Abstract
Telomerase activity is primarily determined by transcriptional regulation of the catalytic subunit, human telomerase reverse transcriptase (hTERT). Several mRNA splice variants for hTERT have been identified, but it is not clear if telomerase activity is determined by the absolute or relative levels of full-length (functional) and variant hTERT transcripts. We have developed an SYBR green-based reverse transcription-quantitative polymerase chain reaction assay for the enumeration of the four common hTERT mRNA variants and correlated these with telomerase activity and telomere length in 24 human melanoma cell lines. All except five of the lines expressed four hTERT transcripts, with an overall significant level of co-occurrence between absolute mRNA levels of full-length α+/β+ hTERT and the three splice variants α-/β+, α+/β-, and α-/β-. On average, α+/β+ made up the majority (48.1%) of transcripts, followed by α+/β- (44.6%), α-/β- (4.4%), and α-/β+ (2.9%). Telomerase activity ranged from 1 to 247 relative telomerase activity and correlated most strongly with the absolute amount of α+/β+ (R = 0.791, P = .000004) and the relative amount of α+/β- (R = -0.465, P = .022). This study shows that telomerase activity in melanoma cells is best determined by the absolute expression of full-length hTERT mRNA and indicates a role for the hTERT β deletion variant in the negative regulation of enzyme activity.
Introduction
Telomerase is a ribonucleoprotein complex that maintains chromosome length by synthesizing and adding repetitive (TTAGGG)n DNA sequences to the ends of telomeres [1]. Its absence from most normal somatic cells is believed to contribute to eventual senescence and limited cellular life span [2], whereas its reactivation in immortalized cells has been associated with the unlimited growth potential required for malignancy [3–5]. In particular, progression of melanoma is known to be accompanied by a steady increase in telomerase activity during the transformation of isolated naevi to metastatic disease [6,7], with virtually all melanoma cell lines exhibiting some degree of telomerase activity [6]. These discriminating properties have made telomerase an attractive target for cancer therapy [8], and there has been much emphasis on methods for detecting and determining the regulatory mechanisms of this important enzyme.
Telomerase is made up of two essential components: a constitutively expressed human telomerase RNA (hTR), which acts as a transcription template [9,10], and a catalytic human telomerase reverse transcriptase (hTERT), whose expression controls enzymatic activity [11,12]. The transcriptional and posttranscriptional regulation of hTERT is complex and remains to be fully elucidated. Studies in human development have revealed the presence of multiple hTERT RNA transcripts occurring in patterns that are both tissue-specific and gestational stage-dependent [13]. Such nonrandom alternative splicing is a common method of genetic regulation in eukaryotes [14], and to date, 10 different splice variants of hTERT have been identified [13,15–17]. The most widely studied variants involve splicing at two main sites: the α splice site, which produces a 36-bp inframe deletion within the conserved reverse transcript motif A; and the β site, which results in a 183-bp deletion and non-sense mutation that truncates the protein, effectively deleting the remaining three reverse transcriptase motifs [13,18]. Splicing at either site can occur independently or in combination to produce three variants from the full-length α+/β+: α-/β+, α+/β-, and α-/β-, which have been shown to occur at approximate proportions of 5%, 1%, 80% to 90%, and 5% to 15%, respectively, within various cancer cell lines [19]. To date, only the α-/β+ variant has been shown to exhibit any regulatory function, acting as a dominant-negative inhibitor of telomerase activity when overexpressed in either normal or tumor cells [20,21]. It is unclear whether the ratio of full length to spliced hTERT is important in determining telomerase activity [22], because some studies have shown that the absolute expression of hTERT is well correlated with telomerase activity [23–27], and still others have found no correlation with either relative or absolute amounts of variant transcripts [28]. The regulatory functions of various hTERT transcripts may well be cell type-specific; however, the many different methods used to quantify hTERT mRNA have made it difficult to interpret these findings.
Most reverse transcription-polymerase chain reaction (RT-PCR)-based assays for hTERT variants use primers that flank the α and β subunits and thus amplify all transcripts in one reaction (Figure 1). The products are then scanned by densitometry to give the relative distribution of hTERT variants in each sample [19,29,30], the results of which are largely biased by the competitive nature of PCR. A few techniques have been developed using dual-labeled probe technology on a real-time PCR platform [31–35]; however, most of these assays use primers designed to the region downstream from the α and β subunits and, therefore, do not discriminate between the deletion variants [31–33]. A commercially available Light Cycler kit (TeloTAGGGG hTERT Quantification Kit; Roche Diagnostics, Basel, Switzerland) has proven popular, but the exact primer sequences are not revealed, thus it is not clear which transcripts are measured. Therefore, results from these types of studies need to be interpreted with caution.
