Abstract
Platelet-derived growth factor BB (PDGF-BB) induced cyclin A expression and CDK2 activity in vascular smooth muscle cells (VSMC). Inhibition of nuclear factors of activated T cell (NFAT) activation by cyclosporin A (CsA) and VIVIT suppressed PDGF-BB-induced cyclin A expression and CDK2 activity, resulting in blockade of VSMC in the G1 phase. In addition, CsA- and VIVIT-mediated inhibition of NFATs and small interfering RNA-targeted down-regulation of cyclin A levels suppressed PDGF-BB-induced VSMC DNA synthesis. PDGF-BB also induced cyclin A mRNA levels in VSMC in an NFAT-dependent manner. Cloning and bioinformatic analysis of rat cyclin A promoter revealed the presence of NFAT-binding elements, and PDGF-BB induced the binding of NFATs to these regulatory sequences in a CsA- and VIVIT-sensitive manner. Chromatin immunoprecipitation analysis showed that NFATc1 binds to the cyclin A promoter in response to PDGF-BB in a VIVIT-sensitive manner. Furthermore, PDGF-BB induced cyclin A promoter-luciferase reporter gene activity in VSMC, and it was inhibited by both CsA and VIVIT. Balloon injury induced cyclin A expression and CDK2 activity in rat carotid arteries, and these responses were also blocked by VIVIT. In addition, VIVIT attenuated balloon injury-induced SMC proliferation, resulting in reduced restenosis. Down-regulation of NFATc1 by its small interfering RNA inhibited PDGF-BB-induced cyclin A expression and DNA synthesis both in rat and human VSMC. Together, these findings demonstrate that the cyclin A-CDK2 complex may be a potential effector of NFATs, specifically NFATc1, in mediating SMC multiplication leading to neointima formation. Therefore, NFATs may be used as target molecules for the development of therapeutic agents against vascular diseases such as restenosis.
Cyclin-dependent kinases (CDKs)2 play an important role in cell cycle progression (1–3). The activities of CDKs are regulated positively by cyclins and negatively by CDK inhibitors (4–12). In fact, CDKs require association with cyclins for their activity (1, 4, 5). For instance, CDK4 requires complex formation with type D cyclins for its activity and so does CDK2 with either cyclin E or cyclin A (13–15). Although the levels of CDKs may or may not change in cycling versus noncycling cells, cyclin levels tend to increase in response to mitogenic stimulants (4, 5). Therefore, the availability of cyclins is a critical factor in the positive regulation of CDKs (4). Sequential activation of cyclin D-CDK4, cyclin E-CDK2, cyclin A-CDK2, and cyclin B1-CDK1 appears to be required for an orderly progression through G1, G1/S transition through S, and G2/M transition phases of the cell cycle (4, 5). Cyclins can be divided into two groups as follows: 1) G1 cyclins, and 2) mitotic cyclins (5). G1 cyclins (type D and E) are important in the modulation of the activity of tumor suppressors such as retinoblastoma protein (Rb) (16). Rb in an active form is under-phosphorylated. It binds to and down-regulates the activity of transcription factors, namely E2Fs, and thereby prevents replicative DNA synthesis (16). G1 cyclin-CDKs (CDK4, CDK6, and CDK2), upon activation, phosphorylate and inactivate Rb. Phosphorylated Rb dissociates from and relieves the inhibitory constraint on E2Fs, which in turn influence transcription of genes required for DNA synthesis (17). On the other hand, the mitotic cyclins, cyclin A and cyclin B, in complex with CDK2 and CDK1, respectively, play an essential role in the progression of the cell through the S phase and G2/M transition phase of the cell cycle (18). Cyclin A also plays a role in G2/M transition via its association with and regulation of CDK1 activity (19). Of the two A-type cyclins known thus far, cyclin A, which is also known as cyclin A2, plays a key role in the regulation of mammalian cell cycle (18, 19). In contrast, cyclin A1, whose expression is restricted to testis, plays a role in spermatogenesis (20, 21). In recent years, it was demonstrated that knock-out mice for any single cyclin or CDK survive normally suggesting redundancy in the signaling events of these molecules fulfilling similar roles (22–24). However, targeting the down-regulation of cyclins along with their partner CDKs leads to blockade of cell proliferation (25, 26). Because vascular smooth muscle growth is an important contributor of proliferative vascular diseases, targeting the disruption of CDK activities via the expression of their inhibitors has been given attention in the development of approaches to prevent these vascular lesions (27–29).
The nuclear factors of activated T cells (NFATs) are a family of transcriptional factors that belong to the Rel group (30). Among the five members of the NFAT family of transcriptional factors cloned and characterized thus far, the activation of NFATc1 to NFATc4 is dependent on a serine/threonine phosphatase, namely calcineurin (30). Although the initial studies showed their presence in immune cells mediating immune responses (31), the subsequent investigations revealed their presence in nonimmune cells, playing a role in various cellular functions, including cardiac hypertrophy, skeletal muscle growth, and osteoclast differentiation (32–35). Furthermore, using gene knock-out mouse models, the importance of NFATs in the development of the cardiovascular system has been demonstrated (36–38). In addition to these functions, work from our laboratory as well as others showed that NFATs are involved in the regulation of cell migration and proliferation (39–45). Furthermore, toward understanding the mechanisms of NFAT involvement in the regulation of VSMC migration, we discovered that these transcriptional factors mediate interleukin-6 expression as one of the effector of molecules of RTK and G-protein-coupled receptor agonist-induced VSMC motility (45). In this study, we have extended our studies toward identifying the mechanisms of NFATs in mediating RTK agonist, PDGF-BB-induced VSMC growth. Here we show that NFATs, specifically NFATc1, mediate the induction of expression of cyclin A, thereby leading to increased CDK2 activity in VSMC and the progression of these cells through the cell cycle in response to PDGF-BB. Furthermore, we demonstrate that BI-induced cyclin A expression and CDK2 activity are also dependent on NFAT activation signaling.
MATERIALS AND METHODS
Reagents—Cyclosporin A (A-195) was bought from Biomol (Plymouth Meeting, PA). Histone H1 was supplied by Calbiochem. Recombinant human PDGF-BB (220-BB) was from R & D Systems Inc. (Minneapolis, MN). Anti-CDK2 (SC-163) and anti-β-tubulin (SC-9104) antibodies and mouse normal serum (SC-45051) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-cyclin A (RB-007-P1 and RB-1548-P0) and anti-PCNA (MS-106-P) antibodies were obtained from NeoMarkers (Fremont, CA). Anti-NFATc1 (MA3-024) antibodies were purchased from Affinity BioReagents (Golden, CO). Monoclonal anti-SMα-actin antibodies (catalog number A2547) were obtained from Sigma. Lipofectamine 2000 reagent, propidium iodide, Taq polymerase, and pCR2.1-TOPO vector were from Invitrogen. RNase A was purchased from Roche Applied Science. T4 polynucleotide kinase, T4 DNA ligase, and the restriction enzymes were obtained from New England Biolabs (Ipswich, MA). Rat NFATc1 siRNA sequences (sense, 5′-CUA CUA AUG AGC AGC GAA AUU-3′; antisense, 5′-UUU CGC UGC UCA UUA GUA GUU-3′), human NFATc1 siRNA sequences (catalog number ON-TARGETplus SMARTpool L-003605-00-0020, human NFATc1, NM_172390), siCONTROL nontargeting siRNA number 2 (catalog number D-0012-02-20) and DharmaFECT 1 transfection reagent (catalog number T-2001-02) were bought from Dharmacon RNAi Technologies (Chicago, IL). [γ-32P]ATP (3000 Ci/mmol) and protein-A-Sepharose (CL-4B) were from Amersham Biosciences. All the primers and oligonucleotides were made by IDT (Coralville, IA).
