Abstract
The low density lipoprotein (LDL) receptor (LDLR) mediates efficient endocytosis of VLDL, VLDL remnants, and LDL. As part of the endocytic process, the LDLR releases lipoproteins in endosomes. The release process correlates with an acid-dependent conformational change in the receptor from an extended, “open” state to a compact, “closed” state. The closed state has an intramolecular contact involving H190, H562, and H586. The current model for lipoprotein release holds that protonation of these histidines drives the conformational change that is associated with release. We tested the roles of H190, H562, and H586 on LDLR conformation and on lipoprotein binding, uptake, and release using variants in which the three histidines were replaced with alanine (AAA variant) or in which the histidines were replaced with charged residues that can form ionic contacts at neutral pH (DRK variant). Contrary to expectation, both the AAA and the DRK variants exhibited normal acid-dependent transitions from open to closed conformations. Despite this similarity, both the AAA and DRK mutations modulated lipoprotein release, indicating that H190, H562, and H586 act subsequent to the conformational transition. These observations also suggest that the intramolecular contact does not drive release through a competitive mechanism. In support of this possibility, mutagenesis experiments showed that β-VLDL binding was inhibited by mutations at D203 and E208, which are exposed in the closed conformation of the LDLR. We propose that H190, H562, and H586 are part of an allosteric mechanism that drives lipoprotein release.
The low density lipoprotein (LDL)2 receptor (LDLR) is a type I transmembrane protein that functions as the principal endocytic receptor for very low density lipoprotein (VLDL), VLDL remnants, and LDL. Lipoproteins bind to the extracellular domain (ectodomain) of the LDLR, which contains a ligand binding domain, an epidermal growth factor (EGF) homology domain, and an O-glycosylated domain (1). The ligand binding domain provides the majority of binding surfaces for lipoproteins (2). This domain consists of seven LDLR type A (LA) repeats, which are small, irregular domains held together by calcium and cystine bridges (3, 4). The EGF homology domain participates in lipoprotein release and consists of two EGF-like domains, six YWTD repeats that form a six-bladed β-propeller, and a third EGF-like repeat (5, 6). Internalization of the LDLR requires the cytoplasmic domain (7, 8).
The LDLR binds to lipoproteins that contain either apolipoprotein B100 (apoB100) or apolipoprotein E (apoE). LDL binds to the LDLR via apoB100, while VLDL and VLDL remnants bind via apoE (9–11). LDL and VLDL compete for binding and interact with overlapping regions of the LDLR (2, 12–14). Both apoB100 and apoE contain clusters of positively charged residues that are required for binding (15–19). These positively charged residues have been proposed to contact acidic residues on the LDLR that are clustered by calcium (20, 21).
Following lipoprotein binding, LDLR-lipoprotein complexes internalize through clathrin-coated pits and traffic to sorting endosomes where lipoprotein release occurs (22, 23). Lipoprotein release requires the EGF homology domain and acidic pH (5). Release occurs at ∼pH 6, which corresponds to the luminal pH of sorting endosomes (5, 23). Coincident with lipoprotein release, the LDLR undergoes a conformational transition from an extended, “open” conformation, which predominates at neutral pH, to a compact, “closed” conformation at acidic pH (24). The crystal structure of the LDLR ectodomain at acidic pH shows that in the closed conformation, the β-propeller region of the EGF homology domain forms an intramolecular contact with LA repeats 4 and 5 (24). At the center of this contact are three histidines (H190, H562, and H586) that form van der Waals contacts and ionic interactions between the β-propeller and the LA repeats. The ionic interactions presumably only form when histidine is positively charged. The imidazole ring of histidine has a pKa of ∼6, suggesting that the three histidines are protonated at endosomal pH.
These observations have led to the current model for acid-dependent lipoprotein release (25). This model proposes that the β-propeller of the EGF homology domain binds to the lipoprotein binding surfaces of LA4 and LA5. Release occurs through dissociation of lipoprotein from the LA repeats followed by binding of the β-propeller to LA4/5. The model also stipulates that the binding of the β-propeller with LA4/5 requires protonation of H190, H562, and H586. Consistent with this model, mutation of all three histidines to alanine inhibits LDL release (26).
The current model makes two key predictions. First, the conformational state of the receptor should depend upon the ability of residues at 190, 562, and 586 to form ionic contacts. Second, lipoproteins should bind to the same surface as the β-propeller.
Here, we tested the current model. We examined the role of the three histidines on receptor conformation and on lipoprotein binding, uptake, and release using LDLR variants in which the three histidines were replaced with alanine or in which the histidines were replaced with charged residues that can maintain ionic interactions at neutral pH. We also used mutagenesis to test whether lipoproteins bind to the same surface on the LDLR as the β-propeller of the EGF homology domain.
EXPERIMENTAL PROCEDURES
Materials—All cell culture reagents were from Invitrogen (Carlsbad, CA). LDLR-/- primary human fibroblasts, human LDL, and rabbit β-migrating VLDL (β-VLDL) were a gift from Michael Brown and Joseph Goldstein (Department of Molecular Genetics, UT Southwestern Medical Center, Dallas, TX). Rabbit polyclonal anti-LDLR (4548) was a gift from Joachim Herz (Department of Molecular Genetics, UT Southwestern Medical Center, Dallas, TX). Mouse monoclonal anti-LDLR (C7) was from Santa Cruz Biotechnology (Santa Cruz, CA). 125I-Bolton-Hunter reagent was from PerkinElmer Life Sciences. Alexa546 succinimidyl ester was from Invitrogen. All other chemicals were from Sigma.
Baculovirus-mediated Protein Expression—Residues 1–699 of the LDLR, LDLR-AAA, and LDLR-DRK were cloned into the pFastBac plasmid (Invitrogen). Recombinant plasmids were used to produce infectious baculoviruses that directed the synthesis of secreted LDLR ectodomains using the Bac-to-Bac system (Invitrogen).
Cell Culture—LDLR-/- primary fibroblasts (isolate 549) were immortalized using the pBabe-puro-hTERT retrovirus (27), which encodes human telomerase. Puromycin-resistant fibroblasts (549T) showed no differences in morphology or growth rate as compared with parental primary fibroblasts, but were no longer limited in the number of cell divisions. In longevity tests, the 549T cells continued to divide normally over the course of 18 months. The behavior of the 549T LDLR-/- fibroblasts was consistent with prior reports showing that ectopic expression of telomerase prevents senescence, but does not induce transformed phenotypes in fibroblasts (28). All fibroblasts were cultured in Medium A (DMEM medium supplemented with 10% (v/v) fetal bovine serum, 20 mm HEPES pH 7.5, 100 units/ml penicillin G, and 100 μg/ml streptomycin. Prior to experimentation, fibroblasts were starved of lipoproteins for 24 h by replacing the culture medium with Medium B (DMEM medium supplemented with 10% (v/v) lipoprotein poor serum, 20 mm HEPES pH 7.5, penicillin G (100 units/ml), and streptomycin (100 μg/ml)).
Gel Filtration—Gel filtration was conducted on a Superdex200 10/30 column attached to an Äkta FPLC (Amersham Biosciences). The column was equilibrated in buffers at pH 6–7 containing 25 mm HEPES, 25 mm maleate, 150 mm NaCl, 1 mm CaCl2, and 0.5% Triton X-100. Triton X-100 was required to maintain the LDLR ectodomains in solution at acidic pH. Samples were equilibrated in the same buffers prior to loading. 0.5-ml fractions were collected, electrophoresed on 5–17% SDS-PAGE gels, transferred to nylon membranes, and immunoblotted for the LDLR using the 4548 polyclonal antibody. Thyroglobulin (85 Å), apoferritin (61 Å), amylase (48 Å), aldolase (45 Å), bovine serum albumin (36 Å), and carbonic anhydrase (20 Å) were used as standards.