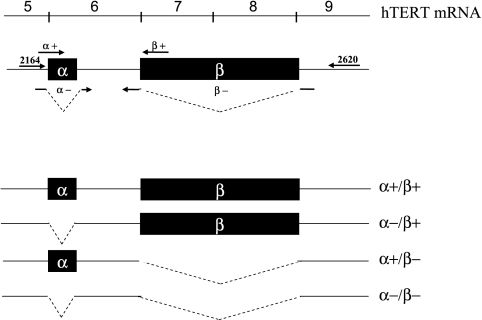
Figure 1.
hTERT mRNA α and β deletion variants. Relevant hTERT exons are numbered and corresponding location of α and β sequences are illustrated by shaded boxes. Deleted sequences are designated by dashed lines. Locations of PCR primers are indicated by arrows and labeled according to the notation used in Tables 1 and 2.
The aim of the present study was to establish whether absolute or relative levels of hTERT variant transcripts determine telomerase activity. We developed an SYBR green-based real-time PCR assay as a more affordable option to labeled probes, with the further advantage of enabling melt curves to confirm the presence of specific transcripts. The assay was then used to determine the absolute and relative hTERT variant expression in a series of melanoma cell lines. In doing so, we have revealed the importance of overall levels of full-length hTERT mRNA in determining telomerase expression, and a possible role for the relative amount of β deletion variant in the regulation of telomerase activity.
Materials and Methods
Cell Lines
A total of 24 human melanoma cell lines were used for this study. ME4405 was a kind gift from Dr. Parmiani (Milan, Italy). MM200 was supplied by Dr. Parsons (Queensland Institute of Medical Research, Brisbane, Australia), and IGR3 was provided by Dr. Hope (Institute of Medical and Veterinary Science, Adelaide, Australia). Clones of the latter two lines along with the remaining lines, which were developed from primary melanoma tissue biopsies after receiving informed patient consent, were kindly provided by the New-castle Melanoma Unit, Calvary Mater Newcastle. HL60 was obtained from the American Type Culture Collection (Manassas, VA) for use as a control. All melanoma cells were cultured in DMEM (JRH Biosciences, Australia), and HL60 was maintained in RPMI 1640 (JRH Biosciences), supplemented with 10% fetal bovine serum at 37°C and 5% CO2.
Telomerase Activity
Telomerase activity was determined using the Telo TAGGG Telomerase PCR ELISAplus kit (Roche, Mannheim, Germany), which is based on the Telomeric Repeat Amplification Protocol assay with a nonradioactive ELISA detection. The procedure was performed on 3 ng of total cell protein in accordance with the manufacturer's instructions and results normalized to the cell line MM200 C3E8. All assays were performed singly and repeated on three separate occasions.
Measurement of Mean Telomere Length
The mean telomere length of each cell line was determined using Southern blot techniques as previously reported [36,37]. Briefly, genomic DNA was isolated and digested with restriction enzymes, RsaI and MspI (Amersham Pharmacia Biotech, Uppsala, Sweden), electrophoresed through a 0.6% agarose gel and transferred to Hybond N+ membrane (Amersham Pharmacia Biotech). A fluorescein-labeled oligonucleotide probe (CCCTAA)3 was hybridized to the membrane and detected using enhanced chemiluminescent detection (Amersham Pharmacia Biotech). Kodak Digital Science documentation software was used to determine the net intensity at 1-kbp intervals along the length of the telomere smear. The mean telomere length was calculated as: ∑(MWi x NIi)/∑(NIi), where MWi = molecular weight (kbp) at interval i and NIi = net intensity (pixels) at interval i [38]. Assessment of the accuracy of the methodology showed a mean ± SE within-day variation of telomere length of 6 ± 1% and between-day variation of 9 ± 2% [36]. For each sample, the average of three mean telomere lengths calculated from three separate blots was used for statistical analysis.
Polymerase Chain Reaction
The full-length hTERT and the three splice variants were amplified from the HL60 cell line by PCR using previously reported primers and methods [16,39]. HL60 was used for optimization because it has been shown to express all four hTERT variants [29]. Products were resolved by agarose gel electrophoresis and identified based on size (Table 1). Bands were extracted from the gel and were purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI). These fragments were used to establish specificity and sensitivity of the qPCR method.
Table 1.
Primers and Expected PCR Products for Amplification of hTERT Splice Variants.
| Forward Primer | Reverse Primer | Annealing Temperature (°C) | Possible Amplicons (bp) |
| 2164 | 2620 | 50–65 (gradient) | α+/β+ = 457 |
| α-/β+ = 421 | |||
| α+/β- = 275 | |||
| α-/β- = 239 | |||
| α+ | β+ | 53 | α+/β+ = 202 |
| α- | β+ | 53 | α-/β+ = 172 |
| α+ | β- | 53 | α+/β- = 189 |
| α- | β- | 58 | α-/β- = 159 |
Establishment of the qPCR Assay
Purified PCR products of each variant were used to optimize the qPCR cycling conditions. Specific primers were designed to the boundary spanning regions of the α and β subunits of hTERT, because this has been shown to be an effective way to detect splice variants without the use of probes [40]. These primers were combined with previously reported primers designed to hybridize to the α (hTERT 2172 [39]) and β subunits (variation of hTERT 2350 [39]) and thus only amplify variants containing these sequences (Table 2). Standard curves were generated, and the minimal detection limit was determined to be one copy per reaction. The specificity of the assay was confirmed by testing each primer pair against the purified PCR products for all variants, and in each case, the primers amplified only the splice variant to which they had been designed, with minimal cross-reactivity.