Cell Culture—Rat VSMC were isolated and subcultured as described previously (42). VSMC were used between 4 and 12 passages. Human aortic smooth muscle cells (HASMC) were bought from Cascade Biologics (Portland, OR). HASMC were grown in medium 231 containing smooth muscle growth supplements, 10 μg/ml gentamycin, and 0.25 μg/ml amphotericin B. Cultures were maintained at 37 °C in a humidified 95% air and 5% CO2 atmosphere. HASMC were quiesced by incubating in medium 231 for 48 h and used to perform the experiments unless otherwise indicated.
Construction of GFP and GFPVIVIT Adenoviral Vectors—Construction of Ad-GFP and Ad-GFPVIVIT was described previously (42).
Western Blot Analysis—Equal amounts of protein from VSMC or tissue extracts were analyzed by Western blotting for the indicated proteins using specific antibodies as described previously (42).
CDK2 Assay—Protein extracts from cells or tissues were prepared using lysis buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 50 mm glycerophosphate, 10 mm NaF, and 1 mm sodium orthovanadate). An equal amount of protein (200 μg) from each sample was immunoprecipitated with anti-CDK2 antibodies (2 μg) overnight at 4 °C followed by incubation with protein-A-Sepharose CL-4B beads for 1 h at room temperature. The immunocomplexes were washed four times with lysis buffer and one time with kinase buffer and incubated in kinase reaction mix containing 25 mm HEPES, pH 7.4, 10 mm MgCl2, 1 mm EGTA, 200 μg/ml histone H1, 1 μCi of [γ-32P]ATP, and 50 μm ATP for 30 min at 30 °C. The kinase reactions were stopped by the addition of SDS-PAGE sample buffer and boiling for 5 min. The reaction products were separated by electrophoresis on 0.1% SDS and 10% polyacrylamide gels. The 32P-labeled histone H1 protein was visualized by autoradiography, and the band intensity was quantified using NIH Image J.
FACS Analysis—Quiescent VSMC that were treated with and without PDGF-BB (20 ng/ml) for 16 h were trypsinized and collected by centrifugation at 1000 rpm for 5 min at 4 °C. After washing with calcium and magnesium-free HBSS containing 1 mm EDTA and 1% (w/v) bovine serum albumin, cells were fixed by adding 2 ml of ice-cold alcohol and collected by centrifugation at 1000 rpm for 5 min at 4 °C. Cells were then rinsed twice with 1 ml of HBSS and suspended in 500 μl of propidium iodide buffer (20 μg of propidium iodide in 1 ml of calcium- and magnesium-free HBSS containing 0.5 mg/ml RNase A) in Falcon polystyrene round bottom tubes (catalog number 352052, BD Biosciences) and incubated at 37 °C for 30 min. Cells were kept in the dark at 4 °C for about 2 h before analyzing for cell cycle progression using LSR II flow cytometer (BD Biosciences). The data were analyzed by Modfit.
RT-PCR—After appropriate treatments, total cellular RNA was isolated from VSMC using TRIzol reagent as per the manufacturer's guidelines (Invitrogen). Reverse transcription was carried out with Superscript III first-strand synthesis system for RT-PCR based on supplier's protocol (Invitrogen). The cDNA was then used as a template for PCR using specific primers. The primers are as follows: rat cyclin A (GenBank™ accession number NM_053702): forward, 5′-GAGCAAAAAGAAACTCAGTGTGAA-3′, and reverse, 5′-CAGACATGGAGGAGAGGAATCTAT-3′; rat β-actin (GenBank™ accession number EF156276): forward, 5′-CGTTGACATCCGTAAAGACC-3′ and reverse, 5′-GATAGAGCCACCAATCCACA-3′; human cyclin A (GenBank™ accession number NM_001237): forward, 5′-TTTAAGGATCTTCCTGTAAAT-3′, and reverse, 5′-ATAGTATGTGGTGATTCAAAA-3′; and human β-actin (GenBank™ accession number NM_001101): forward, 5′-AGCCATGTACGTTGCTAT-3′, and reverse 5′-GATGTCCACGTCACACTTCA-3′. The amplification was carried out on Gene-Amp PCR System 2400 (Applied Biosystems, Foster City, CA), using the following amplification conditions for the above-mentioned genes: for cyclin A, 95 °C for 5 min followed by 30 cycles at 95 °C for 45 s, 58 °C for 45 s, and 72 °C for 50 s with final extension at 72 °C for 7 min; for β-actin, 95 °C for 5 min followed by 30 cycles at 95 °C for 45 s, 55 °C for 45 s, and 72 °C for 30 s with final extension at 72 °C for 7 min. The amplified RT-PCR products were separated on 1.2% (w/v) agarose or 8% native polyacrylamide gels and stained with ethidium bromide, and pictures were captured using AlphaEase Digital Imaging System (Alpha Innotech Corp., San Leandro, CA) and the band intensities were quantified using NIH Image J.
Electrophoretic Mobility Shift Assay—After appropriate treatments, VSMC nuclear extracts were prepared and analyzed for NFAT DNA binding activity as described previously (29). Double-stranded oligonucleotides from -670 to -633 region of rat cyclin A promoter (5′-CTTTATTAGAGGCGAGGAAAAGTTTACTGGAATAC-3′ and 3′-GAAATAATCTCCGCTCCTTTTCAAATGACCTTATG-5′) were used as 32P-labeled probe to measure NFAT DNA binding activities. The nucleotide sequence in boldface type was changed to CTT to make a mutant oligonucleotide probe for testing the specificity of NFAT DNA binding.
Chromatin Immunoprecipitation (ChIP) Assay—ChIP assay was performed on VSMC by using ChIP assay kit following the supplier's protocol (Upstate Biotechnology Inc., Lake Placid, NY). NFATc1-DNA complexes were immunoprecipitated using monoclonal anti-NFATc1 antibodies. Mouse normal serum was used as a negative control. Immunoprecipitated DNA was uncross-linked, subjected to proteinase K digestion, and purified using QIAquick columns (catalog number 28104, Qiagen, Valencia, CA). The purified DNA was used as template for PCR amplification using primers for cyclin A promoter (forward, 5′-GGCATTTTAAATTGTCCCATC-3′, and reverse, 5′-ACTCGTTCGCCTCTCCTGTA-3′) regions flanking the putative NFAT-binding sites. To demonstrate the specificity of NFATc1 binding to the cyclin A promoter, a set of primers (forward, 5′-TAACATTGGTGCTGCCAACTGTCA-3′, and reverse, 5′-GATTTCTGAATCAATTTATTCTTA-3′) in the rat cyclin A cDNA region spanning from 1561 to 1710, i.e. ∼2.5 kb away from the NFAT binding sequence, was used as a negative control. The PCR products were resolved on 1.2% agarose or 8% native polyacrylamide gels and stained with ethidium bromide; pictures were captured using AlphaEase Digital Imaging System (Alpha Innotech Corp., San Leandro, CA), and the band intensities were quantified using NIH Image J.
Cloning of Rat Cyclin A Promoter—Rat cyclin A promoter DNA was amplified from rat genomic DNA by PCR with both forward primer (5′-CAA GTT GGC ATT TTA AAT TGT CCC-3′) and reverse primer (5′-ATC CGT CGG CGG CAG AGC GTG CAA G-3′) that were designed according to the published rat cyclin A promoter (46). The PCR consisted of 200 nm forward and reverse primers, 2.5 units of TaqDNA polymerase, 200 μm dNTPs, 5% (v/v) DMSO, 1.5 mm MgCl2, 10 mm Tris-HCl, pH 8.3, and 50 mm NaCl. The PCR was performed at 94 °C for 2 min followed by 25 cycles at 92 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s with a final extension of 10 min at 72 °C. The PCR-amplified rat cyclin A promoter was cloned into pCR2.1-TOPO vector, and the final clone pF3TOPO was verified by DNA sequencing. The rat cyclin A promoter was released from pCR2.1-TOPO vector by digestion with SacI and XhoI and cloned into the SacI and XhoI sites of pGL3-basic vector. The final construct of rat cyclin A promoter carrying luciferase reporter gene, pF3Luc, was verified by DNA sequencing with RV3 primer (5′-CTA GCA AAA TAG GCT GTC CC-3′).