Introduction of LDLR Variants into LDLR-/- Fibroblasts—LDLR variants were cloned into the pMX-IRES-GFP bicistronic retroviral vector (29). Retroviral vectors were cotransfected with the pAmpho packaging vector (Clontech, Mountain View, CA) into 293T cells to produce infectious, replication-defective retroviruses. Retroviruses were added to LDLR-/- fibroblasts (549T) in the presence of hexadimethrine bromide to facilitate viral entry. Infection rates as assessed by GFP-positive fluorescence were ∼5%. GFP-positive, LDLR-expressing fibroblasts were purified using two rounds of fluorescence-activated cell sorting (FACS) with a MoFlo High Performance Cell Sorter (Dako, Glostrup, Denmark). Purity was ∼90% after the first sort and > 95% after the second sort. Surface LDLR expression was monitored by flow cytometry using anti-LDLR mouse monoclonal antibody, C7 (Santa Cruz Biotechnology, Santa Cruz, CA).
Lipoprotein Binding Assays—Human 125I-LDL and rabbit 125I-β-VLDL binding assays were performed in triplicate using established methods (30). Assays were preformed at 4 °C for 90 min using concentrations of 125I-LDL or 125I-β-VLDL indicated in the figure legends. Results are presented as means ± S.D.
Initial Endocytic Rates—Initial internalization rates were determined as previously described (31, 32). Briefly, cells were incubated with 10 μg/ml 125I-LDL or 5 μg/ml 125I-β-VLDL for 1 h at 4 °C in Medium C (bicarbonate-free MEM supplemented with 20 mm HEPES pH 7.5 and 10% lipoprotein-poor serum). Medium was changed for the times indicated with warm Medium B that also containing either 10 μg/ml 125I-LDL or 5 μg/ml 125I-β-VLDL. Cells were extensively washed with ice-cold PBS and incubated with 1 mg/ml Protease K in Buffer A (PBS + 1 mm EDTA) for 2 h at 4 °C. The cell suspension was then centrifuged at 5000 × g for 10 min over a cushion of 10% sucrose in PBS. The tubes were frozen in liquid nitrogen, cut to separate the cell pellet (internal) from the solution (surface-bound material released by protease K) and counted on a gamma counter. Results are presented as means ± S.D.
Fluorescent Lipoprotein Labeling—DiI (3H-Indolium, 2-(3-(1,3-dihydro-3,3-dimethyl-1-octadecyl-2H-indol-2-ylidene)-1-propenyl)-3,3-dimethyl-1-octadecyl-, perchlorate) was used to label β-VLDL by adding 300 μl of 3 mg/ml DiI suspended in DMSO to 10 mg of β-VLDL in 10 ml of lipoprotein-poor serum with gentle mixing. The suspension was mixed end-over-end for 16 h at 37 °C in the dark. The density of the suspension was increased to 1.019 by the addition of 0.0199 g of KBr per ml of suspension and centrifuged at 120,000 × g for 16 h at 4 °C. DiI-β-VLDL was removed from the top of the tube, dialyzed against PBS, and stored at 4 °C in the dark until use. LDL was labeled using Alexa-546 succinimidyl ester using the manufacturer's recommended protocol (Invitrogen).
Lipoprotein Accumulation—Cells were incubated with either 10 μg/ml Alexa546-LDL or 5 μg/ml DiI-β-VLDL in Medium C for 1 h at 4 °C. The medium was then replaced with warm Medium B containing 10 μg/ml Alexa546-LDL or 5 μg/ml DiI-β-VLDL for the times indicated. Cells were washed with ice-cold PBS, suspended by gentle scraping in PBS and fixed in the presence of 3% paraformaldehyde. Cells were washed with PBS and analyzed by flow cytometry on a FACS Calibur (BD Biosciences, San Jose, CA). Mean fluorescence intensities were recorded for 10,000 events for each experiment.
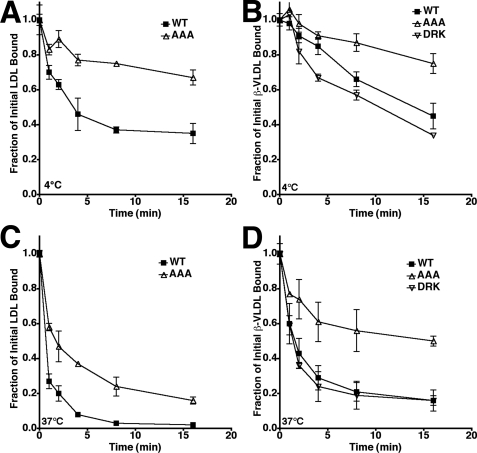
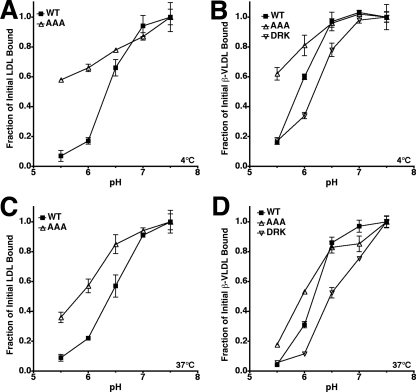
Acid-dependent Release of Cell Surface-bound Lipoprotein—Assays were performed at both 4 and 37 °C. For assays at 4 °C, cells were incubated with either 10 μg/ml 125I-LDL or 5 μg/ml 125I-β-VLDL in Medium C for 1 h at 4 °C. Cells were washed with ice-cold Buffer B (TBS + 1% bovine serum albumin) and incubated with Medium D (bicarbonate-free MEM supplemented with 20 mm HEPES, 20 mm maleate, and 10% lipoprotein-poor serum) at pH 5.5, 6.0, 6.5, 7.0, or 7.5 for 30 min at 4 °C. The cells were washed two times with ice-cold TBS, one time for 10 min with ice-cold Buffer B and two times with ice-cold TBS. Remaining cell-associated, 125I-labeled lipoproteins were liberated by incubation with 0.1 n NaOH and counted on a gamma counter. For assays at 37 °C, cells were preincubated with Medium E, which consisted of Medium B supplemented with 0.45 m sucrose to prevent clathrin-coated pit endocytosis (33). Cells were then incubated with either 10 μg/ml 125I-LDL or 5 μg/ml 125I-β-VLDL in Medium E for 30 min at 37 °C, washed with warm Medium E, and incubated for 30 min at 37 °C with Medium F (bicarbonate-free MEM supplemented with 0.45 m sucrose, 20 mm HEPES, 20 mm maleate, and 10% lipoprotein-poor serum) at pH 5.5, 6.0, 6.5, 7.0, or 7.5. The cells were washed two times with ice-cold TBS, one time for 10 min with ice-cold Buffer B and two times with ice-cold TBS. Remaining cell-associated, 125I-labeled lipoproteins were liberated by incubation with 0.1 n NaOH and counted. All experiments were performed in triplicate and are presented as a fraction of counts in parallel assays in which the acid release step was omitted. Rate constants for release were determined by non-linear curve fitting using a single-phase exponential decay model. Data for the curve fitting was from Fig. 5 with the exception of β-VLDL release at 4 °C, which used release data collected over 30 min (data not shown).
FIGURE 5.
The AAA mutation slows release. Release of prebound 125I-LDL (A and C) or 125I-β-VLDL (B and D) from fibroblasts expressing normal LDLR (WT), LDLR-AAA (AAA), or LDLR-DRK (DRK) in response to pH 5.5 medium was determined at 4 °C (A and B) and 37 °C (C and D) over a 16-min time course. 37 °C trials had 0.45 m sucrose present to prevent coated pit internalization. All experiments were performed in triplicate and are reported as the mean of the fraction of cell-associated lipoprotein remaining ± S.D.
