Abstract
In Bacillus thuringiensis, the hag gene encodes flagellin, the protein responsible for eliciting the immunological reaction in H serotyping. Specific flagellin amino acid sequences have been correlated to specific B. thuringiensis H serotypes, H1 to H67. Ten H serotypes, however, contain three or more antigenic subfactors, labeled a, b, c, d, or e, and have been subdivided into 23 serovars. In the present study, we set out to analyze the sequence diversity of flagellins among serovars from the same H serotypes. We studied the hag genes in 39 B. thuringiensis strains representing the 23 serovars from the 10 H serotypes mentioned above. A serovar and a biovar from an 11th H serotype were also included. The hag genes were amplified and cloned and their nucleotide sequences were determined and translated into amino acid sequences, or the sequences were retrieved directly from GenBank when available. Strains of the H3 serotype contained two or three copies of the fla gene, an ortholog of the hag gene. Strains of the H6 serotype contained three copies. Strains of all other H serotypes each contained a single copy of the hag gene. Alignments of amino acid sequences from all copies in all strains of the H3 serotype revealed short signature sequences, GGAG and SGG, GPDPDDAVKNLT, and DITTTK, that appeared to be specific to the H3c, H3d, and H3e antigenic subfactors, respectively. Similar short signature sequences, GDIT, AFIK, TSAGKA, and SAPSKG, were revealed for H8b, H8c, H20b, and H20c, respectively. Amino acid sequences in the flagellin central variable region were highly conserved among serovars of the H3, H5, H11, and H20 serotypes and much more divergent among serovars of the H4, H10, H18, H24, and H28 serotypes. Two bootstrapped neighbor-joining trees were respectively generated from the alignments of the amino acid sequences translated from all copies of the hag genes in the B. thuringiensis strains of the H3 and H6 serotypes. Sequence identities and relationships were revealed. A third bootstrapped neighbor-joining tree was generated, this one from the alignment of the flagellin amino acid sequences from all the B. thuringiensis strains in the study. Eight clusters, I to VIII, were revealed. Although most clusters contained strains and serovars from the same H serotype, clusters VII and VIII contained serovars from different H serotypes.
Bacillus thuringiensis is a gram-positive, rod-shaped, endospore-forming bacterium known to exhibit specific insecticidal activities. Several screening programs aimed at isolating novel B. thuringiensis strains expressing novel pesticidal activities against economically important pests have been established in the last several decades. It is estimated that by 1999, more than 50,000 B. thuringiensis isolates were kept in various collections worldwide (8).
H serotyping, based on the immunological reaction to the bacterial flagellar antigen, flagellin, has been established as a typing method of choice for the identification and classification of B. thuringiensis strains (1). Today, the widely diverse B. thuringiensis strains are classified into more than 69 H serotypes (3). At least two flagellin genes have been reported to occur in most B. thuringiensis H serotypes: hag (15) and fliC (10). However, up to three flagellin genes, initially referred to as flaA, flaB, and flaC, have been reported for B. thuringiensis serovar alesti (4) (GenBank updated data are available under accession no. X67138). In a recent study (15), we have shown that in B. thuringiensis, like in Escherichia coli (13), the hag-encoded protein is composed of three regions: the conserved N and C termini, referred to as C1 and C2, and a central highly variable region, V. The V region presumably harbors the epitope responsible for eliciting the immunological reaction in H serotyping. More importantly, we have shown that direct correlations between the flagellin hag-encoded amino acid sequences and H serotypes can be established. In addition, B. thuringiensis H serotypes can be grouped phylogenetically based on their flagellin amino acid sequences. This work also opened the door to the classification of novel B. thuringiensis isolates at the H serotype level, even in the absence of antisera. No correlations with the B. thuringiensis fliC-encoded protein could be established (10), and its biological role is still unknown.
Although most H serotypes contain no H antigen variation, 10 H serotypes, namely, H3, H4, H5, H8, H10, H11, H18, H20, H24, and H28, have been shown previously to contain three or more distinct antigenic subfactors (3), labeled a, b, c, d, and e. Consequently, the 69 H serotypes have been further subdivided into at least 82 serological varieties, referred to as serovars.
We report here the molecular basis of antigenic variations at the intra-H serotype level in B. thuringiensis. This information should further refine the correlations between the flagellin hag-encoded amino acid sequences and H serotypes; establish the phylogenetic relationships among B. thuringiensis strains, improve current methods of B. thuringiensis strain classification, and improve the identification of novel B. thuringiensis isolates, even in the absence of antisera, all at the intra-H serotype level; and serve as a reference that may lead to the discovery of novel antigenic subfactors. We have either amplified, cloned, and determined the nucleotide sequences of the flagellin hag genes or retrieved the information directly from GenBank for 39 B. thuringiensis strains that represent 23 serovars from the 10 H serotypes indicated above. An 11th H serotype containing a serovar and a biovar was included in the study. Alignments of the flagellin amino acid sequences have revealed amino acid sequence identities and differences among serovars from the same H serotypes. Key differences in the flagellin V region at the intra-H serotype level are discussed. Finally, a bootstrapped neighbor-joining tree was built to show the phylogenetic relationships among these flagellin amino acid sequences.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The 40 B. thuringiensis strains and the B. thuringiensis serovar alesti strain Bt5 used in this study and their sources and geographical origins are listed in Table 1. These strains represent 24 serovars and 11 H serotypes. A B. thuringiensis serovar is a serological variety of strains that share common flagellar antigens, including, when present, common distinct antigenic subfactors. A pathovar is a pathovariety, and a biovar is a biological variety. The lower ranks of pathovar and biovar designate infracategories based solely on utility attributes. The B. thuringiensis strains were cultured in Luria-Bertani (LB) medium (7) at 30°C.