Luficerase Reporter Gene Assay—VSMC were transduced first with Ad-GFP or Ad-VIVIT and then transfected with cyclin A promoter-luciferase construct using Lipofectamine 2000 reagent. The cells were then quiesced in serum-free Dulbecco's modified Eagle's medium for 36 h. After quiescence, cells were treated with and without PDGF-BB (20 ng/ml) for 8 h. In the CsA experiment, 10 μm CsA was used instead of adenovirus. After the treatments, cells were washed once with ice-cold phosphate-buffered saline and lysed with 200 μl of lysis buffer, and the cell extracts were collected into microcentrifuge tubes. The samples were centrifuged for 2 min at 12,000 × g at 4 °C. The supernatants were transferred into new tubes, and 40 μg of the supernatant was used for measuring luciferase activity using Promega luciferase assay system and Turner Luminometer (TD-20/20) (Turner Designs, Sunnyvale, CA).
Carotid Artery Balloon Injury—All the animal protocols were performed in accordance with the relevant guidelines and regulations approved by the Internal Animal Care & Use Committee of the University of Tennessee Health Science Center. BI was performed essentially as described previously by us (42). Briefly, rats weighing 250–300 g were anesthetized by injecting (intraperitoneally) ketamine (60 mg/kg) and xylazine (5 mg/kg). Under a stereomicroscope, the right common, external, and internal carotid arteries were exposed by a longitudinal midline cervical incision, and blood flow was temporarily interrupted by ligation of the common and internal carotid arteries using vessel clips. External carotid artery was ligated permanently. A 2F Fogarty arterial embolectomy catheter was introduced through an arteriotomy in the external carotid artery just below the ligature and advanced to the common carotid artery. To produce carotid artery injury, the balloon was inflated with saline and passed six times with rotation from just under the proximal edge of the omohyoid muscle to the carotid bifurcation. After this, the balloon was deflated and the catheter was withdrawn. The external carotid artery was ligated with a 6-0 silk suture, and the blood flow was restored by removing the clips at the common and internal carotid arteries. After inspection to ascertain adequate pulsation of the common carotid artery, the surgical incision was closed, and the rats were allowed to recover from anesthesia in a humidified and warmed chamber for 2–4 h. At 3 days or 2 weeks after balloon injury, the animals were sacrificed with an overdose of pentobarbital (200 mg/kg), and the carotid arteries were collected, and either protein was isolated or fixed to make cryosections.
Delivery of Adenoviruses into Injured Arteries—After balloon injury, solutions of 100 μl of Ad-GFP (1010 plaque-forming units/ml) or Ad-VIVIT (1010 plaque-forming units/ml) were infused into the ligated segment of the common carotid artery for 30 min as described previously (42). Viral transductions were evaluated at 1 week post-BI by isolation of arteries followed by either cryostat cross-sectioning and examining for green fluorescence protein expression under Nikon diaphot fluorescence microscope with photometrics CH250 CCD camera (Nikon, Garden City, NY) or by preparing tissue extracts and Western blot analysis of GFP levels using its specific antibodies.
Double Immunofluorescence Staining—After dissecting out, arteries were fixed in 10% formalin and embedded in OCT compound. Cryo-sections (5 μm) were made using Leica Kryostat machine (model CM3050S). After blocking in normal goat serum, the cryo-sections were incubated first with anti-SMCα-actin antibodies (1:500) followed by TRITC-conjugated secondary antibodies. After washing with phosphate-buffered saline and blocking again in normal goat serum, these sections were incubated with anti-cyclin A (1:100) antibodies followed by fluorescein isothiocyanate-conjugated secondary antibodies. Fluorescence was observed under a Zeiss inverted microscope (model Axiovert 200 M).
Histochemistry—The cryo-sections (5 μm) of balloon injured, Ad-GFP, or Ad-VIVIT-transduced carotid arteries were immunostained for PCNA using its specific antibodies or stained with hematoxylin and eosin. The stained sections were observed under a light microscope (model Eclipse 50i, Garden City, NY). SMC proliferation was measured by counting PCNA-positive cells in the intimal and medial regions of each artery using NIH image J. The intimal (I) and medial (M) areas in hematoxylin and eosin-stained sections were measured using NIH image J program, and the I/M ratios and lumen sizes were calculated.
Statistics—All the experiments were repeated several times with similar results. Data are presented as mean ± S.D. The treatment effects were analyzed by Student's t test. p values <0.05 were considered to be statistically significant. In the case of ChIP analysis, electrophoretic mobility shift assay, and Western blotting, one representative set of data is shown.
RESULTS
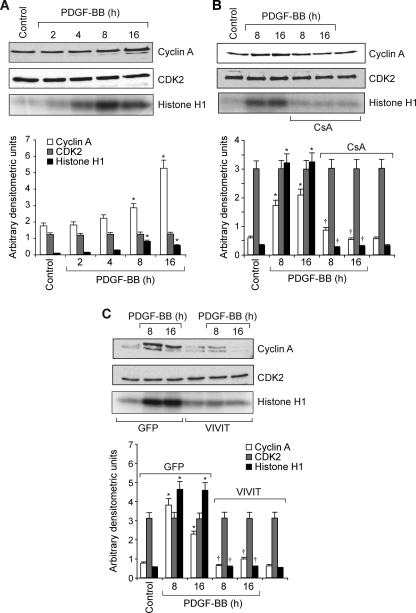
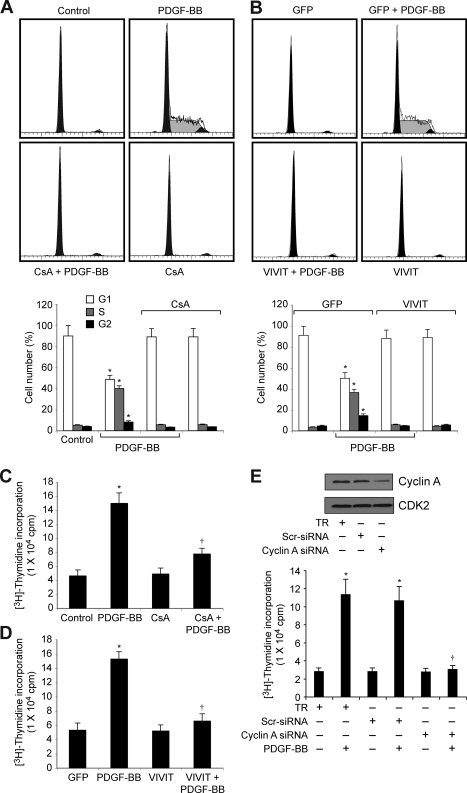
To understand the role of NFATs in vascular wall remodeling, we studied their involvement in cell cycle-dependent gene expression in VSMC in response to PDGF-BB and in the vessel wall in response to BI. PDGF-BB (20 ng/ml) induced cyclin A levels in VSMC in a time-dependent manner with maximum 2–3-fold increases at 8 and 16 h of treatment, respectively (Fig. 1A). On the other hand, it had no effect on CDK2 levels. Because cyclin A forms a complex with and influences CDK2 activity, we next studied the effect of PDGF-BB on the activity of this enzyme. Consistent with its effect on cyclin A levels, PDGF-BB (20 ng/ml) induced CDK2 activity in a time-dependent manner with maximum 3-fold increase at 8 and 16 h (Fig. 1A). Next we tested the effect of CsA on PDGF-BB-induced cyclin A expression. CsA is a potent inhibitor of calcineurin (30). Calcineurin by dephosphorylating facilitates the activation of NFATs (30). CsA (10 μm), while having no effect on the levels of CDK2, completely inhibited PDGF-BB-induced cyclin A expression (Fig. 1B). CsA also blocked PDGF-BB-induced CDK2 activity (Fig. 1B). To confirm these results further, we tested the effect of VIVIT, an NFAT competing peptide for binding to calcineurin (47). Adenovirus-mediated expression of VIVIT also blocked PDGF-BB-induced increases in cyclin A levels and CDK2 activity (Fig. 1C). Because cyclin A/CDK2 plays an important role in cell cycle progression, we examined the effects of CsA and VIVIT on PDGF-BB-induced VSMC cell cycle progression. Upon exposure to PDGF-BB (20 ng/ml) for 16 h, 30–40% of quiescent cells entered into S phase (Fig. 2, A and B). CsA and VIVIT completely blocked PDGF-BB-induced VSMC progression onto S phase (Fig. 2, A and B). To confirm this result, we tested the effect of CsA and VIVIT on PDGF-BB-induced DNA synthesis. As measured by [3H]thymidine incorporation, both CsA and VIVIT completely inhibited PDGF-BB-induced VSMC DNA synthesis (Fig. 2, C and D). Similarly, cyclin A siRNA also blocked PDGF-BB-induced VSMC DNA synthesis (Fig. 2E).