RESULTS
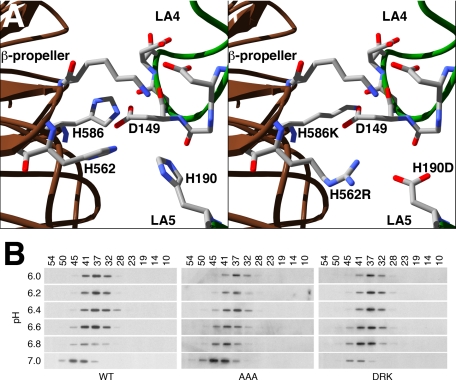
H190, H562, and H586 have been proposed to constitute a pH sensor that controls receptor conformation, thereby regulating lipoprotein binding and release (24–26). To test whether these histidines control the conformational state of the LDLR, we used a baculovirus expression system to produce proteins encompassing the LDLR ectodomain of normal LDLR, of an LDLR variant in which H190, H562, and H586 were replaced with alanine, and of an LDLR variant in which the histidines were replaced with charged residues. If the histidine contacts are required for the closed conformation, then the alanine (AAA) variant should be fixed in the open conformation. By contrast, if the ionic interactions made by the histidines are sufficient to drive adoption of the closed variant, then replacement of the histidines with charged residues that can form ionic contacts at neutral pH should fix the LDLR ectodomain in the closed conformation. The charged variant (DRK) had the following mutations: H190D, H562R, and H586K. The H190D and H562R mutations were designed to form one ionic interaction, while the H586K mutation was designed to form a second interaction with D149 (Fig. 1A). Normal, AAA, and DRK ectodomains were expressed in baculovirus-infected insect cells, equilibrated in buffers of different pH and chromatographed on a superdex 200 gel filtration column using the same buffers. Gel filtration separates proteins based upon their hydrodynamic (Stokes) radius and allows separation of the open and closed states of the LDLR (24). Initial experiments showed that all three ectodomain proteins underwent a conformational transition from a Stokes radius of 43 Å to a radius of 38 Å between pH 7 and pH 6. Further gel filtration experiments within this pH range showed that the pH of half-maximal transition for each ectodomain was ∼pH 6.6 (Fig. 1B). The ability of the LDLR-AAA ectodomain to adopt the closed conformation indicates that the ionic contacts formed by the three histidines at acidic pH are not necessary for adoption of the closed conformation. The ability of the LDLR-DRK to adopt the open conformation indicates that ionic contacts at amino acid positions 190, 562, and 586 are not sufficient for adoption of the closed conformation. Thus, H190, H562, and H586 are neither necessary nor sufficient for the conformational transition associated with lipoprotein release.
FIGURE 1.
Mutations at H190, H562, and H586 have little effect on LDLR ectodomain conformation. H190, H562, and H586 come together at the interface between the β-propeller region of the EGF homology domain and LA repeats 4 and 5. The left panel of A shows the orientation of the three histidines in the closed conformation of the LDLR. The right panel of A shows a model of the interface with the H190D, H562R, and H586K mutations. These mutations were designed to form two ionic contacts: one between K586 and D149, and one between R562 and D190. In panel B, gel filtration was used to determine the hydrodynamic (Stokes) radius of ectodomains from normal (WT), LDLR-AAA (AAA), and LDLR-DRK (DRK) receptors as a function of pH.
While not required for the conformational transition, prior mutational data suggest that H190, H562, and H586 play a significant role in LDLR function. Familial Hypercholesterolemia (FH) is associated with tyrosine substitutions at H190 and H562 (34, 35). Furthermore, triple alanine or tyrosine mutations at H190, H562, and H586 inhibit acid-dependent LDL release from the surface of CHO cells expressing these variants (26). How might the three histidines participate in LDLR function?
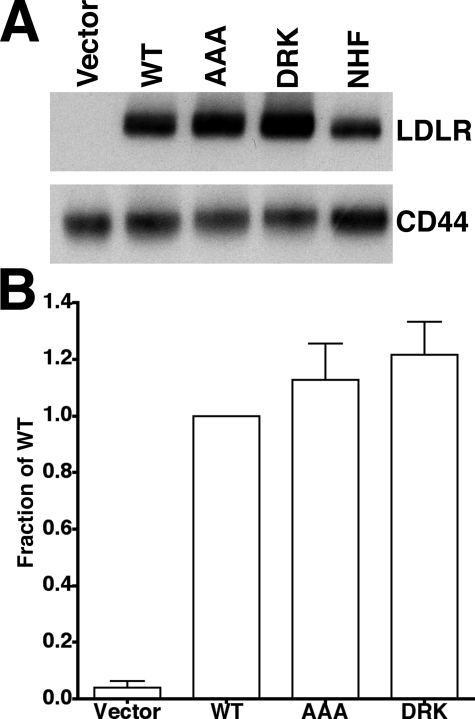
To answer this question, we used fibroblasts that stably express full-length normal LDLR, LDLR-AAA or LDLR-DRK receptors in experiments measuring lipoprotein binding, uptake, and release. Full-length cDNAs for normal LDLR, LDLR-AAA, and LDLR-DRK were introduced into LDLR-/- fibroblasts by retroviral infection. LDLR expression was driven by the 5′-LTR of the retrovirus, which also produced GFP through an internal ribosomal entry site downstream of the coding sequence for the LDLR. GFP-positive cells thus express LDLR, allowing LDLR-expressing cells to be purified by FACS. Infection rates were kept to ∼5% so as to ensure that infected cells had only a single integration event. Consistent with a homogeneous expression level, the variance observed in surface LDLR of infected cells was similar to that seen in normal human fibroblasts expressing the endogenous LDLR (data not shown). Immunoblots of cell lysates showed that LDLR expression was similar in each cell line and that the total LDLR expression level in the infected fibroblasts was only 2–3-fold higher than endogenous LDLR expression in normal human fibroblasts (Fig. 2A). No immature LDLR was observed, indicating that all three variants were glycosylated normally in the Golgi. Flow cytometry using the C7 anti-LDLR antibody showed that cells expressing normal LDLR, LDLR-AAA, and LDLR-DRK had similar numbers of LDLR on their cell surfaces (Fig. 2B). No LDLR was detected in LDLR-/- fibroblasts infected with retroviruses lacking an LDLR gene (Vector). Together, these observations indicate that the AAA and DRK mutations did not impair the expression or surface delivery of the LDLR.
FIGURE 2.
Fibroblasts expressing normal (WT), LDLR-AAA (AAA), or LDLR-DRK (DRK) have similar total and surface receptor expression. A, cell lysates from LDLR-/- fibroblasts that were infected with vector (Vector), normal LDLR (WT), LDLR-AAA (AAA), or LDLR-DRK (DRK) retroviruses or from normal human fibroblasts (NHF) were electrophoresed on 5–17% SDS-PAGE gels, transferred to nylon membranes and immunoblotted for the LDLR. CD44, a transmembrane protein expressed on the cell surface, was used as a loading control. B, the surface expression of LDLR was determined by flow cytometry using the C7 monoclonal antibody to the LDLR. Fluorescence values are presented as a fraction of the WT value.