TABLE 1.
B. thuringiensis strains used in this study
| Serovar | H antigen(s) | Strainb | Country of origin | GenBank accession no. | Gene name(s) in GenBank |
|---|---|---|---|---|---|
| alesti | H3a,3c | Bt5 | X67138 | flaA, flaB, and flaC | |
| H3a,3c | IEBC-T03 001 | France | EF595771 | flaA, flaB, and flaC | |
| H3a,3c | BGSC-4C1 | Czechoslovakia | EF595772 | flaA, flaB, and flaC | |
| H3a,3c | BGSC-4C2 | France | EF595773 | flaA, flaB, and flaC | |
| kurstaki | H3a,3b,3c | IEBC-T03A001 | France | EF595774 | flaA, flaB, and flaC |
| H3a,3b,3c | BGSC-4D2 | Unknown | EF595775 | flaA, flaB, and flaC | |
| H3a,3b,3c | BGSC-4D12 | England | EF595776 | flaA, flaB, and flaC | |
| sumiyoshiensis | H3a,3d | IEBC-T03B001 | Japan | EF595777 | flaC and flaB |
| fukuokaensis | H3a,3d,3e | IEBC-T03C001 | Japan | EF595778 | flaC, flaB1, and flaB2 |
| sotto | H4a,4b | IEBC-T04 001 | Canada | DQ377248 | hag |
| H4a,4b | BGSC-4E1 | United States | EF595779 | hag | |
| H4a,4b | BGSC-4E2 | France | EF595780 | hag | |
| sotto biovar dendrolimus | H4a,4b | IEBC-T04A001 | France | EF595781 | hag |
| kenyae | H4a,4c | IEBC-T04B001 | Kenya | DQ377249 | hag |
| H4a,4c | BGSC-4F1 | England | EF595782 | hag | |
| H4a,4c | BGSC-4F2 | Kenya | EF595783 | hag | |
| H4a,4c | BGSC-4F3 | United States | EF595784 | hag | |
| galleriae | H5a,5b | IEBC-T05 001 | USSR | DQ377226 | hag |
| H5a,5b | BGSC-4G2 | United States | EF595785 | hag | |
| H5a,5b | BGSC-4G4 | England | EF595786 | hag | |
| H5a,5b | BGSC-4G5 | Czechoslovakia | EF595787 | hag | |
| canadensis | H5a,5c | IEBC-T05A001 | Canada | DQ377227 | hag |
| entomocidus | H6 | IEBC-T06 001 | Canada | DQ377250 | hagA, hagB, and hagC |
| entomocidus biovar subtoxicus | H6 | IEBC-T06A001 | Canada | EF595788 | hagA1, hagA2, and hagC |
| morrisoni | H8a,8b | IEBC-T08 001 | United States | DQ377229 | hag |
| morrisoni pathovar tenebrionis | H8a,8b | IEBC-T08 017 | Germany | EF595789 | hag |
| morrisoni pathovar sandiegoa | H8a,8b | EF595790 | hag | ||
| ostriniae | H8a,8c | IEBC-T08A001 | China | DQ377251 | hag |
| nigeriensis | H8b,8d | IEBC-T08B001 | Czechoslovakia | DQ377252 | hag |
| darmstadiensis | H10a,10b | IEBC-T10 001 | Germany | DQ377242 | hag |
| londrina | H10a,10c | IEBC-T10A001 | Brazil | DQ377230 | hag |
| toumanoffi | H11a,11b | IEBC-T11 001 | Germany | DQ377254 | hag |
| kyushuensis | H11a,11c | IEBC-T11A001 | Japan | DQ377255 | hag |
| kumamotoensis | H18a,18b | IEBC-T18 001 | Japan | DQ377293 | hag |
| yosoo | H18a,18c | IEBC-T18A001 | South Korea | DQ377300 | hag |
| yunnanensis | H20a,20b | IEBC-T20 001 | China | DQ377260 | hag |
| pondicheriensis | H20a,20c | IEBC-T20A001 | India | DQ377261 | hag |
| neoleonensis | H24a,24b | IEBC-T24 001 | Mexico | DQ377265 | hag |
| novosibirsk | H24a,24c | IEBC-T24A001 | USSR | EF595791 | hag |
| monterrey | H28a,28b | IEBC-T28 001 | Mexico | DQ377233 | hag |
| jegathesan | H28a,28c | IEBC-T28A001 | Malaysia | DQ377234 | hag |
B. thuringiensis serovar morrisoni pathovar sandiego was purified from M-One, a commercial formulation developed by Mycogen Corp.
IEBC strains are from the International Entomopathogenic Bacillus Centre, Institut Pasteur, Paris, France; BGSC strains are from the Bacillus Genetic Stock Center, Department of Biochemistry, The Ohio State University, Columbus, OH.
E. coli strain TOP10 (Invitrogen Canada, Burlington, Ontario, Canada) was used as a recipient strain for the cloning of the PCR fragments. This strain was cultured overnight at 37°C on LB agar plates for the selection of transformants or in LB broth for cell multiplication, with shaking at 200 rpm. For the selection of transformants, kanamycin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were added to the medium at final concentrations of 50 and 40 μg/ml, respectively.