FIGURE 1.
PDGF-BB induces cyclin A expression and CDK2 activity in NFAT-dependent manner in VSMC. A, quiescent VSMC were treated with and without PDGF-BB (20 ng/ml) for the indicated times, and cell extracts were prepared. An equal amount of protein from control and each treatment was analyzed either for cyclin A and CDK2 levels by Western blotting or for CDK2 activity by immunocomplex kinase assay. B, quiescent VSMC were treated with and without PDGF-BB (20 ng/ml) in the presence and absence of CsA (10 μm) for the indicated times, and cell extracts were prepared and analyzed for cyclin A and CDK2 levels and/or activity as described in A. C, all the conditions were the same as in B except that cells were transduced with either Ad-GFP (control) or Ad-VIVIT with an m.o.i. of 80 and quiesced before treatment with and without PDGF-BB (20 ng/ml) and analyzed for cyclin A and CDK2 levels and/or activity. The bar graphs represent mean ± S.D. values of three independent experiments. *, p < 0.01 versus control or GFP; †, p < 0.01 versus PDGF-BB or GFP + PDGF-BB treatment alone.
FIGURE 2.
Blockade of NFAT activation prevents cell cycle progression of VSMC from G1 to S phase. A, quiescent VSMC were treated with and without PDGF-BB (20 ng/ml) in the presence and absence of CsA (10 μm) for 16 h and subjected to FACS analysis. B, VSMC were transduced with either Ad-GFP or Ad-VIVIT at an m.o.i. of 80, quiesced, treated with and without PDGF-BB (20 ng/ml) for 16 h, and subjected to FACS analysis. Thebar graphs represent mean ± S.D. values of three independent experiments. C, quiescent VSMC were treated with and without PDGF-BB (20 ng/ml) in the presence and absence of CsA (10 μm) for 24 h, and DNA synthesis was measured by [3H]thymidine incorporation. D, conditions were the same as in C except that cells were transduced with either Ad-GFP or Ad-VIVIT at an m.o.i. of 80, and quiesced before subjecting to treatment with PDGF-BB and measuring DNA synthesis. E, top panel, VSMC were transfected with scrambled (Scr) or cyclin A siRNA, and 36 h later cell extracts were prepared, and an equal amount of protein from each condition was analyzed by Western blotting for cyclin A levels. Bottom panel, after transfection with scrambled or cyclin A siRNA, cells were quiesced and treated with and without PDGF-BB (20 ng/ml) for 24 h, and DNA synthesis was measured as described in C.*, p < 0.01 versus control, GFP, or TR; †, p < 0.01 versus PDGF-BB or GFP + PDGF-BB or TR + PDGF-BB treatment alone. TR, transfection reagent.
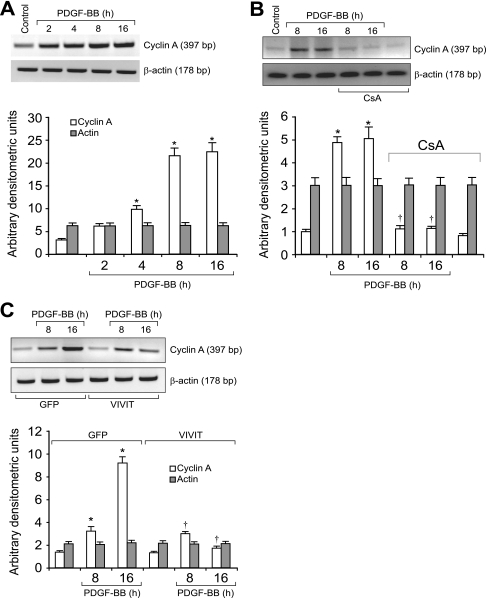
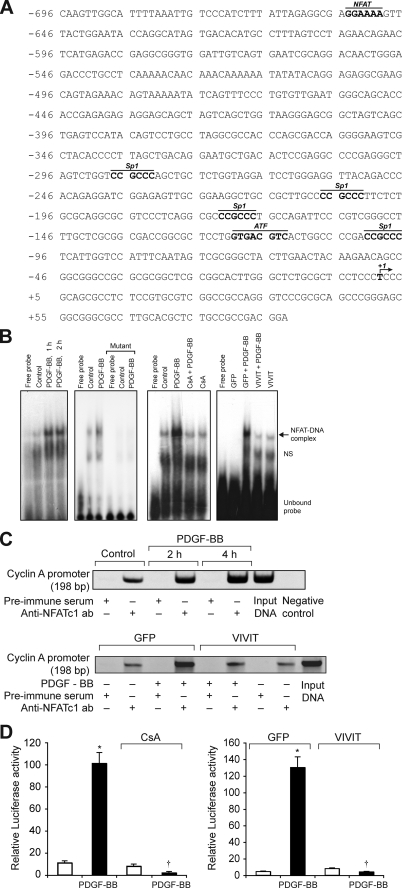
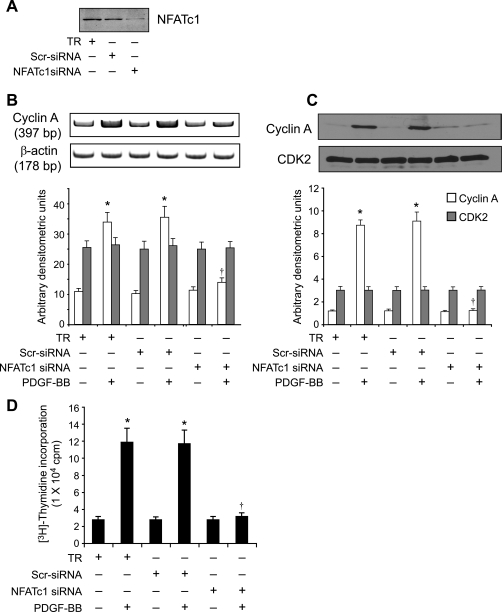
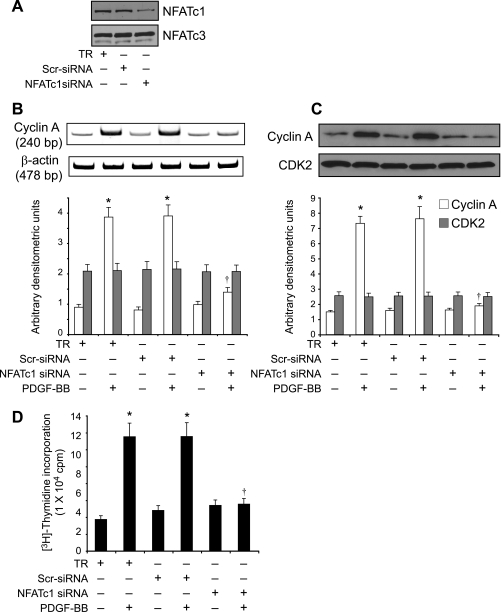
To identify the mechanisms underlying the involvement of NFATs in the regulation of cyclin A expression, we studied the effect of CsA and VIVIT on its mRNA levels. PDGF-BB (20 ng/ml) induced cyclin A mRNA levels in a time-dependent manner with maximum effects at 8 and 16 h (Fig. 3A). CsA and VIVIT inhibited PDGF-BB-induced cyclin A mRNA levels (Fig. 3, B and C). To understand the mechanisms by which NFATs influence cyclin A mRNA levels, we cloned a 696-bp rat cyclin A promoter region using primers designed on a published sequence (46). Bioinformatic analysis of the cloned rat cyclin A promoter revealed the presence of NFAT-binding regulatory sequence spanning from -655 to -650 region (Fig. 4A). To test whether NFATs bind to these elements in response to PDGF-BB, we first performed the time course effect of PDGF-BB on NFAT DNA binding activity using NFAT regulatory sequence present in the cyclin A promoter as a 32P-labeled probe. PDGF-BB (20 ng/ml) induced NFAT DNA binding activity in a time-dependent manner with a maximum 3-fold effect at 1 h using this oligonucleotide sequence as a probe (Fig. 4B). On the other hand, no NFAT DNA binding activity was observed with a 32P-labeled oligonucleotide probe in which the NFAT-binding element was mutated from AGG to TCC. CsA and VIVIT completely inhibited PDGF-BB-induced NFAT DNA binding activity (Fig. 4B). To obtain additional evidence for the binding of NFATs to the cyclin A promoter, we performed ChIP assay using anti-NFATc1 antibodies. ChIP analysis revealed a time-dependent binding of NFATc1 to the cyclin A promoter in VSMC in response to PDGF-BB (Fig. 4C). Blockade of NFAT activation with VIVIT completely inhibited the PDGF-BB-induced binding of NFATc1 to the cyclin A promoter (Fig. 4C). No DNA amplification was observed with a set of primers designed in the cyclin A coding region 2.5 kb away from the NFAT regulatory sequence in the anti-NFATc1 antibody chromatin immunoprecipitants, a result that demonstrates the specificity of NFATc1 binding to cyclin A promoter in response to PDGF-BB. In addition, PDGF-BB (20 ng/ml) induced NFAT-binding element containing 696-bp cyclin A promoter-driven luciferase reporter gene activity, and it was blocked by both CsA and VIVIT (Fig. 4D). To extend the in vitro observations on the role of NFATs in the regulation of cyclin A to in vivo, we next studied the role of NFATs in BI-induced expression of cyclin A. As measured by Western blotting, BI of rat carotid artery induced the expression cyclin A levels at 3 days post-injury, and adenovirus-mediated