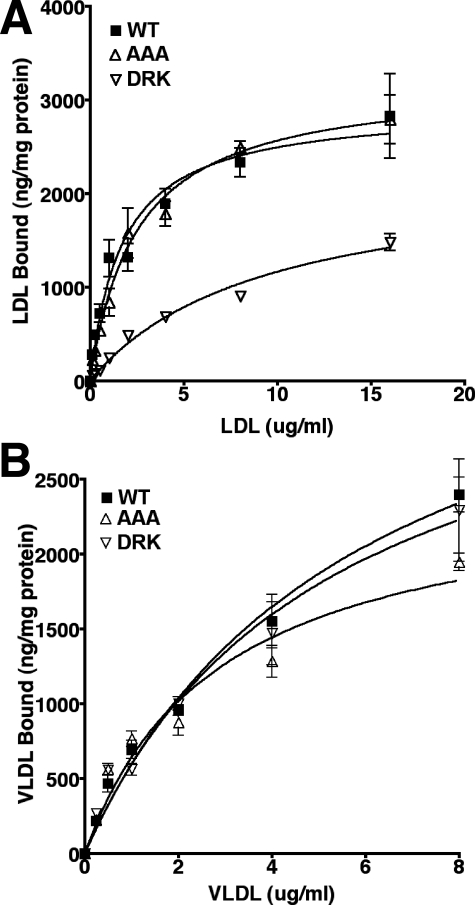
The role of the three histidines in lipoprotein binding was determined by comparing the affinity of 125I-LDL and 125I-β-VLDL for cells expressing normal LDLR, LDLR-AAA, and LDLR-DRK. β-VLDL is a VLDL remnant particle that is commonly used to assess the binding and uptake of VLDL/VLDL remnants by the LDLR (36). Binding was assessed in saturation experiments in which increasing concentrations of 125I-LDL or 125I-β-VLDL were incubated with cells at 4 °C. In these assays, LDLR-AAA cells bound LDL normally; however, LDLR-DRK cells showed reduced ability to bind LDL (Fig. 3A). Scatchard analysis of the binding data revealed that cells expressing the DRK variant had lost high affinity binding activity for LDL (data not shown). By contrast, both the LDLR-AAA cells and LDLR-DRK cells bound β-VLDL normally (Fig. 3B). The ability of LDLR-AAA cells to bind both LDL and β-VLDL normally indicated that the three histidines were not required for lipoprotein binding.
FIGURE 3.
The effect of the AAA and DRK mutations on lipoprotein binding. Saturation binding of 125I-LDL (A) and 125I-β-VLDL (B) was performed using LDLR-/- fibroblasts that were infected with normal LDLR (WT), LDLR-AAA (AAA), or LDLR-DRK (DRK) retroviruses. Experiments were performed in triplicate and data are presented as means ± S.D.
After internalization, LDLR-lipoprotein complexes are transported to endosomes where lipoprotein release occurs. We followed lipoprotein release using two assays: 1) surface assays in which cell-surface bound lipoproteins were released in response to acidic buffers, and 2) cellular assays in which lipoprotein release was followed as a function of lipoprotein internalization and accumulation over time.
In the surface assays, normal LDLR, LDLR-AAA, and LDLR-DRK cells were incubated with 125I-labeled lipoproteins at 4 °C or 37 °C and then shifted to medium at pH 5.5–7.5 in the absence of lipoprotein for 30 min. Release was followed by measuring the amount of 125I-labeled lipoproteins that remained cell associated after the 30-min incubation.
In surface assays at 4 °C, LDLR-AAA cells showed reduced ability to release lipoproteins in response to acidic buffers. Normal LDLR-expressing cells had half-maximal LDL release at pH 6.5, while LDLR-AAA cells had half-maximal release below pH 5.5 (Fig. 4A). The observations with LDLR-AAA cells support the results of Beglova et al. (26) who used flow cytometry to show that at 4 °C, CHO cells expressing the LDLR-AAA have reduced ability to release surface-bound LDL in response to low pH. LDL release by LDLR-DRK cells is not reported because LDL binding by these cells was defective. β-VLDL release from normal LDLR-expressing cells at 4 °C showed half-maximal release at pH 5.8 (Fig. 4B), which is similar to the half-maximal β-VLDL release previously observed in CHO cells expressing normal LDLR (5). LDLR-AAA cells showed reduced ability to release β-VLDL (half-maximal release below pH5.5), while LDLR-DRK cells showed half-maximal release of β-VLDL at pH 6.4.
FIGURE 4.
The AAA mutation hinders acid-dependent lipoprotein release, while the DRK mutation potentiates β-VLDL release in vitro. Release of prebound 125I-LDL (A and C) or 125I-β-VLDL (B and D) from fibroblasts expressing normal LDLR (WT), LDLR-AAA (AAA), or LDLR-DRK (DRK) was measured after 30 min at 4 °C (A and B) or 37°C(C and D) in medium buffered at pH 5.5, 6.0, 6.5, 7.0, or 7.5. In the 37 °C trials, 0.45 m sucrose was included in the medium to prevent clathrin-coated pit internalization. All experiments were performed in triplicate and are reported as the mean of the fraction of cell-associated lipoprotein remaining ± S.D.
Lipoprotein release was also compared in the three cells at 37 °C to test whether the differences observed at 4 °C were consistent with acid-dependent release at physiological temperature. These assays used medium containing 0.45 m sucrose to prevent clathrin-coated pit internalization of lipoproteins (33). At 37 °C, normal LDLR-expressing cells had half-maximal LDL release at pH 6.5, while LDLR-AAA cells had half-maximal release at pH 5.8 (Fig. 4C). β-VLDL release experiments at 37 °C showed half-maximal release points at pH 5.8, pH 6.2, and pH 6.5 for LDLR-AAA, normal LDLR, and LDLR-DRK cells, respectively (Fig. 4D). Together the LDL and β-VLDL release experiments showed that the AAA mutation increased the acid dependence of lipoprotein release, while the DRK mutation reduced the acid dependence of lipoprotein release. These observations also showed that β-VLDL release required a more acidic environment than LDL release.
Surface assays were also used to determine rate constants for lipoprotein release at both 4 °C and 37 °C. 125I-labeled lipoprotein was bound to cells as before and then incubated with pH 5.5 media for 0–16 min (Fig. 5). As with the pH experiments, release was followed as the amount of 125I-labeled lipoprotein that remained cell associated at the end of the incubation. Rates of release were calculated from the resulting curves using a single-phase exponential decay model (Table 1). At 4 °C, the rates of LDL and β-VLDL release from cells expressing normal LDLR were 7.9 × 10-3 s-1 and 9.6 × 10-4 s-1, respectively. These rates are approximately two orders of magnitude faster than the corresponding lipoprotein dissociation rates at neutral pH (37), indicating that acidic pH accelerated lipoprotein dissociation from the normal LDLR. LDLR-AAA cells had slower than normal rates for both LDL and β-VLDL release (Fig. 5 and Table 1). Release by LDLR-AAA cells also plateaued at higher than normal levels for both LDL and β-VLDL. By contrast, LDLR-DRK cells had slightly faster than normal rates of β-VLDL release (Fig. 5, B and D and Table 1). Temperature also played a major role: LDL release rates doubled at 37 °C, while β-VLDL release rates increased by more than 10-fold. These kinetic experiments indicated that the AAA mutation slowed lipoprotein release.
TABLE 1.