Table 2.
Primers Used for hTERT and GAPDH Amplification.
| Primer | Orientation | Sequence | Reference |
| 2164 | Sense | gcctgagctgtactttgtcaa | Krams et al. [39] |
| 2620 | Antisense | cgcaaacagcttgttctccatgtc | Krams et al. [39] |
| α+ | Sense | tgtactttgtcaaggtggatgtg | Krams et al. [39] |
| α- | Sense | ctgagctgtactttgtcaaggac | Lincz et al. (this study) |
| β+ | Antisense | gtacggctggaggtctgtcaa | Variation of [39] |
| β- | Antisense | ggcactggacgtaggacgtgg | Lincz et al. (this study) |
| GAPDHA.69 f | Sense | ctctctgctcctcctgttcgac | Carraro et al. [54] |
| GAPDHA.69 r | Antisense | tgagcgatgtggctcggct | Carraro et al. [54] |
Reverse Transcription-Quantitative Polymerase Chain Reaction
Lysates prepared for telomerase activity analysis (previously mentioned) were pooled for each cell line and RNA extracted using the Aurum Total RNA Mini Kit (Bio-Rad, Hercules, CA) and quantified using the RiboGreen RNA Quantification Kit (Molecular Probes, Eugene, OR) according to the manufacturer's instructions. Reverse transcription reactions were standardized to the lowest RNA concentration, so that equivalent amounts of RNA (112 ng) from each cell line were synthesized into cDNA using Superscript III first-strand cDNA synthesis kit (Invitrogen, Carlsbad, CA). The cDNA equivalent of 5.6 ng of RNA was then amplified by qPCR in 20-µl reactions using Platinum SYBR Green qPCR Supermix-UDG (Invitrogen) with 10 µmol of each appropriate primer. Reactions were run on a Rotor-Gene 3000 (Corbett Life Science, Sydney, Australia) using a four-step cycling program that consisted of the following: 50°C for 2 minutes, 95°C for 2 minutes followed by 60 cycles of 95°C for 10 seconds, 53°C or 58°C for 15 seconds, 72°C for 20 seconds, and data acquisition on the FAM/SYBR channel at 80°C for 10 seconds. This final acquisition was set to eliminate background fluorescence generated below this temperature. For GAPDH, a two-step cycling program was used: 50°C for 2 minutes, 95°C for 2 minutes followed by 60 cycles of 95°C for 10 seconds, and 60°C for 1 minute with data acquisition on the FAM/SYBR channel. A melt curve (57–95°C) was generated at the end of each run to verify specificity.
Data Analysis
Each reaction was performed in duplicate on three separate occasions. Raw fluorescence values were exported and analyzed by Data Analysis for Real-Time PCR, a freely available excel file containing an algorithm that calculates the amplification efficiency of each sample from its amplification profile [41]. This has been validated and found to give comparable results to other methods of quantitation without the need for standard curves [41]. We used the average amplification efficiency for our calculations, because this has been shown to give the most accurate results [42]. Outliers were excluded and the threshold was set based on the mean midpoint (M) of the transformed signal range. Expression values (in arbitrary fluorescence units) were obtained for each hTERT transcript and normalized by dividing them by individual expression values for GAPDH to produce R0. The mean of three experiments was determined for each transcript and expressed as a percentage of the reference cell line, MM200 C3E8 (absolute expression) or as a percentage of the total number of hTERT transcripts for each cell line (relative expression). Spearman's rank and Pearson's correlation coefficients were calculated to assess linear relationships among telomere length, telomerase activity, and hTERT mRNA expression. P values < .05 were considered statistically significant.
Results
The results are summarized in Table 3. Mean telomere length of the cell lines ranged from 2.19 to 7.37 kbp, with an overall average of 3.97 kbp. Telomerase activity was detected in all cells and expressed as a percentage of the MM200 C3E8 reference cell line that was arbitrarily set as 100. Relative telomerase activity (RTA) varied widely (1–247 RTA), with a mean of 112. Similarly, the absolute expression of hTERT mRNA transcripts was highly variable. When the transcripts were expressed as a percentage of the total amount of hTERT mRNA in each cell line, on average, the full-length α+/β+ made up the majority (48.1%) of transcripts, followed closely by the α+/β- variant (44.6%). The α-/β- and dominant-negative α-/β+ variants made up the smallest proportion of transcripts (4.4% and 2.9%, respectively). The average ratio of full-length transcript to spliced transcripts was 1.11 (0.04–2.26).