Amplification, cloning, and sequencing of the fla operon and the hag gene and operon.
Total DNA of the B. thuringiensis strains and recombinant plasmids from the E. coli strain were isolated as described previously (14, 15).
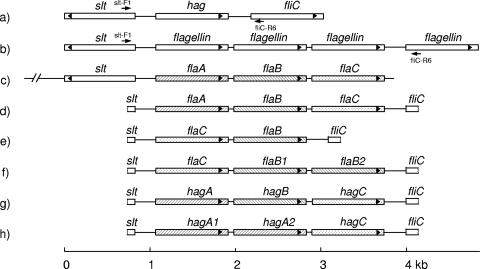
The full genome sequences of B. thuringiensis serovar konkukian strain 97-27 (GenBank accession no. NC_005957), B. thuringiensis strain Al-Hakam (GenBank accession no. NC_008600), and B. cereus strain ATCC 14579 (GenBank accession no. NC_004722) were available from GenBank. In these strains, flagellin genes and flanking genes are organized as follows: slt (encoding a soluble lytic murein transglycosylase), hag, and fliC in both B. thuringiensis strains (Fig. 1a) and slt followed by four flagellin genes in the B. cereus strain (Fig. 1b). A consensus sequence at the 3′ end of slt was used for the design of the following forward primer: slt-F1, 5′-ATATGCAAGCACTTCTTTTACT. The B. thuringiensis fliC gene sequences for 67 H serotypes had been determined previously (10). A consensus sequence at the 5′ end of fliC was used for the design of the following reverse primer: fliC-R6, 5′-ATTHGCDGGATTATCMGAAGC. The respective positions of both primers are shown in Fig. 1a and b. This primer pair, slt-F1/fliC-R6, was used for the amplification of the region between the slt and fliC genes in the B. thuringiensis strains under study (Table 1).
FIG. 1.
Organization of flagellin and neighboring genes. Shown are gene arrangements in B. thuringiensis serovar konkukian strain 97-27 (GenBank accession no. NC_005957) and B. thuringiensis strain Al-Hakam (GenBank accession no. NC_008600) (a), B. cereus strain ATCC 14579 (GenBank accession no. NC_004722) (b), B. thuringiensis serovar alesti strain Bt5 (GenBank accession no. X67138) (c), B. thuringiensis serovars alesti and kurstaki (d), B. thuringiensis serovar sumiyoshiensis (e), B. thuringiensis serovar fukuokaensis (f), B. thuringiensis serovar entomocidus (g), and B. thuringiensis serovar entomocidus biovar subtoxicus (h). Gene names are given. Arrowheads indicate the orientations of the coding regions. The positions and orientations of the primers, slt-F1 and fliC-R6, used in the amplification are given. A scale bar in kilobases is shown below the diagram.
PCR was run for 30 cycles of denaturing at 95°C for 15 s, annealing at 46°C for 30 s, and extension at 72°C for 4 min.
The amplified DNA fragments were cloned into the pCR2.1-TOPO cloning vector using the TOPO TA cloning kit according to the instructions of the manufacturer (Invitrogen Canada). Transformants were selected as described above. Recombinant plasmids were isolated using the alkaline lysis method (11, 14, 15), digested with EcoRI, and visualized on agarose gels to confirm the presence of an inserted fragment.
The nucleotide sequences of cloned fragments were determined by the dideoxynucleotide chain termination method (9) using a capillary array automated DNA sequencer (ABI 3730xl DNA analyzer; Applied Biosystems, Foster City, CA). The sequences of both strands were determined.
Sequence analysis.
The B. thuringiensis hag and fla coding regions and their translated amino acid sequences were determined using the Open Reading Frame (ORF) Finder software from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi). The amino acid sequences were aligned and neighbor-joining trees were constructed (6) and bootstrapped using 1,000 random samples of sites from the alignment, all by employing the ClustalW software (12) from the DNA Data Bank of Japan (DDBJ; http://clustalw.ddbj.nig.ac.jp/top-j.html). The phylogenetic trees were visualized with the TreeView software, version 1.6.1 (5).
Nucleotide sequence accession numbers.
Sequence data have been deposited in the GenBank database under accession no. EF595771 to EF595791.
RESULTS
Diversity of flagellin amino acid sequences.
A total of 41 B. thuringiensis strains representing 24 serovars from 11 H serotypes were included in this study (Table 1). In most cases, strains from same H serotype were selected from distinct geographical origins to avoid the possible duplication of strains. The flagellin gene nucleotide sequence for B. thuringiensis serovar alesti strain Bt5 was readily available from GenBank. A pair of primers (slt-F1/fliC-R6) was used to amplify the flagellin alleles from the remaining 40 B. thuringiensis strains under study.