Rate constants of lipoprotein release
| Lipoprotein | Temperature | WT | AAA | DRK |
|---|---|---|---|---|
| °C | s–1 | s–1 | s–1 | |
| LDL | 4 | 7.9 × 10–3 | 4.5 × 10–3 | N/Da |
| 37 | 2.2 × 10–2 | 9.4 × 10–3 | N/D | |
| β-VLDL | 4 | 9.6 × 10–4 | 1.1 × 10–5 | 1.0 × 10–3 |
| 37 | 1.0 × 10–2 | 7.4 × 10–3 | 1.2 × 10–2 |
N/D, not determined
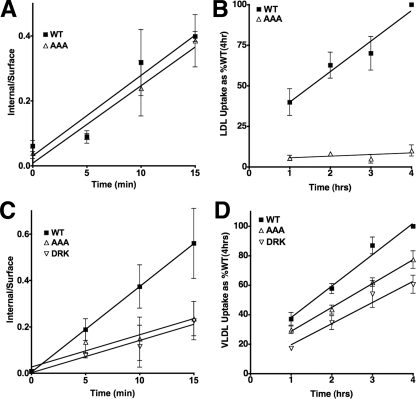
Cellular assays for lipoprotein release combined assays measuring initial rates of lipoprotein internalization with assays measuring steady-state rates of lipoprotein accumulation. Initial rate experiments were conducted over 15 min and measured the ability of cells to internalize 125I-labeled lipoproteins. The lipoprotein accumulation assays used flow cytometry to follow the fluorescent intensity of cells incubated with fluorescently labeled lipoproteins (Alexa546-labeled LDL or DiI-labeled β-VLDL) over a course of 4 h. Alexa546 and DiI dyes are not degraded in lysosomes and provide an easily quantified measure of the amount of lipoprotein that reaches lysosomes (32). Because the LDLR completes ∼5 endocytic cycles per hour (38), if the AAA or DRK mutations prevent release, then lipoprotein should internalize but not accumulate. In this way, the combination of assays for internalization and accumulation provided a measure of the ability of the LDLR to release lipoproteins under cellular conditions.
Cellular assays compared LDL and β-VLDL uptake in normal LDLR, LDLR-AAA and LDLR-DRK cells. LDLR-AAA cells had a normal rate of LDL internalization (Fig. 6A), but a very slow rate of LDL accumulation relative to normal (Fig. 6B). Thus, the AAA mutation disrupted cellular LDL release by the LDLR. In β-VLDL uptake assays, both the LDLR-AAA cells and LDLR-DRK cells showed reduced rates of β-VLDL internalization (Fig. 6C). This reduction likely resulted in the lower level of β-VLDL accumulation that was observed at the 1-h time point in the β-VLDL accumulation assay; however, the rates of β-VLDL accumulation by all three cells over the 4-h time course were similar (Fig. 6D). As compared with LDL, the trafficking of β-VLDL from endosomes to lysosomes is slow (39–41). The increased residence time of LDLR-β-VLDL complexes in endosomes may allow near normal endosomal release of β-VLDL from LDLR-AAA. For LDL by contrast, the LDLR-AAA may exit endosomes prior to release, leading to reduced LDL accumulation.
FIGURE 6.
The effect of the AAA and DRK mutation on lipoprotein release in cellular assays. The ability of fibroblasts expressing normal LDLR (WT), LDLR-AAA (AAA), or LDLR-DRK (DRK) to internalize (A and C) and accumulate (B and D) LDL (A and B), and β-VLDL (C and D) was determined. Internalization assays determined the ratio of internal/surface 125I-LDL (A) or 125I-β-VLDL (C) over 15 min at 37 °C. The accumulation assays determined the amount of Alexa546-LDL (B) or DiI-β-VLDL (D) fluorescence associated with each cell type over 4 h. Data are presented as the percent of normal (WT) at 4 h. All experiments were performed in triplicate and are presented as means ± S.D.
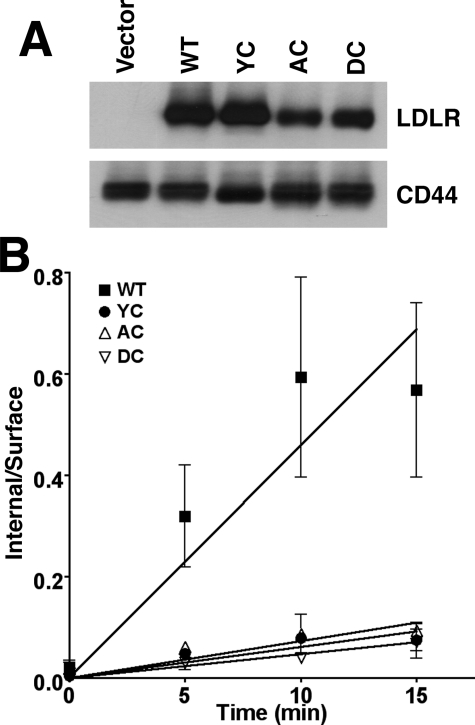
The reduced initial rates of β-VLDL internalization exhibited by the LDLR-AAA and LDLR-DRK cells suggested that clathrin-coated pit internalization of LDLR-β-VLDL complexes was sensitive to mutations at H190, H562 and H586. The cytoplasmic domain of the LDLR has two internalization mechanisms that can support β-VLDL internalization: a well characterized FDNPVY-dependent pathway and a less understood, FDNPVY-independent pathway that only serves for VLDL/VLDL remnant uptake (32, 42, 43). To determine whether the histidine mutations influenced the FDNPVY-dependent pathway, LDLR-/- fibroblasts were infected with retroviruses that encoded LDLR variants that combined the Y807C mutation, which inactivates the FDNPVY807 sequence (7), with either the AAA (LDLR-AC) or DRK (LDLR-DC) mutations. All receptors expressed normally in these fibroblasts (Fig. 7A). Comparison of the initial rates of internalization showed that LDLR-Y807C, LDLR-AC, and LDLR-DC fibroblasts had similar rates of β-VLDL internalization (Fig. 7B). These observations suggested that mutations in H190, H562, and H586 influenced FDNPVY-dependent β-VLDL internalization.
FIGURE 7.
The reduction in β-VLDL internalization by the AAA and DRK mutations involves the FDNPVY sequence. A shows immunoblots of cell lysates from LDLR-/- fibroblasts infected with retroviruses expressing no LDLR (Vector), normal LDLR (WT), LDLR-Y807C (YC), LDLR-Y807C+AAA (AC), or LDLR-Y807C+DRK (DC). B shows the internal/surface ratios of β-VLDL endocytosis at the indicated times at 37 °C. Experiments were performed in triplicate, and the data are presented as means ± S.D.
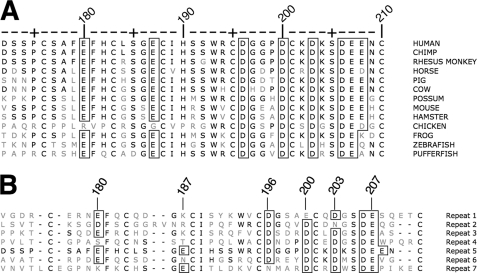
Together, the assays for lipoprotein binding, uptake and release show that H190, H562, and H586 participate in β-VLDL internalization and in lipoprotein release. To address how H190, H562, and H586 participate in release, we employed mutagenesis to identify the residues on LA5 that are required for normal lipoprotein binding with the goal of determining whether lipoproteins bind to the same surface as the EGF homology domain. Experiments focused on LA5 because this repeat contains H190 and is the only repeat that is required for both LDL and β-VLDL binding (2, 13). The lipoprotein binding sites on LA5 likely involve acidic residues because mutations of basic residues in apoB100 and apoE impair binding of LDL and VLDL to the LDLR (15–19). LA5 has eight acidic residues that are conserved between species (Fig. 8A). Residues D196, D200, D206, and E207 chelate calcium; E180 is largely buried; and E187, D203, and E208 are exposed on the LA5 surface (24, 44). Four of the acidic residues are part of two negatively charged clefts that have been implicated in lipoprotein binding (45). Cleft 1 contains D196 and D200, while cleft 2 contains D203 and E208. E187 is near both cleft 1 and cleft 2, and may be able to participate in interactions at either cleft. In the closed conformation, the β-propeller of the EGF homology domain binds to cleft 1, while cleft 2 is exposed. Sequence comparison of LA repeats in human LDLR show that D196 and D200 are conserved between different LA repeats, while E187 and E208 are not (Fig. 8B), suggesting E187 and E208 may be involved in LA5-specific functions.