transduction of VIVIT completely suppressed this effect (Fig. 5A). Similarly, BI induced CDK2 activity, and this response as well was blocked by VIVIT (Fig. 5A). To test whether BI-induced NFAT-mediated cyclin A expression occurs in SMC, we performed double immunofluorescence staining for SMα-actin and cyclin A co-localization. Substantial co-localization of SMα-actin and cyclin A was observed in the arteries in response to BI, and adenovirus-mediated expression of VIVIT reduced this co-immunostaining by about 90% suggesting that NFAT-dependent increases in cyclin A expression occur in SMC of balloon-injured arteries (Fig. 5B). Consistent with cyclin A/CDK2 activity, BI induced PCNA expression, and it was blocked by VIVIT (Fig. 6A). Because blockade of NFAT activation signaling suppressed cyclin A/CDK2 activity resulting in reduced PCNA expression, we further tested the effect of VIVIT on neointima formation, as SMC proliferation is an important contributor of injury-induced vessel wall remodeling. Substantial neointima formation was observed at 2 weeks post-BI and, consistent with our earlier observations (42), adenovirus-mediated transduction of VIVIT attenuated this response by about 40% resulting in decreased I/M ratio and increased lumen size (Fig. 6B). To identify the NFAT family member mediating PDGF-BB-induced cyclin A expression and DNA synthesis in VSMC, we tested the role of NFATc1 using siRNA approach. Down-regulation of NFATc1 levels by its siRNA blocked PDGF-BB-induced cyclin A expression at both mRNA and protein levels and DNA synthesis in rat VSMC (Fig. 7, A–D). To extend these observations to human vascular disease, we next studied the role of NFATc1 in the regulation of cyclin A expression in HASMC. Down-regulation of NFATc1 levels by its siRNA also suppressed PDGF-BB-induced cyclin A expression and DNA synthesis in HASMC as well (Fig. 8, A–D).
FIGURE 3.
PDGF-BB induces the expression of cyclin A mRNA in NFAT-dependent manner in VSMC. A, quiescent VSMC were treated with and without PDGF-BB (20 ng/ml) for the indicated times, and total cellular RNA was isolated. An equal amount of RNA was subjected to RT-PCR analysis of cyclin A and β-actin mRNA levels using their respective primers. B, quiescent VSMC were treated with and without PDGF-BB (20 ng/ml) in the presence and absence of CsA (10 μm) for the indicated times, and total cellular RNA was isolated and analyzed for cyclin A and β-actin mRNA levels as described in A. C, conditions were the same as in B except that cells were transduced with either Ad-GFP or Ad-VIVIT at an m.o.i. of 80 and quiesced before subjecting to PDGF-BB treatment and RT-PCR analysis. The bar graphs represent mean ± S.D. values of three independent experiments. *, p < 0.01 versus control or GFP; †, p < 0.01 versus PDGF-BB, GFP + PDGF-BB treatment alone.
FIGURE 4.
PDGF-BB induces cyclin A promoter-driven luciferase reporter gene activity in NFAT-dependent manner in VSMC. A, rat cyclin A promoter region showing the potential transcription factor-binding motifs. B, 1st panel from left, quiescent VSMC were treated with and without PDGF-BB (20 ng/ml) for the indicated times, and nuclear extracts were prepared. An equal amount of nuclear protein from control and each treatment was analyzed for DNA binding activity using 32P-labeled NFAT consensus sequences from rat cyclin A promoter as a probe. 2nd panel, 32P-labeled consensus or mutant NFAT probes in the DNA binding assays using control and 1 h-PDGF-BB (20 ng/ml)-treated nuclear extracts. 3rd panel, all the conditions were the same as in 1st panel except that cells were treated with and without PDGF-BB (20 ng/ml) in the presence and absence of CsA (10 μm) for 1 h, and nuclear extracts were prepared and analyzed for NFAT DNA binding activity. 4th panel, conditions were the same as in 3rd panel except that cells were transduced with either Ad-GFP or Ad-VIVIT at an m.o.i. of 80 and quiesced before subjecting to treatment with PDGF-BB, preparation of nuclear extracts, and measuring NFAT DNA binding activity. C, top panel, chromatin immunoprecipitation assay was performed with control and various time periods of PDGF-BB-treated VSMC using monoclonal anti-NFATc1 antibodies, and the resulting DNA fragments were subjected to PCR amplification using primers spanning the NFAT consensus sequences from rat cyclin A promoter. Bottom panel, VSMC were transduced with either Ad-GFP or Ad-VIVIT at an m.o.i. of 80, quiesced, treated with and without PDGF-BB (20 ng/ml) for 2 h, and subjected to ChIP assay as described in the top panel. D, left panel, VSMC that were transfected with cyclin A promoter-luciferase construct and quiesced were treated with and without PDGF-BB (20 ng/ml) in the presence and absence of CsA (10 μm) for 8 h, and cell extracts were prepared and assayed for luciferase activity. Right panel, all the conditions were the same as in the left panel except that cells were transduced first with Ad-GFP or Ad-VIVIT at an m.o.i. of 80 and then transfected with luciferase reporter plasmid DNA followed by quiescence and treatment with and without PDGF-BB and measuring luciferase activity. *, p < 0.01 versus control or GFP; †, p < 0.01 versus PDGF-BB or GFP + PDGF-BB treatment alone.
FIGURE 5.