Table 3.
Summary of Results for 24 Melanoma Cell Lines.
| Analysis | Mean | Range | |
| Telomere length (kbp)* | 3.97 | 2.19–7.37 | |
| RTA (% of reference cell line) | 112 | 1–247 | |
| Absolute hTERT transcript mRNA | α+/β+ | 116 | 3–430 |
| (% of reference cell line) | |||
| R0 | 6.1 x 10-4 | 1.6 x 10-5 to 2.2 x 10-3 | |
| α-/β+ | 115 | 0–520 | |
| R0 | 4.0 x 10-5 | 0.0 to 1.9 x 10-4 | |
| α+/β- | 96 | 15–247 | |
| R0 | 4.3 x 10-4 | 7.3 x. 10-5 to 1.6 x 10-3 | |
| α-/β- | 60 | 0–271 | |
| R0 | 7.7 x 10-5 | 0.0 to 7.9 x 10-4 | |
| Relative hTERT transcript mRNA | α+/β+ | 48.1 | 3.94–69.3 |
| (% of all transcripts) | α-/β+ | 2.9 | 0.0–8.2 |
| α+/β- | 44.6 | 21.9–96.1 | |
| α-/β- | 4.4 | 0.0–21.8 | |
| Ratio α+/β+:α-/β+, α+/β-, α-/β- | 1.11 | 0.04–2.26 |
n = 22.
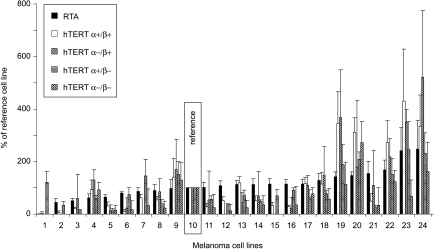
Individually, the full-length hTERT α+/β+ transcript and its α+/β- variant were detected in all cell lines (Figure 2). Only six cell lines did not express all of the variants; of these, three did not express either of the α deletion variants α-/β+ and α-/β-, two lacked only the α-/β+ transcript, whereas only one failed to express the double-deletion variant α-/β-.
Figure 2.
RTA and absolute expression of hTERT mRNA transcripts in melanoma cell lines. Levels of RTA and each variant mRNA are expressed as a percentage of the reference cell line, MM200 C3E8, arbitrarily designated as 100%, and error bars indicate SDs between three separate experiments.
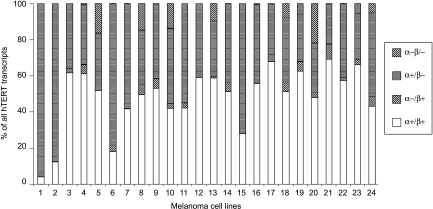
There was high disparity in the relative proportions of hTERT mRNA variants among different cell lines (Figure 3). The full-length transcript was predominantly expressed in most cell lines, whereas the α+/β- was the main transcript in six lines.
Figure 3.
Relative expression of hTERT mRNA transcripts in melanoma cell lines. Levels of each variant mRNA are expressed as a percentage of the total four hTERT transcripts per cell line.
Absolute Expression of the Full-length hTERT Transcript Is the Best Predictor of Telomerase Activity
The absolute level of each individual hTERT variant was positively correlated with telomerase activity, with the full-length message showing the strongest association (R = 0.791, P = .000004; Table 4). The relative amount of full-length hTERT was also positively correlated with telomerase activity, although this was not statistically significant (R = 0.403, P = .051), whereas the ratio of full-length hTERT to variant hTERT was significantly correlated with telomerase activity (R = 0.414, P = .044). The relative expression of α+/β- was negatively correlated with telomerase activity (R = .0.465, P = .022), whereas there was no significant correlation between telomerase activity and relative expression of either of the α deletion variants (α-/β+, α-/β-).
Table 4.
Correlations with Absolute and Relative Expression of hTERT Variants.
| Telomere Length | RTA | Absolute | ||||
| α+/β+ | α-/β+ | α+/β- | ||||
| Absolute | α+/β+ | -0.266 (0.231) | 0.791 (0.000) | |||
| α-/β+ | -0.231 (0.299) | 0.603 (0.002) | 0.731 (0.000) | |||
| α+/β- | -0.276 (0.214) | 0.535 (0.007) | 0.559 (0.004) | 0.309 (0.141) | ||
| α-/β- | -0.257 (0.249) | 0.614 (0.001) | 0.825 (0.000) | 0.749 (0.000) | 0.547 (0.006) | |
| Relative | ||||||
| α+/β+ | α-/β+ | α+/β- | ||||
| Relative | α+/β+ | -0.059 (0.793) | 0.403 (0.051) | |||
| α-/β+ | -0.247 (0.268) | 0.371 (0.074) | 0.389 (0.061) | |||
| α+/β- | 0.143 (0.526) | -0.465 (0.022) | -0.810 (0.000) | -0.479 (0.018) | ||
| α-/β- | -0.232 (0.298) | 0.320 (0.127) | 0.218 (0.305) | 0.411 (0.046) | -0.632 (0.001) | |
| Ratio | α+/β+: | |||||
| α-/β+, | 0.047 (0.837) | 0.414 (0.044) | ||||
| α+/β-, | ||||||
| α-/β- | ||||||
| RTA | -0.233 (0.297) | |||||
Numbers given are correlation coefficients with associated P values italicized in parenthesis. Statistically significant values are highlighted in bold.