FIGURE 8.
Sequence comparisons. A compares the amino acid sequence of LA5 from 12 species. E180, D196, D200, D203, D206, and E207 are conserved in all species. E208 is conserved in 8 of the 12 B compares the amino acid sequence of human LA5 with the other six LA repeats of human LDLR. D206 E207 are conserved; E180, D196, D200, and D203 are somewhat conserved; while E187 and E208 are conserved between different repeats. Boxed residues are acidic residues that are conserved with human Residue numbering is for human LA5.
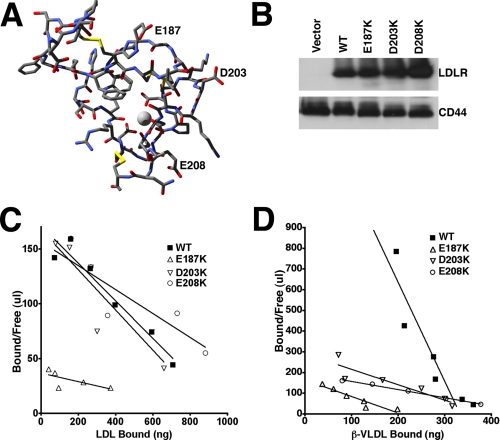
We tested whether cleft 2 was involved in lipoprotein binding by introducing into LDLR-/- fibroblasts LDLR variants that had lysine substitutions at E187, D203, or E208 (Fig. 9A) and measuring lipoprotein binding affinity. LDLR-E187K, LDLR-D203K and LDLR-E208K were introduced into LDLR-/- fibroblasts using retroviral infection. Cells expressing these variants had similar levels of LDLR expression (Fig. 9B). In 125I-LDL binding assays, cells expressing LDLR-D203K or LDLR-E208K had normal LDL binding affinity, while cells expressing LDLR-E187K had a 5-fold loss in LDL binding affinity (Fig. 9C). By contrast, in 125I-β-VLDL binding assays, cells expressing LDLR-E187K, LDLR-D203K, or LDLR-E208K showed a 6-fold, 6-fold, or 10-fold reduction in β-VLDL affinity, respectively (Fig. 9D). These observations indicate that E187 is involved in both LDL and β-VLDL binding, while D203 and E208 are only involved in β-VLDL binding. The effects on β-VLDL binding by the D203K and E208K mutations implicate cleft 2 as the binding site for apoE.
FIGURE 9.
The role of E187, D203, and E208 on lipoprotein binding. A shows the structure of LA5 in ball and stick representation. Carbon is colored dark gray; nitrogen is blue; oxygen is red; and sulfur is yellow. Calcium is shown as a light-gray sphere. E187, D203, and E208 are labeled. B shows immunoblots of cell lysates from LDLR-/- fibroblasts that were infected with retroviruses expressing no LDLR (Vector), normal LDLR (WT), LDLR-E187K, LDLR-D203K, or LDLR-E208K. The upper portion of B shows the immunoblot for LDLR, while the lower portion shows the immunoblot for CD44, which was used as a loading control. C shows the Scatchard plot of 125I-LDL binding to the cell surface of the indicated fibroblasts, while D shows the Scatchard plot of 125I-β-VLDL binding.
DISCUSSION
The current model for lipoprotein release by the LDLR holds that H190, H562, and H586 serve as a pH sensor, which, when protonated, facilitates the association of the β-propeller of the EGF homology domain with lipoprotein binding surfaces on LA4/5 (24–26). The results of this study show that these histidines have little impact on the acid-dependent transition between the open and closed state of the LDLR (Fig. 1). Thus, H190, H562, and H586 do not constitute the pH sensor.
Our results also suggest that the EGF homology domain does not drive release though competition with lipoproteins. Previous kinetic experiments of lipoprotein dissociation at neutral pH showed that the dissociation rate constants for HDLc (an apoE-containing lipoprotein) and LDL from normal LDLR are 1.7 × 10-5 s-1 and 6.3 × 10-5 s-1, respectively (37). The dissociation rates for β-VLDL are likely similar to those for HDLc, because both lipoproteins bind to the LDLR via apoE and both have similar binding affinity for the LDLR (12, 37, 46). Under acidic conditions, the lipoprotein dissociation rates increase (Fig. 5 and Table 1); however, acidic buffers are unable to drive β-VLDL release from LDLRs lacking an EGF homology domain (5), indicating that the EGF homology domain accelerates lipoprotein release in response to acidic conditions. Competitive mechanisms of inhibition influence ligand binding but do not accelerate the rate of ligand dissociation. Thus, the kinetics of lipoprotein dissociation argues that the EGF homology domain does not act in a competitive manner and suggests that the β-propeller region forms an allosteric interaction with LA4/5.
An allosteric interaction implies that lipoproteins bind to a different site on LA4/5 than the EGF homology domain. Consistent with this possibility, the LDLR-AAA ectodomain displayed a normal acid-dependent conformational change (Fig. 1); however, cells expressing the LDLR-AAA variant had defective lipoprotein release (Figs. 4, 5, 6). These observations suggest that adoption of the closed conformation is not sufficient to drive release. If the β-propeller can form the intramolecular contact but not drive lipoprotein release, then the lipoprotein binding sites must be distinct from the binding site for the β-propeller.
Subsequent mutagenesis experiments support the possibility of distinct binding sites for lipoproteins and the EGF homology domain. Replacement of D203 or E208 with lysine impaired the ability of the LDLR to bind to β-VLDL (Fig. 9), suggesting that D203 and E208 contact basic residues of apoE. D203 and E208 are part of cleft 2, which is exposed in the closed conformation. Significantly, LDL binding affinity was not reduced, indicating that D203 and E208 are not involved in LDL binding and that the D203K and E208K mutations do not have global effects on the folding of LA5.
Allosteric interactions frequently drive conformational changes that alter protein function. Comparison of the crystal structure of LA5 alone (44) and LA5 as part of the crystal structure of the LDLR ectodomain (24) suggests that LA5 has an altered conformation in the closed conformation. In the closed conformation, H190 interacts with backbone atoms of N148 of LA4 and the imidazole ring of H562 of the EGF homology domain. These interactions appear to pull H190 toward the junction between the β-propeller and LA4, distorting the strand containing H190 in LA5. One consequence of this distortion is that the side chain of E187 points in opposite directions in the two structures. E187 is near both cleft 1, which interacts with the β-propeller, and cleft 2, which we propose binds to apoE. Thus, H562, H190, and E187 may act as part of an allosteric switch. Consistent with this possibility, the E187K mutation reduced both LDL and β-VLDL binding (Fig. 9). The E187K mutation is also a naturally occurring Familial Hypercholesterolemia mutation (FH-Jerusalem) that is associated with an 85–95% loss of LDLR function (47). The E187K mutation is unlikely to have a global effect on receptor folding because, while this mutation slows folding of LA5 (48, 49), we did not observe immature LDLR in cells expressing the LDLR-E187K (Fig. 9), indicating that the LDLR-E187K passed quality control in the endoplasmic reticulum and received normal glycosylation in the Golgi.
We propose the following allosteric model for lipoprotein release by the LDLR (Fig. 10A). At neutral pH, the receptor is in the open conformation and the EGF homology domain is distant from LA4/5. The absence of the EGF homology domain allows LA5 to form functional binding sites for lipoproteins. These binding sites involve E187, D203, and E208 for the apoE interaction, and E187 for apoB100 binding. When the receptor experiences low pH in endosomes, the EGF homology domain engages LA4/5. This engagement involves H190, H562, and H586, which drive a conformational change in LA5 that disrupts the lipoprotein binding sites on LA5, thereby facilitating release. It should be noted that the EGF homology also contacts LA4, and this interaction may have further allosteric effects on lipoprotein dissociation.