Blockade of NFAT activation suppresses balloon injury-induced cyclin A expression and CDK2 activity in the arteries. Soon after balloon injury, the rats received adenovirus expressing either GFP or VIVIT by infusion into the injured arteries. Three days after balloon injury, rats were sacrificed, and the injured right common carotid arteries and uninjured left common carotid arteries were dissected out, and either tissue extracts or sections were made. A, an equal amount of protein from uninjured and balloon-injured Ad-GFP or Ad-VIVIT-transduced arteries was analyzed for cyclin A andβ-tubulin expression and CDK2 activity by Western blotting and immunocomplex kinase assay, respectively, as described in Fig. 1 legend. The bar graph represents mean ± S.D. values of six animals in three groups. B, double immunofluorescence staining of balloon injured Ad-GFP or Ad-VIVIT transduced carotid artery sections for SMα-actin and cyclin A. *, p < 0.01 versus UI; †, p < 0.01 versus GFP-BI. UI, uninjured.
FIGURE 6.
Blockade of NFAT activation suppresses BI-induced SMC proliferation and neointima formation. A, all the treatment conditions were the same as in Fig. 5 except that after BI, rats were sacrificed; arteries were isolated and fixed, and SMC proliferation was measured by PCNA staining as described under “Materials and Methods.” Top panel shows the representative pictures of balloon-injured Ad-GFP or Ad-VIVIT-transduced carotid artery sections that were stained for PCNA. The bar graph in the bottom panel shows the quantitative analysis of the PCNA-positive cells counted in 10 randomly selected fields of the immunostained sections of the balloon-injured Ad-GFP or Ad-VIVIT-transduced arteries. B, all treatment conditions were the same as in A, except that 2 weeks after BI, rats were sacrificed; arteries were isolated and fixed, and cross-sections were made and stained with hematoxylin and eosin. After morphometry analysis, the I/M ratios and lumen sizes were calculated. Top panel shows the representative pictures of the balloon-injured Ad-GFP or Ad-VIVIT-transduced carotid artery cross-sections that were stained with hematoxylin and eosin. The bar graphs in the bottom panel show the quantitative analysis of the I/M ratios and lumen sizes of the balloon-injured Ad-GFP or Ad-VIVIT-transduced rat carotid arteries. *, p < 0.05 versus GFP-BI (n = 6).
FIGURE 7.
Down-regulation of NFATc1 by its siRNA blocks PDGF-BB-induced cyclin A expression and DNA synthesis in rat VSMC. A, VSMC were transfected with scrambled (Scr) control or NFATc1 siRNA oligonucleotides, and 36 h later cell extracts were prepared, and an equal amount of protein from each condition was analyzed by Western blotting for NFATc1 levels using its specific antibodies. B and C, after transfection with scrambled control or NFATc1 siRNA oligonucleotides and quiescence, cells were treated with and without PDGF-BB (20 ng/ml) for 8 h, and either RNA or proteins were isolated and analyzed for cyclin A mRNA (B) and protein (C) levels by RT-PCR and Western blot analysis, respectively, using its specific primers and antibodies. D, all the conditions were the same as in B except that after quiescence cells were treated with and without PDGF-BB (20 ng/ml) for 24 h, and DNA synthesis was measured by [3H]thymidine incorporation. *, p < 0.01 versus TR; †, p < 0.01 versus TR + PDGF-BB treatment alone.
FIGURE 8.
Down-regulation of NFATc1 by its siRNA blocks PDGF-BB-induced cyclin A expression and DNA synthesis in human VSMC. A, VSMC were transfected with scrambled control or NFATc1 siRNA oligonucleotides, and 36 h later cell extracts were prepared, and an equal amount of protein from each condition was analyzed by Western blotting for NFATc1 levels using its specific antibodies. The blot was reprobed with anti-NFATc3 antibodies for specificity. B and C, after transfection with scrambled control or NFATc1 siRNA oligonucleotides and quiescence, cells were treated with and without PDGF-BB (20 ng/ml) for 8 h, and either RNA or proteins were isolated and analyzed for cyclin A mRNA (B) and protein (C) levels by RT-PCR and Western blot analysis, respectively, using its specific primers and antibodies. D, all the conditions were the same as in B except that after quiescence cells were treated with and without PDGF-BB (20 ng/ml) for 24 h, and DNA synthesis was measured by [3H]thymidine incorporation. *, p < 0.01 versus TR; †, p < 0.01 versus TR + PDGF-BB treatment alone.
DISCUSSION
The major observations of the present study are as follows. 1) PDGF-BB induced the expression of cyclin A resulting in increased CDK2 activity in VSMC. 2) Inhibition of NFAT activation signaling by both pharmacological and genetic approaches suppressed PDGF-BB-induced cyclin A expression and CDK2 activity in VSMC. 3) Interference with NFAT activation signaling also blocked the progression of VSMC from G1 to S phase of the cell cycle. 4) Cloning and bioinformatic analysis of rat cyclin A promoter revealed the presence of NFAT-binding elements and PDGF-BB-induced NFAT binding activity to promoter elements in CsA- and VIVIT-sensitive manner. 5) ChIP analysis revealed that NFATc1 binds to cyclin A promoter in cells in a time-dependent and NFAT activation signaling-dependent manner in response to PDGF-BB. 6) Interference with NFAT activation by CsA and VIVIT also inhibited PDGF-BB-induced NFAT-binding site containing 700 bp cyclin A promoter-driven luciferase reporter gene activity in VSMC. 7) BI of rat carotid artery induced cyclin A expression and its partner CDK2 activity, resulting in increased SMC proliferation, and these responses were completely suppressed by adenovirus-mediated transduction of VIVIT. 8) BI-induced substantial neointima formation at 2 weeks of post-BI and adenovirus-mediated transduction of VIVIT attenuated this effect by about 40%. 9) Down-regulation of NFATc1 via its siRNA inhibited PDGF-BB-induced cyclin A expression and DNA synthesis both in RASMC and HASMC. Together, these observations provide mechanistic evidence for the role of NFATs, specifically NFATc1, in injury-induced vascular wall remodeling.
Previous studies from our laboratory as well as others have shown that activation of NFATs is required for VSMC proliferation (39, 42, 48). The studies from our laboratory also showed a role for NFATs in the regulation of VSMC migration (45). Toward understanding the mechanisms of NFATs in the regulation of cell migration, we reported that interleukin-6 is one of the downstream effector molecules of NFATs in mediating both RTK and G-protein-coupled receptor agonist-induced VSMC migration (45). In this study, we identified cyclin A, a cell cycle-dependent molecule, as a downstream effector of NFATs in mediating VSMC proliferation in response to a potent vascular mitogen, PDGF-BB. The observations that VIVIT blocks BI-induced expression of cyclin A and its partner CDK2 activity in rat carotid arteries further support the role of NFATs in the regulation of cell cycle genes even in vivo. In addition, the decreased cyclin A/CDK2 activity by VIVIT in balloon-injured arteries correlated with diminished DNA synthesis as measured by a reduction in the expression of PCNA, a cofactor for DNA polymerases (49). The decreased PCNA levels in balloon-injured and VIVIT-transduced arteries further correlated with a reduction in SMC number and neointima formation. The finding that BI-induced increases in cyclin A expression occur in SMC as revealed by co-localization of cyclin A with smooth muscle α-actin and that inhibition of NFAT activation signaling by adenovirus-mediated expression of VIVIT suppressed this co-localization further strengthens the role of NFATs in the regulation of VSMC growth in vivo as well. Because ChIP analysis revealed that NFATc1 binds to cyclin A promoter in VSMC in response to PDGF-BB, and siRNA-mediated down-regulation of NFATc1 suppressed PDGF-BB-induced cyclin A expression and DNA synthesis both in rat and human VSMC, it is likely that this member of the NFAT family of transcriptional factors is involved in the regulation of VSMC proliferation. In this regard, it should be pointed out that forced expression of a constitutively active NFATc1 in murine 3T3-L1 fibroblasts leads to their transformation (41). NFATc1 has also been shown to play a role in vascular endothelial growth factor-induced endothelial cell proliferation (40). Furthermore, conditional expression of NFATc1 enhanced pancreatic β-cell proliferation (43). In addition to these observations, disruption of NFATc1 led to impaired myocardial development with defects in valve formation (36, 37). Despite the role of NFATs in the regulation of growth and development, particularly of cardiovascular systems, the underlying mechanisms are not known. In this regard, this study provides a direct evidence for the role of NFATs in the regulation of cyclin A, which plays a crucial role in the regulation of cell proliferation. A large body of data indicates that cyclin A/CDK2 is involved in the progression of a cell through the S phase of the cell cycle (18, 19). In this aspect, the involvement of NFATs in the regulation of cyclin A/CDK2 activity tightly links their capacity to regulation of cell proliferation. However, one study showed that NFATc2-/- lymphocytes grow faster than wild type cells and express higher levels of cyclins (50). NFATc2 has also been shown to repress cyclin A and CDK4 expression in T cells (51, 52). These observations led to the assumption that NFATs negatively regulate cyclins/CDKs and thereby cell proliferation. However, based on these observations as well as the present findings, it appears that different members of the NFAT family of transcription factors are involved in differential regulation of cyclins, with NFATc1 and -c2 as positive and negative regulators, respectively. Recently, it was reported that constitutive expression of cyclin A in the myocardium leads to a regenerative response after infarction and prevents heart failure (53). Furthermore, a combinatorial loss of CDK4 and CDK2 led to embryonic lethality with heart defects (54). Because the knock out of NFATc1 led to defects in cardiac development (36, 37) and the present findings revealed that NFATs modulate the expression of cyclin A in VSMC, it is possible that the NFAT-cyclin A signaling axis may be an important player in the development of the cardiovascular system.