A High Level of Co-occurrence Exists between hTERT Variants
Despite some cell lines completely lacking expression of particular variants, there was an overall significant level of co-occurrence between absolute mRNA levels of the full length hTERT α+/β+ transcript and the three splice variants α-/β+ (R = 0.731), α+/β- (R = 0.559), and α-/β- (R = 0.825). Between variants, there was correlation between absolute expression of all hTERT transcripts except for α-/β+ and α+/β-. When expressed as relative proportions of hTERT, the amount of α+/β- variant was significantly inversely related to that of all other transcripts.
Telomere Length Was Not Correlated to Telomerase Activity
Telomere length was not correlated to telomerase activity (R = -0.233, P = .297) or to absolute or relative expression of any of the individual hTERT transcripts.
Discussion
The only comprehensive method of hTERT splice variant quantification to emerge so far has been a technique developed by Ohyashiki et al. [35], which uses TaqMan primer/probe sets to determine both absolute and relative transcript numbers for each of the four main hTERT variants. A similar approach by Mavrogiannou et al. [34] also seems to be highly specific for each variant but perhaps not as quantitatively reliable because it does not use a reference gene. Ours is the first report of an SYBR green-based assay that can distinguish between the four hTERT variant transcripts, providing a more economical option for such studies.
Unlike results reported for other cell types, the hTERT α+/β+ variant accounted for the largest fraction of hTERTmessage in this series of melanoma cells [19,34]. Our data confirms similar results from the only other investigation of these variants in a series of 52 melanoma lesions, which also found that expression of the full-length transcript was generally equal or slightly higher than the spliced variants, with a prevalence toward expression of the β deletion variant [22]. Taken together, these results suggest that the relative proportions of variants in melanoma cells do not follow the pattern described previously for other immortalized human cells, including mammary, prostate, renal, and non-small lung carcinoma cell lines [19]. In particular, we found higher-than-expected proportions of the full-length α+/β+, similar to a recent report on 6 cell lines and 28 non-small cell lung cancer tissue samples, which found that the α+/β+ made up approximately 50% of the total transcripts in these cells [34]. These authors also used a quantitative real-time PCR analysis that was specific for each variant and attributed their findings to this more sensitive approach. Thus, it is not clear if our findings are unique to melanoma cells or simply due to the different methods used in individual studies.
We found a strong level of co-expression of all three deletion variants with the full-length transcript, suggesting that up-regulation of hTERT transcription affects all variants to a similar extent. This phenomenon has also been reported for other cell types, where copy numbers of full-length hTERT mRNA correlated with all but the α deletion variant in acute leukemia cells [35] and specifically with the β deletion variant in cirrhotic liver [25] and melanoma lesions [22]. Chromosomal imbalances are common in melanoma, and in particular, the sequences on 5p15.33, which harbor the gene for hTERT, have been found to be overrepresented in 33% of cases [43]. This could result in dysregulation of the gene and may account for the widespread overexpression of all variants. The present study shows that it is this absolute increase in transcription of hTERT mRNA that is most important for determining telomerase activity.
Since the discovery that the α deletion variant has dominantnegative regulatory influences on telomerase [20,21], some studies have focused on this variant [44]. Only one has specifically addressed the β deletion variant and found that overexpression does not cause down-regulation of telomerase activity in immortalized lung fibroblasts, prostate carcinoma, or non-small cell lung cancer cell lines [20]. It is plausible that telomerase regulation by its splice variants is cell type-specific, as suggested by the precise patterns of splicing that occur in particular cell types during development [13]. It is conceivable that these same patterns become reactivated during tumorigenesis and, if so, could have profound implications for the development of therapeutic telomerase inhibitors.
In the present study, telomere length was not related to telomerase activity or hTERT gene expression, suggesting that the maintenance of chromosome length may not be the primary function of telomerase in these neoplastic cells. This theory is supported by studies showing that inhibition of telomerase can induce apoptosis of cancer cells in a manner that is independent of both p53 and telomere shortening [45–47]. Furthermore, therapeutic induction of differentiation in some cancer cells is accompanied by a reduction in telomerase activity [48–51], and recent gene expression profiling has revealed that this is more likely to be a causative rather than consequential factor of cell maturation [52]. These alternative roles for telomerase may be regulated through the splicing of hTERT, and further investigation into thesemechanisms will require the use of sensitive quantitation methods.