FIGURE 10.
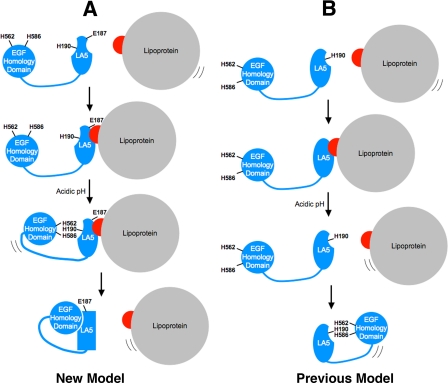
Model of lipoprotein release by the LDLR. In the depicted models, the LDLR is shown in blue, lipoprotein is in gray, and apolipoprotein is in red. Only the EGF homology domain and LA5 of the LDLR are depicted. In the proposed allosteric model (A), lipoproteins bind to the LDLR via E187, D203, and E208 (apoE) or via E187 (apoB100) (1st and 2nd panels of A). Upon acidification, the EGF homology domain contacts LA5 (3rd panel of A), which drives a conformational change in LA5 that involves H190, H562, and H586. The conformational change in LA5 disrupts the binding sites for apolipoproteins, thereby driving release (4th panel of A). This new model stands in contrast to the previous model (panel B). In the previous model, lipoproteins bound to the same surface as the EGF homology domain (top two panels of B). The previous model also proposed that, as lipoprotein dissociated, the EGF homology domain replaced lipoprotein via an interaction that required protonation of H190, H562, and H586 (bottom two panels of B).
This new allosteric model differs with the previous model (Fig. 10B) in two respects. First, the previous model proposed that the β-propeller bound to the same surface on LA4/5 as lipoproteins. This aspect of the previous model necessitated release of lipoproteins prior to engagement by the β-propeller. As discussed above, the kinetics of lipoprotein release (Table 1) do not support this idea. The new model proposes that lipoproteins and the β-propeller bind to separate surfaces on the LDLR. Second, the previous model stipulated that association of the β-propeller with LA4/5 required ionic interactions involving H190, H562, and H586. The gel filtration data (Fig. 1) shows that these three histidines do not control the transition from the open to the closed conformational state of the LDLR. The new model proposes that the closed conformational state is necessary but not sufficient for release and that H190, H562, and H586 are part of an allosteric mechanism of lipoprotein release.
Interestingly, our data also show that mutations at H190, H562, and H586 reduce the rate of β-VLDL internalization (Fig. 6). How the three histidines facilitate β-VLDL uptake is not clear; however, the effect of these mutations on β-VLDL internalization was FDNPVY-dependent (Fig. 7), suggesting that the three histidines promote the association or activity of adaptor proteins that bind to the FDNPVY sequence of the LDLR. Dab-2 and the autosomal recessive hypercholesterolemia protein (ARH) are two adaptors that bind to the FDNPVY sequence and to components of the endocytic machinery of clathrin-coated pits (50, 51). Dab-2 or ARH are required for FDNPVY-dependent LDLR internalization (52, 53). Interestingly, lipoprotein binding to the LDLR promotes the association of ARH with the plasma membrane (54), suggesting that lipoprotein binding is coupled to adaptor protein engagement. Consistent with this possibility, we have recently shown that lipoproteins promote the coated-pit targeting and internalization of the LDLR (32). The mechanism by which H190, H562, and H586 might participate in the coupling of lipoprotein binding with internalization is currently under investigation.
Acknowledgments
We thank Angela Mobley for FACS assistance, Michael Brown and Joseph Goldstein for LDL and β-VLDL, Brenda Pallares for administrative support, and Jonathan Cohen and Richard Anderson for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant HL085218. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: LDL, low density lipoprotein; LDLR, low density lipoprotein receptor; VLDL, very low density lipoprotein; EGF, epidermal growth factor; apoB100, apolipoprotein B100; apoE, apolipoprotein E; LA, LDLR type A repeat; DiI, 3H-Indolium, 2-(3-(1,3-dihydro-3,3-dimethyl-1-octadecyl-2H-indol-2-ylidene)-1-propenyl)-3,3-dimethyl-1-octadecyl-perchlorate; PBS, phosphate-buffered saline; FACS, fluorescent-activated cell sorting; GFP, green fluorescent protein.
References
- 1.Yamamoto, T., Davis, C. G., Brown, M. S., Schneider, W. J., Casey, M. L., Goldstein, J. L., and Russell, D. W. (1984) Cell 3927 -38 [DOI] [PubMed] [Google Scholar]
- 2.Esser, V., Limbird, L. E., Brown, M. S., Goldstein, J. L., and Russell, D. W. (1988) J. Biol. Chem. 26313282 -13290 [PubMed] [Google Scholar]
- 3.Daly, N. L., Scanlon, M. J., Djordjevic, J. T., Kroon, P. A., and Smith, R. (1995) Proc. Natl. Acad. Sci. U. S. A. 926334 -6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.North, C. L., and Blacklow, S. C. (2000) Biochemistry 392564 -2571 [DOI] [PubMed] [Google Scholar]
- 5.Davis, C. G., Goldstein, J. L., Sudhof, T. C., Anderson, R. G., Russell, D. W., and Brown, M. S. (1987) Nature 326760 -765 [DOI] [PubMed] [Google Scholar]
- 6.Jeon, H., Meng, W., Takagi, J., Eck, M. J., Springer, T. A., and Blacklow, S. C. (2001) Nat. Struct. Biol. 8 499-504 [DOI] [PubMed] [Google Scholar]
- 7.Davis, C. G., Lehrman, M. A., Russell, D. W., Anderson, R. G., Brown, M. S., and Goldstein, J. L. (1986) Cell 4515 -24 [DOI] [PubMed] [Google Scholar]
- 8.Lehrman, M. A., Goldstein, J. L., Brown, M. S., Russell, D. W., and Schneider, W. J. (1985) Cell 41 735-743 [DOI] [PubMed] [Google Scholar]
- 9.Bradley, W. A., and Gianturco, S. H. (1986) J. Lipid Res. 2740 -48 [PubMed] [Google Scholar]
- 10.Schneider, W. J., Kovanen, P. T., Brown, M. S., Goldstein, J. L., Utermann, G., Weber, W., Havel, R. J., Kotite, L., Kane, J. P., Innerarity, T. L., and Mahley, R. W. (1981) J. Clin. Investig. 681075 -1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, M. S., Kovanen, P. T., and Goldstein, J. L. (1981) Science 212628 -635 [DOI] [PubMed] [Google Scholar]
- 12.Brown, M. S., and Goldstein, J. L. (1974) Proc. Natl. Acad. Sci. U. S. A. 71 788-792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell, D. W., Brown, M. S., and Goldstein, J. L. (1989) J. Biol. Chem. 26421682 -21688 [PubMed] [Google Scholar]
- 14.Fisher, C., Abdul-Aziz, D., and Blacklow, S. C. (2004) Biochemistry 431037 -1044 [DOI] [PubMed] [Google Scholar]
