvement in LP-BER. While the presence of LP-BER in mammalian mitochondria is the common conclusion in all three studies, we have a disagreement with the other two studies in the details. Our results strongly suggest the presence of FEN-1 in the mitochondria, but it does not appear to play a major role in LP-BER activity. We observed only a small effect of FEN-1 deficiency on LP-BER activity. More importantly, we could not detect FEN-1 in the mitochondrial repair complex that is competent to carry out LP-BER. Furthermore, we observed preferential cleavage of a 4-nt segment from an 8-nt flap by the unidentified 5′-exo/endonuclease while FEN-1 cleaved the 8-nt flap in agreement with its known activity. It is thus likely that several 5′-exo/endonucleases present in the mammalian mitochondria could participate in LP-BER. The presence of additional 5′-exo/endonucleases in the human mitochondria was also suggested by Liu et al. (37). Hence the discrepancy in the conclusions could be due to variable levels of the nucleases in different cell types, and this issue could be settled after comprehensive characterization of these key enzymes.
Acknowledgments
We thank Alan E. Tomkinson (University of Maryland) and Binghui Shen (City of Hope National Medical Center) for generous gifts of recombinant DNA ligase 3 and FEN-1-specific antibody, respectively. We also thank Tapas Hazra and Corey Theriot for useful suggestions. The secretarial assistance of Wanda Smith is much appreciated.
This work was supported, in whole or in part, by National Institutes of Health intramural research funds (to W. C. C.). This work was also supported by United States Public Health Service Grants P01 AG10514 and R01 CA53791 (to S. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: mt, mitochondria; LP, long patch; BER, base excision repair; THF, tetrahydrofuran; AP, abasic; dRP, deoxyribose-phosphate; ROS, reactive oxygen species; SN, single nucleotide; FEN-1, flap endonuclease 1; EndoG, endonuclease G; lig1, DNA ligase 1; lig3, DNA ligase 3; pol β, DNA polymerase β; pol δ(ε), DNA polymerase δ(ε); pol γ, DNA polymerase γ; PBS, phosphate-buffered saline; nt, nucleotides; siRNA, small interfering RNA.
References
- 1.Robin, E. D., and Wong, R. (1988) J. Cell. Physiol. 136507 –513 [DOI] [PubMed] [Google Scholar]
- 2.Anderson, S., Bankier, A. T., Barrell, B. G., de Bruijn, M. H., Coulson, A. R., Drouin, J., Eperon, I. C., Nierlich, D. P., Roe, B. A., Sanger, F., Schreier, P. H., Smith, A. J., Staden, R., and Young, I. G. (1981) Nature 290457 –465 [DOI] [PubMed] [Google Scholar]
- 3.Richter, C., Park, J. W., and Ames, B. N. (1988) Proc. Natl. Acad. Sci. U. S.A. 856465 –6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandavilli, B. S., Santos, J. H., and Van Houten, B. (2002) Mutat. Res. 509127 –151 [DOI] [PubMed] [Google Scholar]
- 5.Sung, J. S., and Demple, B. (2006) FEBS J. 2731620 –1629 [DOI] [PubMed] [Google Scholar]
- 6.Richter, C. (1995) Int. J. Biochem. Cell Biol. 27647 –653 [DOI] [PubMed] [Google Scholar]
- 7.Lindahl, T., and Andersson, A. (1972) Biochemistry 113618 –3623 [DOI] [PubMed] [Google Scholar]
- 8.Roginskaya, M., Bernhard, W. A., Marion, R. T., and Razskazovskiy, Y. (2005) Radiat. Res. 16385 –89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dizdaroglu, M., Jaruga, P., Birincioglu, M., and Rodriguez, H. (2002) Free Rad. Biol. Med. 321102 –1115 [DOI] [PubMed] [Google Scholar]
- 10.Clayton, D. A., Doda, J. N., and Friedberg, E. C. (1974) Proc. Natl. Acad. Sci. U. S.A. 712777 –2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogenhagen, D. F., Pinz, K. G., and Perez-Jannotti, R. M. (2001) Progr. Nucleic Acid Res. Mol. Biol. 68257 –271 [DOI] [PubMed] [Google Scholar]