Finally, it is becoming clear that alternative splicing of pre-mRNA is a common process, affecting at least 74% of human genes and effectively increasing the coding potential of the genome [53]. Missplicing of cellular genes often occurs in cancer, and deciphering the regulatory mechanisms governing splice site selection is a rapidly emerging area of research [53]. Cells such as the melanoma lines described herein should provide valuable tools to further this field of knowledge.
Acknowledgments
The authors thank Peter Hersey for providing the melanoma cell lines and the Lions Club of Cessnock for supporting the Hunter Medical Research Institute.
Abbreviations
- hTERT
human telomerase reverse transcriptase
- RTA
relative telomerase activity
- hTR
human telomerase RNA
Footnotes
This project was funded by grants from the Hunter Medical Research Institute and the Calvary Mater Newcastle. Purchase of the Rotor-Gene was made possible by a Coalfields Cancer Support Group Equipment Grant.
References
- 1.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 2.Wright WE, Shay JW. The two-stage mechanism controlling cellular senescence and immortalization. Exp Gerontol. 1992;27:383–389. doi: 10.1016/0531-5565(92)90069-c. [DOI] [PubMed] [Google Scholar]
- 3.Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JH, Meyerson M, Weinberg RA. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 4.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 5.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 6.Glaessl A, Bosserhoff AK, Buettner R, Hohenleutner U, Landthaler M, Stolz W. Increase in telomerase activity during progression of melanocytic cells from melanocytic naevi to malignant melanomas. Arch Dermatol Res. 1999;291:81–87. doi: 10.1007/s004030050387. [DOI] [PubMed] [Google Scholar]
- 7.Miracco C, Pacenti L, Santopietro R, Laurini L, Biagioli M, Luzi P. Evaluation of telomerase activity in cutaneous melanocytic proliferations. Hum Pathol. 2000;31:1018–1021. doi: 10.1053/hupa.2000.9779. [DOI] [PubMed] [Google Scholar]
- 8.Kelland L. Targeting the limitless replicative potential of cancer: the telomerase/telomere pathway. Clin Cancer Res. 2007;13:4960–4963. doi: 10.1158/1078-0432.CCR-07-0422. [DOI] [PubMed] [Google Scholar]
- 9.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 10.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 11.Nakayama J, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Ide T, Ishikawa F. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat Genet. 1998;18:65–68. doi: 10.1038/ng0198-65. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 13.Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 1998;58:4168–4172. [PubMed] [Google Scholar]
- 14.Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 15.Saeboe-Larssen S, Fossberg E, Gaudernack G. Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC Mol Biol. 2006;7:26. doi: 10.1186/1471-2199-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 17.Hisatomi H, Ohyashiki K, Ohyashiki JH, Nagao K, Kanamaru T, Hirata H, Hibi N, Tsukada Y. Expression profile of a gamma-deletion variant of the human telomerase reverse transcriptase gene. Neoplasia. 2003;5:193–197. doi: 10.1016/S1476-5586(03)80051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ulaner GA, Hu JF, Vu TH, Oruganti H, Giudice LC, Hoffman AR. Regulation of telomerase by alternate splicing of human telomerase reverse transcriptase (hTERT) in normal and neoplastic ovary, endometrium and myometrium. Int J Cancer. 2000;85:330–335. [PubMed] [Google Scholar]
- 19.Yi X, Shay JW, Wright WE. Quantitation of telomerase components and hTERTmRNA splicing patterns in immortal human cells. Nucleic Acids Res. 2001;29:4818–4825. doi: 10.1093/nar/29.23.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi X, White DM, Aisner DL, Baur JA, Wright WE, Shay JW. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia. 2000;2:433–440. doi: 10.1038/sj.neo.7900113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colgin LM, Wilkinson C, Englezou A, Kilian A, Robinson MO, Reddel RR. The hTERTalpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia. 2000;2:426–432. doi: 10.1038/sj.neo.7900112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villa R, Porta CD, Folini M, Daidone MG, Zaffaroni N. Possible regulation of telomerase activity by transcription and alternative splicing of telomerase reverse transcriptase in human melanoma. J Invest Dermatol. 2001;116:867–873. doi: 10.1046/j.1523-1747.2001.01343.x. [DOI] [PubMed] [Google Scholar]