- 15.Knott, T. J., Rall, S. C. J., Innerarity, T. L., Jacobson, S. F., Urdea, M. S., Levy-Wilson, B., Powell, L. M., Pease, R. J., Eddy, R., Nakai, H., Byers, M., Priestley, L. M., Robertson, E., Rall, L. B., Betsholtz, C., Shows, T. B., Mahley, R. W., and Scott, J. (1985) Science 23037 -43 [DOI] [PubMed] [Google Scholar]
- 16.Yang, C. Y., Chen, S. H., Gianturco, S. H., Bradley, W. A., Sparrow, J. T., Tanimura, M., Li, W. H., Sparrow, D. A., DeLoof, H., Rosseneu, M., Lee, F. S., Gu, Z. W., Gotto, A. M., and Chan, L. (1986) Nature 323738 -742 [DOI] [PubMed] [Google Scholar]
- 17.Weisgraber, K. H., Innerarity, T. L., Harder, K. J., Mahley, R. W., Milne, R. W., Marcel, Y. L., and Sparrow, J. T. (1983) J. Biol. Chem. 25812348 -12354 [PubMed] [Google Scholar]
- 18.Mahley, R. W., Innerarity, T. L., Pitas, R. E., Weisgraber, K. H., Brown, J. H., and Gross, E. (1977) J. Biol. Chem. 2527279 -7287 [PubMed] [Google Scholar]
- 19.Weisgraber, K. H., Innerarity, T. L., and Mahley, R. W. (1978) J. Biol. Chem. 2539053 -9062 [PubMed] [Google Scholar]
- 20.Brown, M. S., and Goldstein, J. L. (1986) Science 23234 -47 [DOI] [PubMed] [Google Scholar]
- 21.Fisher, C., Beglova, N., and Blacklow, S. C. (2006) Mol. Cell 22277 -283 [DOI] [PubMed] [Google Scholar]
- 22.Anderson, R. G., Brown, M. S., and Goldstein, J. L. (1977) Cell 10 351-364 [DOI] [PubMed] [Google Scholar]
- 23.Maxfield, F. R., and McGraw, T. E. (2004) Nat. Rev. Mol. Cell Biol. 5 121-132 [DOI] [PubMed] [Google Scholar]
- 24.Rudenko, G., Henry, L., Henderson, K., Ichtchenko, K., Brown, M. S., Goldstein, J. L., and Deisenhofer, J. (2002) Science 2982353 -2358 [DOI] [PubMed] [Google Scholar]
- 25.Beglova, N., and Blacklow, S. C. (2005) Trends Biochem. Sci. 30309 -317 [DOI] [PubMed] [Google Scholar]
- 26.Beglova, N., Jeon, H., Fisher, C., and Blacklow, S. C. (2004) Mol. Cell 16 281-292 [DOI] [PubMed] [Google Scholar]
- 27.Counter, C. M., Hahn, W. C., Wei, W., Caddle, S. D., Beijersbergen, R. L., Lansdorp, P. M., Sedivy, J. M., and Weinberg, R. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 9514723 -14728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morales, C. P., Holt, S. E., Ouellette, M., Kaur, K. J., Yan, Y., Wilson, K. S., White, M. A., Wright, W. E., and Shay, J. W. (1999) Nat. Genet. 21 115-118 [DOI] [PubMed] [Google Scholar]
- 29.Liu, X., Sun, Y., Constantinescu, S. N., Karam, E., Weinberg, R. A., and Lodish, H. F. (1997) Proc. Natl. Acad. Sci. U. S. A. 9410669 -10674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein, J. L., Basu, S. K., and Brown, M. S. (1983) Methods Enzymol. 98241 -260 [DOI] [PubMed] [Google Scholar]
- 31.Lombardi, P., Mulder, M., van der Boom, H., Frants, R. R., and Havekes, L. M. (1993) J. Biol. Chem. 26826113 -26119 [PubMed] [Google Scholar]
- 32.Michaely, P., Zhao, Z., Li, W. P., Garuti, R., Huang, L. J., Hobbs, H. H., and Cohen, J. C. (2007) EMBO J. 263273 -3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heuser, J. E., and Anderson, R. G. (1989) J. Cell Biol. 108389 -400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopkins, P. N., Wu, L. L., Stephenson, S. H., Xin, Y., Katsumata, H., Nobe, Y., Nakajima, T., Hirayama, T., Emi, M., and Williams, R. R. (1999) J. Hum. Genet. 44 364-367 [DOI] [PubMed] [Google Scholar]
- 35.Sun, X. M., Patel, D. D., Webb, J. C., Knight, B. L., Fan, L. M., Cai, H. J., and Soutar, A. K. (1994) Arterioscler. Thromb. 1485 -94 [DOI] [PubMed] [Google Scholar]
- 36.Goldstein, J. L., Ho, Y. K., Brown, M. S., Innerarity, T. L., and Mahley, R. W. (1980) J. Biol. Chem. 2551839 -1848 [PubMed] [Google Scholar]
- 37.Pitas, R. E., Innerarity, T. L., Arnold, K. S., and Mahley, R. W. (1979) Proc. Natl. Acad. Sci. U. S. A. 762311 -2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown, M. S., Anderson, R. G., Basu, S. K., and Goldstein, J. L. (1982) Cold Spring Harb. Symp. Quant. Biol. 46713 -721 [DOI] [PubMed] [Google Scholar]
- 39.Jones, N. L., Saunders, J. A., and Mallory, R. R. (2000) J. Lipid Res. 411823 -1831 [PubMed] [Google Scholar]
- 40.Tabas, I., Myers, J. N., Innerarity, T. L., Xu, X. X., Arnold, K., Boyles, J., and Maxfield, F. R. (1991) J. Cell Biol. 1151547 -1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabas, I., Lim, S., Xu, X. X., and Maxfield, F. R. (1990) J. Cell Biol. 111929 -940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen, W. J., Goldstein, J. L., and Brown, M. S. (1990) J. Biol. Chem. 2653116 -3123 [PubMed] [Google Scholar]
- 43.Blasiole, D. A., Oler, A. T., and Attie, A. D. (2008) J. Biol. Chem. 28311374 -11381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fass, D., Blacklow, S., Kim, P. S., and Berger, J. M. (1997) Nature 388691 -693 [DOI] [PubMed] [Google Scholar]
- 45.Prevost, M., and Raussens, V. (2004) Proteins 55874 -884 [DOI] [PubMed] [Google Scholar]
- 46.Mahley, R. W., and Innerarity, T. L. (1977) J. Biol. Chem. 2523980 -3986 [PubMed] [Google Scholar]
- 47.Hobbs, H. H., Brown, M. S., and Goldstein, J. L. (1992) Hum. Mutat. 1 445-466 [DOI] [PubMed] [Google Scholar]
- 48.Blacklow, S. C., and Kim, P. S. (1996) Nat. Struct. Biol. 3758 -762 [DOI] [PubMed] [Google Scholar]
- 49.Arias-Moreno, X., Arolas, J. L., Aviles, F. X., Sancho, J., and Ventura, S. (2008) J. Biol. Chem. 28313627 -13637 [DOI] [PubMed] [Google Scholar]
- 50.Mishra, S. K., Watkins, S. C., and Traub, L. M. (2002) Proc. Natl. Acad. Sci. U. S. A. 9916099 -16104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He, G., Gupta, S., Yi, M., Michaely, P., Hobbs, H. H., and Cohen, J. C. (2002) J. Biol. Chem. 27744044 -44049 [DOI] [PubMed] [Google Scholar]
- 52.Maurer, M. E., and Cooper, J. A. (2006) J. Cell Sci. 1194235 -4246 [DOI] [PubMed] [Google Scholar]
- 53.Keyel, P. A., Mishra, S. K., Roth, R., Heuser, J. E., Watkins, S. C., and Traub, L. M. (2006) Mol. Biol. Cell 174300 -4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sirinian, M. I., Belleudi, F., Campagna, F., Ceridono, M., Garofalo, T., Quagliarini, F., Verna, R., Calandra, S., Bertolini, S., Sorice, M., Torrisi, M. R., and Arca, M. (2005) J. Biol. Chem. 28038416 -38423 [DOI] [PubMed] [Google Scholar]