- 12.Larsen, N. B., Rasmussen, M., and Rasmussen, L. J. (2005) Mitochondrion 589 –108 [DOI] [PubMed] [Google Scholar]
- 13.Almeida, K. H., and Sobol, R. W. (2007) DNA Rep. 6695 –711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortini, P., and Dogliotti, E. (2007) DNA Rep. 6398 –409 [DOI] [PubMed] [Google Scholar]
- 15.Hegde, M. L., Hazra, T. K., and Mitra, S. (2008) Cell Res. 1827 –47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longley, M. J., Prasad, R., Srivastava, D. K., Wilson, S. H., and Copeland, W. C. (1998) Proc. Natl. Acad. Sci. U. S.A. 9512244 –12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto, Y., Kim, K., and Bogenhagen, D. F. (1994) Mol. Cell. Biol. 146187 –6197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto, Y., and Kim, K. (1995) Science (NY) 269699 –702 [DOI] [PubMed] [Google Scholar]
- 19.Pinz, K. G., and Bogenhagen, D. F. (1998) Mol. Cell. Biol. 181257 –1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domena, J. D., and Mosbaugh, D. W. (1985) Biochemistry 247320 –7328 [DOI] [PubMed] [Google Scholar]
- 21.Schulke, N., Sepuri, N. B., Gordon, D. M., Saxena, S., Dancis, A., and Pain, D. (1999) J. Biol. Chem. 27422847 –22854 [DOI] [PubMed] [Google Scholar]
- 22.Gordon, D. M., Wang, J., Amutha, B., and Pain, D. (2001) Biochem. J. 356207 –215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramana, C. V., Boldogh, I., Izumi, T., and Mitra, S. (1998) Proc. Natl. Acad. Sci. U. S.A. 955061 –5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das, A., Wiederhold, L., Leppard, J. B., Kedar, P., Prasad, R., Wang, H., Boldogh, I., Karimi-Busheri, F., Weinfeld, M., Tomkinson, A. E., Wilson, S. H., Mitra, S., and Hazra, T. K. (2006) DNA Rep. 51439 –1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izumi, T., and Mitra, S. (1998) Carcinogenesis 19525 –527 [DOI] [PubMed] [Google Scholar]
- 26.Szczesny, B., Hazra, T. K., Papaconstantinou, J., Mitra, S., and Boldogh, I. (2003) Proc. Natl. Acad. Sci. U. S. A. 10010670 –10675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szczesny, B., and Mitra, S. (2005) Mechanisms Ageing Dev. 1261071 –1078 [DOI] [PubMed] [Google Scholar]
- 28.Stucki, M., Jonsson, Z. O., and Hubscher, U. (2001) J. Biol. Chem. 2767843 –7849 [DOI] [PubMed] [Google Scholar]
- 29.Stierum, R. H., Dianov, G. L., and Bohr, V. A. (1999) Nucleic Acids Res. 273712 –3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piersen, C. E., Prasad, R., Wilson, S. H., and Lloyd, R. S. (1996) J. Biol. Chem. 27117811 –17815 [DOI] [PubMed] [Google Scholar]
- 31.Bouex, P., Sabourin, M., Chaignepain, S., Castroviejo, M., and Laquel-Robert, P. (2002) Biochim. Biophys. Acta 157472 –84 [DOI] [PubMed] [Google Scholar]
- 32.Laquel-Robert, P., Sellem, C. H., Sainsard-Chanet, A., and Castroviejo, M. (2007) Biochim. Biophys. Acta 1770527 –542 [DOI] [PubMed] [Google Scholar]
- 33.Holt, I. J., Lorimer, H. E., and Jacobs, H. T. (2000) Cell 100515 –524 [DOI] [PubMed] [Google Scholar]
- 34.Kaguni, L. S. (2004) Annu. Rev. Biochem. 73293 –320 [DOI] [PubMed] [Google Scholar]
- 35.Barzilay, G., Mol, C. D., Robson, C. N., Walker, L. J., Cunningham, R. P., Tainer, J. A., and Hickson, I. D. (1995) Nat. Struct. Biol. 2561 –568 [DOI] [PubMed] [Google Scholar]
- 36.Akbari, M., Visnes, T., Krokan, H. E., and Otterlei, M. (2008) DNA Rep. 7605 –616 [DOI] [PubMed] [Google Scholar]
- 37.Liu, P., Qian, L., Sung, J. S., de Souza-Pinto, N. C., Zheng, L., Bogenhagen, D. F., Bohr, V. A., Wilson, D. M., 3rd, Shen, B., and Demple, B. (2008) Mol. Cell. Biol. 284975 –4987 [DOI] [PMC free article] [PubMed] [Google Scholar]