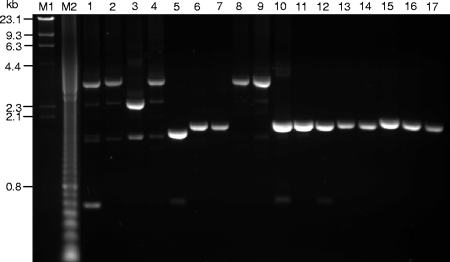
The region between the 3′ end of the slt gene and the 5′ end of the fliC gene was amplified by PCR (Fig. 1a and b), which yielded a major 3.3-kb fragment from each of nine strains of serovars alesti, kurstaki, fukuokaensis, entomocidus, and entomocidus biovar subtoxicus and a major 2.4-kb fragment from the single strain of serovar sumiyoshiensis. All other strains each yielded a major fragment ranging in size from 1.5 to 1.8 kb. A subset of these results is shown in Fig. 2. The nucleotide sequences of all 40 major amplicons were determined.
FIG. 2.
Agarose gel electrophoresis analysis of the products from the amplification of the flagellin genes from selected B. thuringiensis serovars by using the slt-F1/fliC-R6 primer pair. Lanes: 1, serovar alesti BGSC-4C1; 2, serovar kurstaki IEBC-T03A001; 3, serovar sumiyoshiensis IEBC-T03B001; 4, serovar fukuokaensis IEBC-T03C001; 5, serovar sotto IEBC-T04 001; 6, serovar kenyae IEBC-T04B001; 7, serovar galleriae IEBC-T05 001; 8, serovar entomocidus IEBC-T06 001; 9, serovar entomocidus biovar subtoxicus IEBC-T06A001; 10, serovar morrisoni IEBC-T08 001; 11, serovar nigeriensis IEBC-T08B001; 12, serovar darmstadiensis IEBC-T10 001; 13, serovar toumanoffi IEBC-T11 001; 14, serovar kumamotoensis IEBC-T18 001; 15, serovar yunnanensis IEBC-T20 001; 16, serovar neoleonensis IEBC-T24 001; and 17, serovar monterrey IEBC-T28 001. Molecular size markers in lanes M1 and M2 were lambda DNA digested with HindIII and a 100-bp DNA ladder, respectively.
All nucleotide sequences were analyzed for the presence of ORFs. In the H3 serotype, strains from serovars alesti, kurstaki, and fukuokaensis had three ORFs each and the strain from serovar sumiyoshiensis had two ORFs. Each ORF was highly homologous to either flaA, flaB, or flaC in serovar alesti strain Bt5 (4) (GenBank accession no. X67138). The designations flaA, flaB, flaB1, flaB2, and flaC (Fig. 1c, d, e, and f) were assigned based on a phylogenetic analysis of the translated amino acid sequences, as described below (see Fig. 4). Interestingly, the flaA, flaB, and flaC genes were separated by two nearly identical spacers of 65 nucleotides. In the H6 serotype, strains from serovar entomocidus and its biovar subtoxicus each had three ORFs. Each ORF was highly homologous to the flagellin hag gene (15). The ORFs in serovar entomocidus were named hagA, hagB, and hagC (Fig. 1g), whereas those in serovar entomocidus biovar subtoxicus were named hagA1, hagA2, and hagC (Fig. 1 h) after a phylogenetic analysis of the translated amino acid sequences, as explained below (see Fig. 5). The nucleotide sequences obtained for all strains of the other nine H serotypes each had only one ORF. These ORFs were orthologous copies of the B. thuringiensis hag gene (15) and were named accordingly.
FIG. 4.
Bootstrapped neighbor-joining tree of B. thuringiensis H3 serotype strains generated from the alignment of flagellin allelic amino acid sequences. Groupings are indicated by the capital letters A, B, and C. Bootstrap values higher than 50% are indicated. The horizontal bar represents a difference in amino acids of 10%.
FIG. 5.
Bootstrapped neighbor-joining tree of B. thuringiensis H6 serotype strains generated from the alignment of flagellin allelic amino acid sequences. Groupings are indicated by the capital letters A, B, and C. Bootstrap values higher than 50% are indicated. The horizontal bar represents a difference in amino acids of 10%.
All fla and hag nucleotide sequences were translated into amino acid sequences. The 41 B. thuringiensis strains studied here yielded a total of 62 FlaA, FlaB, FlaC, HagA, HagB, HagC, and Hag amino acid sequences. All 62 amino acid sequences were aligned (see section SI in the supplemental material). The N-terminal sequences of the FlaA and FlaB proteins had been determined experimentally by Lövgren et al. (4) and were used here in the identification of the start site common to all flagellin amino acid sequences. The conserved C1 and C2 regions and the variable V region were readily identified as defined previously (15). The C1, V, and C2 regions corresponded to amino acid positions 1 to 131, 132 to 311, and 312 to 395, respectively.
Analysis of flagellin amino acid sequences at the intra-H serotype level. (i) H3 serotype.
The flagellin FlaA, FlaB, and FlaC amino acid sequences of four serovar alesti strains (Bt5, IEBC-T03 001, BGSC-4C1, and BGSC-4C2), three serovar kurstaki strains (IEBC-T03A001, BGSC-4D2, and BGSC-4D12), one sumiyoshiensis strain (IEBC-T03B001), and one fukuokaensis strain (IEBC-T03C001) were aligned (see section SII in the supplemental material). The C1, V, and C2 regions are delimited at positions 1 and 131, 132 and 199, and 200 and 283, respectively. Comparison among all 26 aligned FlaA, FlaB, and FlaC sequences revealed 16 and 7 positions with amino acid substitutions in the C1 and C2 regions, respectively. A total of 33 positions with amino acid substitutions and gaps in the V regions were revealed.