It was reported that cyclin A is a target gene for a number of transcription factors, including AP-1 (activator protein-1) and ATF-2 (activating transcription factor-2) in mediating growth in many cell types in response to various growth factors (55, 56). Many studies showed that NFATs interact with other transcriptional factors such as AP-1, EGR-1 (early growth response protein-1), and Sp1 (stimulating protein 1) in the regulation of cytokine and cell division kinase inhibitor genes (57–59). Although electrophoretic mobility shift assay and ChIP analysis revealed that NFATc1 binds to cyclin A promoter in VSMC, it is quite possible that this interaction could be mediated via a mechanism involving its interaction with other transcription factors such Sp1 or ATF-2 whose binding sites are also present in the cyclin A promoter region. Future studies are required to address whether NFATc1 interacts with these transcription factors in the regulation of cyclin A gene in mediating PDGF-BB-induced VSMC proliferation.
In summary, these results demonstrate for the first time that NFATs, particularly NFATc1, target cyclin A/CDK2 in the regulation of VSMC multiplication and thereby in restenosis. In view of its role in VSMC migration and proliferation, NFATc1 may be a potential target for the development of anti-restenotic agents.
This work was supported, in whole or in part, by National Institutes of Health Grant HL069908 (to G. N. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CDK, cyclin-dependent kinase; VSMC, vascular smooth muscle cell; SMC, smooth muscle cell; PDGF-BB, platelet-derived growth factor BB; siRNA, small interfering RNA; ChIP, chromatin immunoprecipitation; SMC, smooth muscle cell; Rb, retinoblastoma protein; PCNA, proliferating cell nuclear antigen; Ad-GFP, adenovirus-green fluorescent protein; RT, reverse transcription; FACS, fluorescence-activated cell sorter; HASMC, human aortic smooth muscle cell; HBSS, Hanks' balanced salt solution; m.o.i., multiplicity of infection; TRITC, tetramethylrhodamine isothiocyanate; NFAT, nuclear factors of activated T cell; CsA, cyclosporin A; BI, balloon injury; CsA, cyclosporin A; RTK, receptor tyrosine kinase; SMα-actin, smooth muscle α-actin.
References
- 1.Pavletich, N. P. (1999) J. Mol. Biol. 287 821-828 [DOI] [PubMed] [Google Scholar]
- 2.Morgan, D. O. (1995) Nature 374 131-134 [DOI] [PubMed] [Google Scholar]
- 3.Nigg, E. A. (1995) BioEssays 17 471-480 [DOI] [PubMed] [Google Scholar]
- 4.Ohtsubo, M., and Roberts, J. M. (1993) Science 259 1908-1912 [DOI] [PubMed] [Google Scholar]
- 5.Sherr, C. J. (1993) Cell 73 1059-1065 [DOI] [PubMed] [Google Scholar]
- 6.Harper, J. W., Adami, G. R., Wei, N., Keyomarsi, K., and Elledge, S. J. (1993) Cell 75 805-816 [DOI] [PubMed] [Google Scholar]
- 7.Sherr, C. J., and Roberts, J. M. (1995) Genes Dev. 9 1149-1163 [DOI] [PubMed] [Google Scholar]
- 8.Xiang, Y., Hannon, G. J., Zhang, H., Casso, D., Kobayashi, R., and Beach, D. (1993) Nature 366 701-704 [DOI] [PubMed] [Google Scholar]
- 9.Hengst, L., and Reed, S. (1998) Genes Dev. 12 3882-3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waga, S., Hannon, G. J., Beach, D., and Stillman, B. (1994) Nature 369 574-578 [DOI] [PubMed] [Google Scholar]
- 11.Kato, J. Y., Matsuoka, M., Polyak, K., Massauge, J., and Sherr, C. J. (1994) Cell 79 487-496 [DOI] [PubMed] [Google Scholar]
- 12.Toyoshima, H., and Hunter, T. (1994) Cell 78 67-74 [DOI] [PubMed] [Google Scholar]
- 13.Kato, J. Y., Matsushime, H., Hiebert, S. W., Ewen, M. E., and Sherr, C. J. (1993) Genes Dev. 7 331-342 [DOI] [PubMed] [Google Scholar]
- 14.Koff, A., Giordano, A., Desai, D., Yamashita, K., Harper, J. W., Elledge, S., Nishimoto, T., Morgan, D. O., Franza, B. R., and Roberts, J. M. (1992) Science 257 1689-1694 [DOI] [PubMed] [Google Scholar]
- 15.Sunters, A., Thomas, D. P., Yeudall, W. A., and Grigoriadis, A. E. (2004) J. Biol. Chem. 279 9882-9891 [DOI] [PubMed] [Google Scholar]
- 16.Weinberg, R. A. (1995) Cell 81 323-330 [DOI] [PubMed] [Google Scholar]
- 17.Johnson, D. G., Schwarz, J. K., Cress, W. D., and Nevins, J. R. (1993) Nature 365 349-352 [DOI] [PubMed] [Google Scholar]
- 18.Nurse, P. (1994) Cell 79 547-550 [DOI] [PubMed] [Google Scholar]
- 19.Sever-Chroneos, Z., Angus, S. P., Fribourg, A. F., Wan, H., Todorov, I., Knudsen, K. E., and Knudsen, E. S. (2001) Mol. Cell. Biol. 21 4032-4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang, R., Morosetti, R., and Koeffler, H. P. (1997) Cancer Res. 57 913-920 [PubMed] [Google Scholar]
- 21.Ravnik, S. E., and Wolgemuth, D. J. (1999) Dev. Biol. 207 408-418 [DOI] [PubMed] [Google Scholar]
- 22.Murray, A. W. (2004) Cell 16 221-234 [DOI] [PubMed] [Google Scholar]
- 23.Sherr, C. J., and Roberts, J. M. (2004) Genes Dev. 18 2699-2711 [DOI] [PubMed] [Google Scholar]
- 24.Kozar, K., Ciemerych, M. A., Rebel, V. I., Shigematsu, H., Zagozdzon, A., Sicinska, E., Geng, Y., Yu, Q., Bhattacharya, S., Bronson, R. T., Akashi, K., and Sicinski, P. (2004) Cell 118 477-491 [DOI] [PubMed] [Google Scholar]
- 25.Chen, W., Lee, J., Cho, S. Y., and Fine, H. A. (2004) Cancer Res. 64 3949-3957 [DOI] [PubMed] [Google Scholar]
- 26.Gondeau, C., Gerbal-Chaloin, S., Bello, P., Aldrian-Herrada, G., Morris, M. C., and Divita, G. (2005) J. Biol. Chem. 280 13793-13800 [DOI] [PubMed] [Google Scholar]