- 23.Barclay JY, Morris AG, Nwokolo CU. HTERT mRNA partially regulates telomerase activity in gastric adenocarcinoma and adjacent normal gastric mucosa. Dig Dis Sci. 2005;50:1299–1303. doi: 10.1007/s10620-005-2776-5. [DOI] [PubMed] [Google Scholar]
- 24.Shimojima M, Komine F, Hisatomi H, Shimizu T, Moriyama M, Arakawa Y. Detection of telomerase activity, telomerase RNA component, and telomerase reverse transcriptase in human hepatocellular carcinoma. Hepatol Res. 2004;29:31–38. doi: 10.1016/j.hepres.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Kotoula V, Hytiroglou P, Pyrpasopoulou A, Saxena R, Thung SN, Papadimitriou CS. Expression of human telomerase reverse transcriptase in regenerative and precancerous lesions of cirrhotic livers. Liver. 2002;22:57–69. doi: 10.1046/j.0106-9543.2001.01594.x. [DOI] [PubMed] [Google Scholar]
- 26.Sun PM, Wei LH, Luo MY, Liu G, Wang JL, Mustea A, Konsgen D, Lichtenegger W, Sehouli J. The telomerase activity and expression of hTERT gene can serve as indicators in the anti-cancer treatment of human ovarian cancer. Eur J Obstet Gynecol Reprod Biol. 2007;130:249–257. doi: 10.1016/j.ejogrb.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 27.Satra M, Tsougos I, Papanikolaou V, Theodorou K, Kappas C, Tsezou A. Correlation between radiation-induced telomerase activity and human telomerase reverse transcriptase mRNA expression in HeLa cells. Int J Radiat Biol. 2006;82:401–409. doi: 10.1080/09553000600800090. [DOI] [PubMed] [Google Scholar]
- 28.Barclay JY, Morris A, Nwokolo CU. Telomerase, hTERT and splice variants in Barrett's oesophagus and oesophageal adenocarcinoma. Eur J Gastroenterol Hepatol. 2005;17:221–227. doi: 10.1097/00042737-200502000-00014. [DOI] [PubMed] [Google Scholar]
- 29.Liu WJ, Zhang YW, Zhang ZX, Ding J. Alternative splicing of human telomerase reverse transcriptase may not be involved in telomerase regulation during all-trans-retinoic acid-induced HL-60 cell differentiation. J Pharmacol Sci. 2004;96:106–114. doi: 10.1254/jphs.fp0030600. [DOI] [PubMed] [Google Scholar]
- 30.Zaffaroni N, Villa R, Pastorino U, Cirincione R, Incarbone M, Alloisio M, Curto M, Pilotti S, Daidone MG. Lack of telomerase activity in lung carcinoids is dependent on human telomerase reverse transcriptase transcription and alternative splicing and is associated with long telomeres. Clin Cancer Res. 2005;11:2832–2839. doi: 10.1158/1078-0432.CCR-04-1293. [DOI] [PubMed] [Google Scholar]
- 31.Yajima T, Yagihashi A, Kameshima H, Furuya D, Kobayashi D, Hirata K, Watanabe N. Establishment of quantitative reverse transcription-polymerase chain reaction assays for human telomerase-associated genes. Clin Chim Acta. 2000;290:117–127. doi: 10.1016/s0009-8981(99)00188-6. [DOI] [PubMed] [Google Scholar]
- 32.Bieche I, Nogues C, Paradis V, Olivi M, Bedossa P, Lidereau R, Vidaud M. Quantitation of hTERT gene expression in sporadic breast tumors with a real-time reverse transcription-polymerase chain reaction assay. Clin Cancer Res. 2000;6:452–459. [PubMed] [Google Scholar]
- 33.Buttitta F, Pellegrini C, Marchetti A, Gadducci A, Cosio S, Felicioni L, Barassi F, Salvatore S, Martella C, Coggi G, et al. Human telomerase reverse transcriptase mRNA expression assessed by real-time reverse transcription polymerase chain reaction predicts chemosensitivity in patients with ovarian carcinoma. J Clin Oncol. 2003;21:1320–1325. doi: 10.1200/JCO.2003.09.065. [DOI] [PubMed] [Google Scholar]
- 34.Mavrogiannou E, Strati A, Stathopoulou A, Tsaroucha EG, Kaklamanis L, Lianidou ES. Real-time RT-PCR quantification of human telomerase reverse transcriptase splice variants in tumor cell lines and non-small cell lung cancer. Clin Chem. 2007;53:53–61. doi: 10.1373/clinchem.2006.073015. [DOI] [PubMed] [Google Scholar]
- 35.Ohyashiki JH, Hisatomi H, Nagao K, Honda S, Takaku T, Zhang Y, Sashida G, Ohyashiki K. Quantitative relationship between functionally active telomerase and major telomerase components (hTERT and hTR) in acute leukaemia cells. Br J Cancer. 2005;92:1942–1947. doi: 10.1038/sj.bjc.6602546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakoff JA, De Waal E, Garg M, Denham J, Scorgie F, Enno A, Lincz LF, Ackland SP. Telomere length in haemopoietic stem cells can be determined from that of mononuclear blood cells or whole blood. Leuk Lymphoma. 2002;43:2017–2020. doi: 10.1080/1042819021000015970. [DOI] [PubMed] [Google Scholar]