Amino acid sequence comparisons among all flagellin copies revealed short antigenic subfactor-specific signature sequences (Fig. 3). GGAG at positions 137 to 140 in FlaA and SGG at positions 137 to 139 in FlaC of serovars alesti (H3a,3c) and kurstaki (H3a,3b,3c) appeared to be specific to the H3c antigenic subfactor. Likewise, GPDPDDAVKNLT at positions 178 to 189 in FlaB of serovar sumiyoshiensis (H3a,3d) and FlaB2 of serovar fukuokaensis (H3a,3d,3e) appeared to be specific to the H3d antigenic subfactor. Finally, DITTTK at positions 169 to 174 in FlaB1 of serovar fukuokaensis appeared to be specific to the H3e antigenic subfactor. No short sequence putatively specific to the H3b antigenic subfactor was detected.
FIG. 3.
Fla central variable (V) region amino acid sequence alignment for the H3 serotype strains. Boxed letters correspond to short antigenic subfactor-specific signature sequences. Bold letters represent distinct amino acids in the particular positions. Numbers above the sequences indicate the positions of amino acids in the whole-protein sequence alignment.
A bootstrapped neighbor-joining tree was constructed from the alignment of the amino acid sequences translated from the fla alleles (Fig. 4). Three major groups, designated A, B, and C, were revealed. The gene names flaA, flaB, flaB1, flaB2, and flaC for serovars alesti, kurstaki, sumiyoshiensis, and fukuokaensis were assigned in accordance with the grouping of the translated amino acid sequences with the corresponding FlaA, FlaB, and FlaC sequences of serovar alesti strain Bt5. Group A comprised all seven FlaA sequences from all four serovar alesti and three serovar kurstaki strains. Group B comprised all 10 FlaB sequences. Here, the serovar fukuokaensis FlaB2 was grouped with the serovar sumiyoshiensis FlaB. The fukuokaensis FlaB1, although in group B, was more distant. Group C comprised all nine FlaC sequences. Here, the serovar fukuokaensis FlaC sequence appeared to be indistinguishable from the serovar sumiyoshiensis FlaC sequence. No FlaA sequences were found in serovars fukuokaensis and sumiyoshiensis. These data suggest that serovars fukuokaensis and sumiyoshiensis are phylogenetically very close. Indeed, they were both isolated from Kyûshû Island in Japan.
(ii) H4 serotype.
The flagellin (Hag protein) amino acid sequences from the four serovar sotto (H4a,4b) and four serovar kenyae (H4a,4c) strains were aligned (see section SIII in the supplemental material). The sequences of the four serovar sotto strains were identical. The sequences of the four serovar kenyae strains were highly conserved, with only three amino acid substitutions within the V regions. Sequence alignment comparison among all eight strains revealed only four positions with amino acid substitutions in the C1 regions and identical sequences in the C2 regions. However, a total of 43 positions with amino acid substitutions in the V regions were revealed. The first 40 amino acids at the N termini of the V regions were highly conserved, with only two positions with amino acid substitutions. A total of 41 positions with amino acid substitutions over the next 134 positions were revealed. Presumably, some of these substitutions led to the different H4b and H4c antigenic subfactors.
(iii) H5 serotype.
The alignment of the flagellin amino acid sequences of the four serovar galleriae (H5a,5b) strains revealed 100% identity (see section SIV in the supplemental material). When these sequences were aligned with the sequence from serovar canadensis (H5a,5c), five amino acid substitutions were revealed, all in the V regions. Here also, it can be presumed that some, perhaps all, substitutions gave rise to the different H5b and H5c antigenic subfactors.
(iv) H6 serotype.
The number of publicly available strains of the H6 serotype is very limited, and no antigenic subfactors are known. Here, only the two strains from B. thuringiensis serovar entomocidus and its biovar subtoxicus were analyzed.
The three flagellin amino acid sequences of serovar entomocidus and the three flagellin amino acid sequences of serovar entomocidus biovar subtoxicus were aligned (see section SV in the supplemental material). The C1, V, and C2 regions are delimited at positions 1 and 131, 132 and 189, and 190 and 273, respectively. The alignment of the amino acid sequences of HagA from serovar entomocidus and HagA1 and HagA2 from its biovar subtoxicus revealed only three positions with amino acid substitutions in the C1 regions. The V and C2 regions were identical. The HagC amino acid sequences were identical. Comparison among all six aligned sequences revealed nine and four positions with amino acid substitutions in the C1 and C2 regions, respectively. A total of 27 positions with amino acid substitutions and gaps in the V regions were revealed.
A bootstrapped neighbor-joining tree was constructed from the alignment of the amino acid sequences translated from the six hag alleles (Fig. 5). Three major groups, designated A, B, and C, were revealed. The gene names hagA1, hagA2, and hagC for serovar entomocidus biovar subtoxicus were assigned in accordance with the grouping of the translated amino acid sequences with the corresponding HagA and HagC sequences of serovar entomocidus. Group A comprised three closely related flagellins, one from serovar entomocidus, HagA, and two from serovar entomocidus biovar subtoxicus, HagA1 and HagA2. Group C contained the HagC from serovar entomocidus and the HagC from serovar entomocidus biovar subtoxicus. The two HagC sequences appeared to be indistinguishable. Group B comprised only the HagB from serovar entomocidus. This group was distant from the two other groups. No HagB was found in serovar entomocidus biovar subtoxicus.
Given the high number of amino acid differences between the two strains, it will not be surprising, should additional strains be discovered, that antigenic subfactors will be introduced.