- 27.Chang, M. W., Barr, E., Lu, M. M., Barton, K., and Leiden, J. M. (1995) J. Clin. Investig. 96 2260-2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, Z. Y., Simari, R. D., Perkins, N. D., San, H., Gordon, D., Nabel, G. J., and Nabel, E. G. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 7905-7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, D., Krasinski, K., Chen, D., Sylvester, A., Chen, J., Nisen, P. D., and Andres, V. (1997) J. Clin. Investig. 99 2334-2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan, P. G., Chen, L., Nardone, J., and Rao, A. (2003) Genes Dev. 17 2205-2232 [DOI] [PubMed] [Google Scholar]
- 31.Boise, L. H., Petryniak, B., Mao, X., June, C. H., Wang, C. Y., Lindsten, T., Bravo, R., Kovary, K., Leiden, J. M., and Thompson, C. B. (1993) Mol. Cell. Biol. 13 1911-1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molkentin, J. D., Lu, J. R., Antos, C. L., Markham, B., Richardson, J., Robbins, J., Grant, S. R., and Olson, E. N. (1998) Cell 93 215-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boss, V., Abbott, K. L., Wang, X. F., Pavlath, G. K., and Murphy, T. J. (1998) J. Biol. Chem. 273 19664-19671 [DOI] [PubMed] [Google Scholar]
- 34.Horsley, V., Friday, B. B., Matteson, S., Kegley, K. M., Gephart, J., and Pavlath, G. K. (2001) J. Cell Biol. 153 329-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishida, N., Hayashi, K., Hoshijima, M., Ogawa, T., Koga, S., Miyatake, Y., Kumegawa, M., Kimura, T., and Takeya, T. (2002) J. Biol. Chem. 277 41147-41156 [DOI] [PubMed] [Google Scholar]
- 36.de la Pompa, J. L., Timmerman, L. A., Takimoto, H., Yoshida, H., Elia, A. J., Samper, E., Potter, J., Wakeham, A., Marengere, L., Langile, B. L., Crabtree, G. R., and Mak, T. W. (1998) Nature 392 182-186 [DOI] [PubMed] [Google Scholar]
- 37.Ranger, A. M., Grusby, M. J., Hodge, M. R., Gravallese, E. M., de la Brousse, C. F., Hoet, T., Mickanin, C., Baldwin, H. S., and Glimcher, L. H. (1998) Nature 392 186-190 [DOI] [PubMed] [Google Scholar]
- 38.Graef, I. A., Chen, F., Chen, L., Kuo, A., and Crabtree, G. R. (2001) Cell 105 863-875 [DOI] [PubMed] [Google Scholar]
- 39.Yellaturu, C. R., Ghosh, S. K., Rao, R. K., Jennings, L. K., Hassid, A., and Rao, G. N. (2002) Biochem. J. 368 183-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson, E. N., Lee, Y. M., Sander, T. L., Rabkin, E., Schoen, F. J., Kaushal, S., and Bischoff, J. (2003) J. Biol. Chem. 278 1686-1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neal, J. W., and Clipstone, N. A. (2003) J. Biol. Chem. 278 17246-17254 [DOI] [PubMed] [Google Scholar]
- 42.Liu, Z., Zhang, C., Dronadula, N., Li, Q., and Rao, G. N. (2005) J. Biol. Chem. 280 14700-14708 [DOI] [PubMed] [Google Scholar]
- 43.Heit, J. J., Apelqvist, A. A., Gu, X., Winslow, M. M., Neilson, J. R., Crabtree, G. R., and Kim, S. K. (2006) Nature 443 345-349 [DOI] [PubMed] [Google Scholar]
- 44.Jauliac, S., Lopez-Rodriguez, C., Shaw, L. M., Brown, L. F., Rao, A., and Toker, A. (2002) Nat. Cell Biol. 4 540-544 [DOI] [PubMed] [Google Scholar]
- 45.Liu, Z., Dronadula, N., and Rao, G. N. (2004) J. Biol. Chem. 279 41218-41226 [DOI] [PubMed] [Google Scholar]
- 46.Shimizu, M., Nomura, Y., Suzuki, H., Ichikawa, E., Takeuchi, A., Suzuki, M., Nakamura, T., Nakajima, T., and Oda, K. (1998) Exp. Cell Res. 239 93-103 [DOI] [PubMed] [Google Scholar]
- 47.Aramburu, J., Yaffe, M. B., Lopez-Rodriguez, C., Cantley, L. C., Hogan, P. G., and Rao, A. (1999) Science 285 2129-2133 [DOI] [PubMed] [Google Scholar]
- 48.Yu, H., Sliedregt-Bol, K., Overkleeft, H., van der Marel, G. A., van Berkel, T. J., and Biessen, E. A. (2006) Arterioscler. Thromb. Vasc. Biol. 26 1531-1537 [DOI] [PubMed] [Google Scholar]
- 49.Moldovan, G. L., Pfander, B., and Jentsch, S. (2007) Cell 129 665-679 [DOI] [PubMed] [Google Scholar]
- 50.Caetano, M. S., Vieira-de-Abreu, A., Teixeira, L. K., Werneck, M. B., Barcinski, M. A., and Viola, J. P. (2002) FASEB J. 16 1940-1942 [DOI] [PubMed] [Google Scholar]
- 51.Carvalho, L. D., Teixeira, L. K., Carrossini, N., Caldeira, A. T., Ansel, K. M., Rao, A., and Viola, J. P. (2007) Cell Cycle 6 1789-1795 [DOI] [PubMed] [Google Scholar]
- 52.Baksh, S., Widlund, H. R., Frazer-Abel, A. A., Du, J., Fosmire, S., Fisher, D. E., DeCaprio, J. A., Modiano, J. F., and Burakoff, S. J. (2002) Mol. Cell 10 1071-1081 [DOI] [PubMed] [Google Scholar]
- 53.Cheng, R. K., Asai, T., Tang, H., Dashoush, N. H., Kara, R. J., Costa, K. D., Naka, Y., Wu, E. X., Wolgemuth, D. J., and Chaudhry, H. W. (2007) Circ. Res. 100 1741-1748 [DOI] [PubMed] [Google Scholar]
- 54.Berthet, C., Klarmann, K. D., Hilton, M. B., Suh, H. C., Keller, J. R., Kivokawa, H., and Kaldis, P. (2006) Dev. Cell. 10 563-573 [DOI] [PubMed] [Google Scholar]
- 55.Katabami, M., Donninger, H., Hommura, F., Leaner, V. D., Kinoshita, I., Chick, J. F., and Birrer, M. J. (2005) J. Biol. Chem. 280 16728-16738 [DOI] [PubMed] [Google Scholar]
- 56.Beier, F., Taylor, A. C., and Lu Valle, P. (2000) J. Biol. Chem. 275 12948-12953 [DOI] [PubMed] [Google Scholar]
- 57.Jhonson, B. V., Bert, A. G., Rvan, G. R., Condina, A., and Cockerill, P. N. (2004) Mol. Cell. Biol. 24 7914-7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Decker, F. L., Sherka, C., and Zipfel, P. F. (1998) J. Biol. Chem. 273 26923-26930 [DOI] [PubMed] [Google Scholar]
- 59.Santini, M. P., Talora, C., Seki, T., Bolgan, L., and Dotto, G. P. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 9575-9580 [DOI] [PMC free article] [PubMed] [Google Scholar]