- 37.Wynn RF, Cross MA, Hatton C, Will AM, Lashford LS, Dexter TM, Testa NG. Accelerated telomere shortening in young recipients of allogeneic bone-marrow transplants. Lancet. 1998;351:178–181. doi: 10.1016/S0140-6736(97)08256-1. [see comments] [DOI] [PubMed] [Google Scholar]
- 38.Vaziri H, Schachter F, Uchida I, Wei L, Zhu X, Effros R, Cohen D, Harley CB. Loss of telomeric DNA during aging of normal and trisomy 21 human lymphocytes. Am J Hum Genet. 1993;52:661–667. [PMC free article] [PubMed] [Google Scholar]
- 39.Krams M, Claviez A, Heidorn K, Krupp G, Parwaresch R, Harms D, Rudolph P. Regulation of telomerase activity by alternate splicing of human telomerase reverse transcriptase mRNA in a subset of neuroblastomas. Am J Pathol. 2001;159:1925–1932. doi: 10.1016/S0002-9440(10)63039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandenbroucke II, Vandesompele J, Paepe AD, Messiaen L. Quantification of splice variants using real-time PCR. Nucleic Acids Res. 2001;29:E68. doi: 10.1093/nar/29.13.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cikos S, Bukovska A, Koppel J. Relative quantification of mRNA: comparison of methods currently used for real-time PCR data analysis. BMC Mol Biol. 2007;8:113. doi: 10.1186/1471-2199-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pirker C, Holzmann K, Spiegl-Kreinecker S, Elbling L, Thallinger C, Pehamberger H, Micksche M, Berger W. Chromosomal imbalances in primary and metastatic melanomas: over-representation of essential telomerase genes. Melanoma Res. 2003;13:483–492. doi: 10.1097/00008390-200310000-00007. [DOI] [PubMed] [Google Scholar]
- 44.Spiropoulou T, Ferekidou L, Angelopoulou K, Stathopoulou A, Talieri M, Lianidou ES. Effect of antineoplastic agents on the expression of human telomerase reverse transcriptase beta plus transcript in MCF-7 cells. Clin Biochem. 2004;37:299–304. doi: 10.1016/j.clinbiochem.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 45.Folini M, Brambilla C, Villa R, Gandellini P, Vignati S, Paduano F, Daidone MG, Zaffaroni N. Antisense oligonucleotide-mediated inhibition of hTERT, but not hTERC, induces rapid cell growth decline and apoptosis in the absence of telomere shortening in human prostate cancer cells. Eur J Cancer. 2005;41:624–634. doi: 10.1016/j.ejca.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Brambilla C, Folini M, Gandellini P, Daprai L, Daidone MG, Zaffaroni N. Oligomer-mediated modulation of hTERT alternative splicing induces telomerase inhibition and cell growth decline in human prostate cancer cells. Cell Mol Life Sci. 2004;61:1764–1774. doi: 10.1007/s00018-004-4062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S, Rosenberg JE, Donjacour AA, Botchkina IL, Hom YK, Cunha GR, Blackburn EH. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and antitelomerase short-interfering RNA. Cancer Res. 2004;64:4833–4840. doi: 10.1158/0008-5472.CAN-04-0953. [DOI] [PubMed] [Google Scholar]
- 48.Das A, Banik NL, Ray SK. Retinoids induced astrocytic differentiation with down regulation of telomerase activity and enhanced sensitivity to taxol for apoptosis in human glioblastoma T98G and U87MG cells. J Neurooncol. 2008;87:9–22. doi: 10.1007/s11060-007-9485-1. [DOI] [PubMed] [Google Scholar]
- 49.Kunisada M, Budiyanto A, Bito T, Nishigori C, Ueda M. Retinoic acid suppresses telomerase activity in HSC-1 human cutaneous squamous cell carcinoma. Br J Dermatol. 2005;152:435–443. doi: 10.1111/j.1365-2133.2005.06471x. [DOI] [PubMed] [Google Scholar]
- 50.Purev E, Soprano DR, Soprano KJ. Effect of all-trans retinoic acid on telomerase activity in ovarian cancer cells. J Exp Clin Cancer Res. 2004;23:309–316. [PubMed] [Google Scholar]
- 51.Liu L, Berletch JB, Green JG, Pate MS, Andrews LG, Tollefsbol TO. Telomerase inhibition by retinoids precedes cytodifferentiation of leukemia cells and may contribute to terminal differentiation. Mol Cancer Ther. 2004;3:1003–1009. [PubMed] [Google Scholar]
- 52.Bagheri S, Nosrati M, Li S, Fong S, Torabian S, Rangel J, Moore DH, Federman S, Laposa RR, Baehner FL, et al. Genes and pathways downstream of telomerase in melanoma metastasis. Proc Natl Acad Sci USA. 2006;103:11306–11311. doi: 10.1073/pnas.0510085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- 54.Carraro G, Albertin G, Forneris M, Nussdorfer GG. Similar sequencefree amplification of human glyceraldehyde-3-phosphate dehydrogenase for real time RT-PCR applications. Mol Cell Probes. 2005;19:181–186. doi: 10.1016/j.mcp.2004.11.004. [DOI] [PubMed] [Google Scholar]