(v) H8 serotype.
A total of five H8 serotype strains which cover four antigenic subfactors (H8a, H8b, H8c, and H8d) were analyzed. The alignment of flagellin Hag amino acid sequences of the three serovar morrisoni (H8a,8b) strains revealed 100% identity (see section SVI in the supplemental material). The orthologous sequence from serovar ostriniae (H8a,8c) showed six amino acid substitutions, all in the V region. When the three serovar morrisoni sequences were aligned with the orthologous sequence from serovar nigeriensis (H8b,8d), 54 amino acid substitutions were revealed, 1 in the C1 regions, 52 in the V regions, and 1 in the C2 regions. A gap of four amino acids in the V regions was also revealed. When the serovar ostriniae and nigeriensis sequences were aligned, 59 amino acid substitutions were revealed, 1 in the C1 regions, 57 in the V regions, and 1 in the C2 regions. A gap of four amino acids was present in the V regions. A comparison among all five sequences revealed 59 positions with amino acid substitutions, 1 in the C1 regions, 57 in the V regions, and 1 in the C2 regions. A gap of four amino acids was present in the V regions. Interestingly enough, however, two short amino acid sequences, GDIT and ADIK, at positions 278 to 281, appeared to be specific to the H8b and H8c antigenic subfactors, respectively. Additional H8 serotype strains are needed to confirm this specificity.
(vi) H10, H11, H18, H20, H24, and H28 serotypes.
The numbers of publicly available strains of the H10, H11, H18, H20, H24, and H28 serotypes are very limited, and only three antigenic subfactors, a, b, and c, are known for each H serotype. Only two strains with different combinations of antigenic subfactors were analyzed for each serotype.
(a) H10 serotype.
The flagellin Hag C1 and C2 regions in serovars darmstadiensis (H10a,10b) and londrina (H10a,10c) were identical. A total of 22 amino acid substitutions in the V regions were revealed (see section SVII in the supplemental material).
(b) H11 serotype.
The flagellin Hag C1 and C2 regions in serovars toumanoffi (H11a,11b) and kyushuensis (H11a,11c) were identical. A total of four amino acid substitutions in the V regions were revealed (see section SVIII in the supplemental material).
(c) H18 serotype.
Only two strains of the H18 serotype were analyzed: one each of B. thuringiensis serovars kumamotoensis (H18a,18b) and yosoo (H18a,18c). No amino acid substitution in the C1 regions was found. Two substitutions in the C2 regions were detected. A total of 27 substitutions and a gap of two amino acids in the V regions were revealed (see section SIX in the supplemental material).
(d) H20 serotype.
The flagellin Hag C1 regions in serovars yunnanensis (H20a,20b) and pondicheriensis (H20a,20c) were very similar. A single amino acid substitution in the C1 regions was revealed, at position 21. The C2 regions were identical. A total of nine amino acid substitutions in the V regions were revealed (see section SX in the supplemental material). Interestingly, most of these substitutions were found at positions 220 to 225. Two short amino acid sequences, TSAGKA and SAPSKG, appeared to be specific to the H20b and H20c antigenic subfactors, respectively. Additional strains are needed to confirm this specificity.
(e) H24 serotype.
The flagellin Hag C1 and C2 regions in serovars neoleonensis (H24a,24b) and novosibirsk (H24a,24c) were very similar. Two amino acid substitutions in the C1 regions, at positions 21 and 104, and two in the C2 regions, at positions 299 and 326, were revealed. A total of 51 amino acid substitutions and a single gap in the V regions were revealed (see section SXI in the supplemental material).
(f) H28 serotype.
The flagellin Hag C1 regions in serovars monterrey (H28a,28b) and jegathesan (H28a,28c) were very similar, with only three amino acid substitutions, at positions 93, 105, and 119, and the C2 regions were identical. A total of 20 amino acid substitutions in the V regions were revealed (see section SXII in the supplemental material).
Comparisons of V regions among serovars from the same H serotypes have revealed a limited number of amino acid substitutions for the H11 and H20 serotypes and a higher number of amino acid substitutions for the H10, H18, H24, and H28 serotypes. For the latter, it is likely that should additional strains be discovered, additional antigenic subfactors will be introduced.
Phylogenetic analysis of flagellin among H serotypes.
A bootstrapped neighbor-joining tree was constructed from the alignment of 41 translated flagellin amino acid sequences. A single amino acid sequence, translated from the most upstream flagellin gene, was selected for each strain of the H3 and H6 serotypes (Fig. 1). These gene products were FlaA for serovars alesti and kurstaki, FlaC for serovars sumiyoshiensis and fukuokaensis, HagA for serovar entomocidus, and HagA1 for serovar entomocidus biovar subtoxicus (Fig. 1; also see section SXIII in the supplemental material). The Hag amino acid sequences were used for all other strains. Forty-one strains covering 24 serovars from 11 H serotypes were included. Figure 6 shows the phylogenetic relationships among different serovars of the same H serotypes and among different serovars from different H serotypes. The phylogenetic tree contained two major branches, which encompassed eight distinct clusters, I to VIII. Clusters contained strains that showed flagellin amino acid sequence identities of more than 85%. The first major branch contained clusters I and II, and the second major branch contained clusters III to VIII. Strains and serovars from the same H serotypes were found closely grouped together in the same cluster. In some cases, different H serotypes were grouped in the same cluster. Cluster I encompassed the two strains of the H6 serotype. Cluster II contained all nine strains of the H3 serotype. These nine strains could be further subdivided into two subgroups: a first subgroup that contained the four serovar alesti (H3a,3c) and three serovar kurstaki (H3a,3b,3c) strains and a second subgroup that contained serovars fukuokaensis (H3a,3d,3e) and sumiyoshiensis (H3a,3d). Cluster III encompassed the eight strains of the H4 serotype. Here also, the cluster could be subdivided into two subgroups. A first subgroup contained all four serovar sotto (H4a,4b) strains, including the strain of the serovar sotto biovar dendrolimus, and a second subgroup contained all four serovar kenyae (H4a,4c) strains. Levels of amino acid sequence differences between the two serovars were within 6%. Cluster IV contained the two H18 serotype strains: the serovar kumamotoensis (H18a,18b) and serovar yosoo (H18a,18c) strains. Cluster V encompassed the two H11 serotype strains: the serovar kyushuensis (H11a,11c) and serovar toumanoffi (H11a,11b) strains. Serovars in this cluster were closely grouped together. Levels of amino acid sequence differences between these serovars were within 0.5%. Cluster VI contained all five H5 serotype strains: the serovar canadensis (H5a,5c) and the four serovar galleriae (H5a,5b) strains. Cluster VII contained two H serotypes, H10 and H28. Serovars of the same H serotype were closely grouped together. Levels of amino acid sequence differences between serovars of same H serotype and between the two H serotypes were within 4 and at least 7.5%, respectively. Cluster VIII contained three H serotypes, H20, H24, and H8. The two H20 serovars, pondicheriensis (H20a,20c) and yunnanensis (H20a,20b), formed a distinct subgroup. The two H24 serovars, novosibirsk (H24a,24c) and neoleonensis (H24a,24b), and one H8 serovar, nigeriensis (H8b,8d), formed a second distinct subgroup. The two H24 serovars were not as closely grouped as other serovars from same H serotypes have been. Amino acid sequence divergence between the two serovars was 7.5%. It is interesting that although the serovar neoleonensis strain and the serovar nigeriensis strain belong to different H serotypes, their flagellin amino acid sequences were similar enough to be grouped together here. The level of amino acid sequence difference between the two serovars was 1.3%. The other four H8 serotype strains formed a third distinct subgroup. The three serovar morrisoni (H8a,8b) strains appeared to be indistinguishable. Serovar ostiniae (H8a,8c) was located very closely.
FIG. 6.
Bootstrapped neighbor-joining tree of B. thuringiensis strains generated from the alignment of flagellin amino acid sequences. Clusters are indicated by roman numerals I through VIII. For the H3 and H6 serotypes, only the amino acid sequence translated from the most upstream flagellin gene was used in the generation of this tree. Bootstrap values higher than 50% are indicated. The horizontal bar represents a difference in amino acids of 10%.
DISCUSSION
Of the 69 known B. thuringiensis H serotypes, 10 had long since been shown to contain three or more antigenic subfactors and had been further subdivided into 23 serovars. We have shown here that these serovars, with the exceptions of serovars of the H3 and H6 serotypes, each contained a single hag sequence. The H3 serotype includes four serovars. Serovar sumiyoshiensis was shown here to harbor two paralogous hag sequences, and serovars alesti, kurstaki, and fukuokaensis each harbor three. Likewise, in the H6 serotype, serovar entomocidus and its biovar subtoxicus were shown to harbor three paralogous hag sequences. The high levels of homology among the paralogous copies suggest that they originated from the duplication of an ancestral hag gene. Whether all copies are expressed is unknown but will be addressed below.
Short antigenic subfactor signature sequences, presumably specific to H3c, H3d, and H3e, were discovered. No such sequence was found for H3b. Other short antigenic subfactor signature sequences specific to H8b, H8c, H20b, and H20c were also discovered. It would be interesting to design specific primers based on these signature sequences to determine whether a rapid method based on specific PCR amplification could be developed for the identification of strains of these H serotypes at the serovar level.
In the bacterial flagellum, the filament is assumed to be composed of multiple subunits of a single protein, flagellin. Although this is most likely true for the majority of B. thuringiensis strains—those with a single hag gene—our findings, based on the presence of short signature sequences in different flagellin copies, seem to indicate that multiple subunits of up to three flagellin proteins may be assembled into a flagellar filament in the B. thuringiensis H3 serotype. Interestingly, Lövgren et al. (4) have demonstrated the expression of flaA and flaB in B. thuringiensis serovar alesti, suggesting that both corresponding proteins are synthesized. Certainly, additional work is necessary to confirm that multiple subunits of both flagellins, here FlaA and FlaB, and perhaps a third, FlaC, are indeed assembled into a filament. The story may be different for the H6 serotype. It also harbors multiple flagellin genes, but no antigenic subfactors are known, and thus, no short signature sequences in different flagellin copies have been discovered. Whether two or three copies are expressed is unknown.
Sequence alignment comparisons revealed that flagellin amino acid sequences from different serovars of the H5, H11, and H20 serotypes were highly conserved and that those from different serovars of the H4, H10, H18, H24, and H28 serotypes were highly divergent. Perhaps surprisingly, despite this high level of divergence among flagellin amino acid sequences, only three antigenic subfactors, a, b, and c, are known for each H serotype in the latter group. It is worth pointing out that the number of publicly available strains of the H4, H10, H18, H24, and H28 serotypes is small. For example, the collection of B. thuringiensis and B. sphaericus at the Institut Pasteur (2) contained more than 3,000 B. thuringiensis strains. For the H28 serotype, the collection contained only two serovar monterrey strains and five serovar jegathesan strains. It is likely that should additional strains be discovered, additional antigenic subfactors may be introduced. A total of five and four antigenic subfactors in the H3 and H8 serotypes, respectively, are known. Here, the antigenic subfactors were introduced based on serological cross saturation tests, as the number of strains discovered and characterized kept growing. Certainly, the discovery of additional strains may still lead to the introduction of additional antigenic subfactors.
Six of the eight clusters revealed in the phylogenetic analysis (Fig. 6) contained strains and serovars of a single H serotype. Clusters VII and VIII were more heterogeneous and contained strains and serovars of two and three H serotypes, respectively. Certainly, in these clusters, strains and serovars from the same H serotypes were found in closer proximity than those from different H serotypes, yet different H serotypes were grouped into the same clusters because their flagellin amino acid sequences showed more than 85% identity, indicating their close phylogenetic relationships.
In two recent studies, we showed that all B. thuringiensis strains harbor at least two flagellin genes, hag (15) and fliC (10). We also showed that the hag gene encodes the protein responsible for eliciting the immunological reaction under the conditions used for the H serotyping of B. thuringiensis strains. The role of fliC was unknown. We suggested that the so-called hag gene, used for the H serotyping of B. thuringiensis strains and serovars and also of B. cereus sensu lato species and strains, and the hag ortholog fla, used for the H3 serotype, be renamed fliC in full accordance with the revised E. coli nomenclature so that orthologous sequences would be directly comparable. We also suggested that the gene initially designated fliC be renamed accordingly. The gene names used in the present study were chosen based on names previously used in the scientific literature for flagellin genes in B. thuringiensis and other Bacillaceae. We suggest that these genes be renamed in full accordance with the revised E. coli nomenclature by an international committee.
We are now planning to follow up this work by focusing on the identification of the epitopes and the role of the paralogous flagellin gene copies in the B. thuringiensis H3 serotype.
Supplementary Material
Acknowledgments
We thank Daniel R. Zeigler from the Bacillus Genetic Stock Center, The Ohio State University, Columbus, and Huguette de Barjac, Marguerite M. Lecadet, Emmanuel Frachon, and Jean-François Charles, formerly from the International Entomopathogenic Bacillus Centre, Institut Pasteur, Paris, France, for kindly providing us with most bacterial strains used in this study.
Footnotes
Published ahead of print on 27 June 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.de Barjac, H., and A. Bonnefoi. 1962. Essai de classification biochimique et sérologique de 24 souches de Bacillus du type B. thuringiensis. Entomophaga 7:5-31. [Google Scholar]
- 2.Lecadet, M.-M. 1998. Collection of Bacillus thuringiensis and Bacillus sphaericus (classified by H serotypes). Catalogue of strains no. 1, unité des bactéries entomopathogènes. International Entomopathogenic Bacillus Centre, Institut Pasteur, Paris, France.
- 3.Lecadet, M.-M., E. Frachon, V. Cosmao Dumanoir, H. Ripouteau, S. Hamon, P. Laurent, and I. Thiéry. 1999. Updating the H-antigen classification of Bacillus thuringiensis. J. Appl. Microbiol. 86:660-672. [DOI] [PubMed] [Google Scholar]
- 4.Lövgren, A., M. Y. Zhang, A. Engström, and R. Landén. 1993. Identification of two expressed flagellin genes in the insect pathogen Bacillus thuringiensis subsp.alesti. J. Gen. Microbiol. 139:21-30. [DOI] [PubMed] [Google Scholar]
- 5.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 6.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 7.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 8.Sanchis, V., J. Chafaux, and D. Lereclus. 1996. Amélioration biotechnologique de Bacillus thuringiensis: les enjeux et les risques. Ann. Inst. Pasteur Actual. 7:271-284. [Google Scholar]
- 9.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soufiane, B., D. Xu, and J.-C. Côté. 2007. Flagellin (FliC) protein sequence diversity among Bacillus thuringiensis does not correlate with H serotype diversity. Antonie van Leeuwenhoek 92:449-461. [DOI] [PubMed] [Google Scholar]
- 11.Stephen, D., C. Jones, and J. P. Schofield. 1990. A rapid method for isolating high quality plasmid DNA suitable for DNA sequencing. Nucleic Acids Res. 18:7463-7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, L., D. Rothemund, H. Curd, and P. R. Reeves. 2003. Species-wide variation in the Escherichia coli flagellin (H-antigen) gene. J. Bacteriol. 185:2936-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu, D., and J.-C. Côté. 2003. Phylogenetic relationships between Bacillus species and related genera inferred from comparison of 3′ end 16S rDNA and 5′ end 16S-23S ITS nucleotide sequences. Int. J. Syst. Evol. Microbiol. 53:695-704. [DOI] [PubMed] [Google Scholar]
- 15.Xu, D., and J.-C. Côté. 2006. Sequence diversity of the Bacillus thuringiensis, and B. cereus sensu lato, flagellin (H antigen) protein: comparison with H serotype diversity. Appl. Environ. Microbiol. 72:4653-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.